Abstract
The retinoid X receptor (RXR) is a key regulator in multiple signaling pathways because it can form either a homodimer with itself or a heterodimer with members of the class I nuclear receptors. The RXR-containing dimers regulate transcription by recruiting coactivators or corepressors to the target promoters. The binding of coactivators to RXR is mediated through a hydrophobic pocket formed in part by the C-terminal activation helix (AF-2). However, little is known about interactions of corepressors with RXR and its roles in transcriptional repression. Here we show that the repression activity of RXR correlates with its binding to the corepressor silencing mediator for retinoid and thyroid hormone receptors (SMRT). This intrinsic repression activity is masked by the AF-2 helix, which antagonizes SMRT binding. Inhibition of SMRT binding by the AF-2 helix requires specific amino acid sequences and the helical structure. Furthermore, the SMRT-binding site on RXR is independent of helix 11 but overlaps with the coactivator-binding pocket. On the basis of these results, we propose a structural model to help understand the molecular mechanism of corepressor recruitment by RXR.
The retinoid X receptors (RXRα, -β, and -γ) form heterodimers with members of the class 1 nuclear receptors (NRs). These include the retinoic acid receptors (RARs), thyroid hormone receptors, vitamin D3 receptor, and peroxisome proliferator-activated receptors (PPARs) (1), as well as several orphan receptors such as the liver X receptors (2), pregnane X receptor (3), and constitutively activated receptor (4). RXRs can also form homodimers and bind 9-cis-retinoic acid to regulate target gene expression (5). Thus, RXRs play important roles in multiple signaling pathways, including cell growth, differentiation, homeostasis, metabolism, and development (5, 6).
Like other members of the NR superfamily, RXR is made up of an N-terminal activation domain, a central DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD). The LBD is a multifunction domain, capable of ligand binding, dimerization, transcription activation, and interactions with transcriptional cofactors. The ability of LBD to activate transcription is controlled by a C-terminal helix termed AF-2 helix or helix 12. Ligand binding triggers a conformational change of the AF-2 helix, which forms part of a “charge clamp” needed for recruiting coactivators (7). In the absence of ligands, some NRs can also repress gene expression by interaction with transcriptional corepressors such as the nuclear receptor corepressor (N-CoR) (8) and the silencing mediator for retinoid and thyroid hormone receptors (SMRT) (9–11). The ligand-induced conformational change causes dissociation of the corepressors, allowing the receptor to interact with coactivators. These ligand-initiated events are key steps in hormone-regulated gene expression that affect cell growth and differentiation (7, 12).
The importance of the AF-2 helix in regulating coactivator and corepressor binding is clearly illustrated by crystal structures of apo- (unliganded) and holo- (ligand-bound) LBDs of several NRs (13–19). These crystal structures reveal that LBD fold into a common three-layered α-helical sandwich that contains a hydrophobic cavity for ligand binding. In the unliganded form, the AF-2 helix of RXR extends downward from the LBD and forms intermolecular interactions with the coactivator pocket of its neighboring monomer, thus preventing coactivator binding (16, 20). The binding of 9-cis-retinoic acid to RXR initiates a series of intramolecular interactions that cause dissociation of the AF-2 helix from the coactivator pocket and stabilization of the AF-2 helix in a position that permits coactivator recruitment (15, 21). The coactivator LxxLL motif is then positioned into the coactivator pocket by a pair of “charged clamps” donated by K284 of helix 3 and E453 of the AF-2 helix (15, 18).
The corepressor SMRT contains two separate NR-interacting domains, termed ID1 and ID2 (22). Within each ID, an LxxLL-like corepressor motif (also called CoRNR box or LxxxIxxxI/L motif) is responsible for interactions with NRs (23–25). Stoichiometry studies have shown that each of the two IDs interacts with a single NR in a NR dimer (26, 27). Thus, it is conceivable that the complex between a corepressor and a NR dimer is stabilized by interactions mediated by both ID1 and ID2 domains. The sequence similarity between the corepressor motif and the coactivator motif suggests a conserved mechanism for NR interactions with coactivators and corepressors.
The molecular basis of cofactor interactions with RXR is important for understanding NR-signaling pathway because RXR is involved in heterodimerization with multiple NRs. However, the mechanism of corepressor binding to RXR remains elusive. It has been shown that RXR interacts with SMRT and N-CoR, and that RXR AF-2 helix inhibits the interactions (28). However, the molecular basis of AF-2 helix-mediated inhibition of corepressor binding remains unknown. Furthermore, it has been suggested that RXR interacts with corepressors through the helix 11 region (29), contrasting with the idea that the corepressor motif binds to NRs by a similar mechanism as the coactivator motif (23–25). Thus, to better understand the molecular mechanism of the RXR/corepressor interactions, we characterized the binding between RXR and SMRT. Based on these biochemical studies and existing crystal structures, we propose a structural model of the RXRα/SMRT-ID2 complex to understand their molecular interactions.
Materials and Methods
Plasmids and Chemicals.
The human retinoid X receptor-alpha (hRXRα, NR2B1) (30) and its derivatives and mutants were created by standard molecular cloning techniques, including restriction digestion with endonucleases (New England BioLabs or Roche Molecular Biochemicals), Pfu PCR (Stratagene), and Quick-Change site-directed mutagenesis (Stratagene), followed by end-joining ligation with T4 DNA ligase (Roche Molecular Biochemicals). Derivatives of SMRT (9, 10) were created similarly. GST-cSMRT has been reported (9). All mutations were verified by DNA sequencing. All constructs used for in vitro transcription/translation and transient transfection were in the pCMX vector (31). Other details for plasmids are included in figure legends or available on request. Glutathione S-transferase (GST) fusion protein purification was conducted as described (32) with glutathione-conjugated agarose beads (Sigma).
GST Pull-Down Assay.
Purified GST fusion protein (≈5 μg) was incubated with 5 μl of [35S]methionine-labeled protein with moderate shaking at 4°C overnight in binding buffer (20 mM Hepes, pH 7.7/75 mM KCl/0.1 mM EDTA/2.5 mM MgCl2/0.05% Nonidet P-40/1 mM DTT/1 mg/ml BSA). 35S-Labeled probes were generated by Quick-coupled in vitro transcription/translation reactions (Promega), and precleared with GST at room temperature for 5 min. The GST and GST fusion protein were blocked with BSA at room temperature for 5 min before use in the pull-down assay. The bound 35S-labeled protein was washed three times with binding buffer and beads collected by centrifugation. The bound protein was eluted in SDS sample buffer, subjected to SDS/PAGE, and detected by autoradiography.
Gel Electrophoresis Mobility-Shift Assay.
The sequence of the DR5 element used for RAR/RXR gel shift assays was AGCTTAAGAGGTCACCGAAAGGTCACTCGCAT. The double-stranded DR5 was end-labeled with [32P]dCTP by standard Klenow fill-in reaction. The purified probe was incubated with 35S-labeled receptors in binding buffer containing 7.5% glycerol, 20 mM Hepes (pH 7.5), 2 mM DTT, 0.1% Nonidet P-40, 1 μg poly(dI-dC), and 100 mM KCl. GST-cSMRT was eluted from glutathione agarose beads with 10 mM reduced glutathione and added to the binding reaction. The DNA-protein complex was formed on ice for 1 h and resolved on a 5% native polyacrylamide gel, which was subsequently dried and subjected to autoradiography.
Cell Culture and Transient Transfection.
HEK293 and CV-1 cells were maintained in DMEM supplemented with 10% FBS (GIBCO) and 5 μg/μl gentamycin at 37°C/5% CO2. Cells were plated for transfection in DMEM supplemented with 10% resin-charcoal-stripped FBS in 12-well plates 1 day before transfection. HEK293 cells were transfected by using the standard calcium phosphate precipitation method. Twelve hours after transfection, cells were washed with PBS and refed fresh media. After 24 h, cells were harvested for β-galactosidase and luciferase activities. Luciferase activity was determined with a MLX plate luminometer (Dynex Technologies, Chantilly, VA) and normalized relative to the cotransfected β-galactosidase activity.
Molecular Modeling of RXRα/SMRT-ID2 Complex.
The model for the RXRα/SMRT complex was built from the x-ray structure of SMRT bound to PPARα and GW6471 (33), and the x-ray structure of the tetrameric form of RXRα (16). The PPARα/SMRT-ID2 structure was first superimposed onto the RXR structure to minimize the rms deviation of backbone atoms in the core of the LBD; this superimposition brought the core RXRα helices into approximate coincidence with the corresponding PPARα helices, and effectively superimposed their corepressor-binding sites. The SMRT peptide was transferred from the PPARα structure into the RXRα structure, and then examined graphically. The conformation of the L+1 residue of SMRT was adjusted to improve its fit against RXRα Trp-305 by using the program INSIGHT-II (Accelrys, San Diego, CA). The RXRα/SMRT model structure was then refined by energy minimization by using the MVP program (34), where the RXRα protein was held fixed during the minimization, except for six residues lying near the SMRT peptide interface.
Results
RXR Interacts with the SMRT-ID2 Domain.
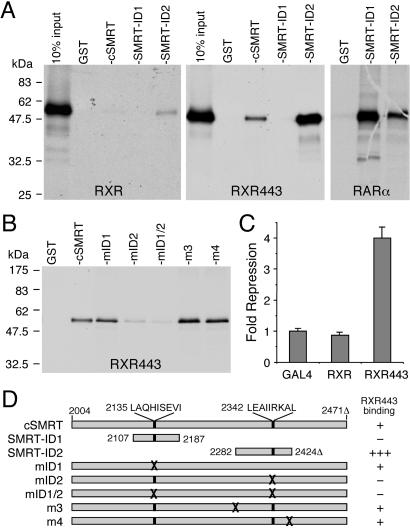
To assess the ability of RXR to recruit corepressors, we determined the binding of the SMRT-ID2 domain to RXR by GST-pull-down assay. Fig. 1A shows that the full-length RXR binds only weakly to SMRT in the absence of ligand. However, deletion of the AF-2 helix at amino acid 443 (RXR443) significantly increases the RXR bindings to SMRT-ID2 and cSMRT, which contains both ID1 and ID2 domains. No interactions of RXR with the SMRT-ID1 are detectable, although under similar conditions the SMRT-ID1 binds strongly to RAR (Fig. 1A). These results are consistent with previous studies (9, 22, 35), and further suggest that RXR has a strong preference for SMRT-ID2.
Figure 1.
RXR binds to the SMRT-ID2 and AF-2 helix inhibits the interaction. (A) GST-pull-down assays showing bindings of 35S-RXR, 35S-RXR443, and 35S-RAR to GST-cSMRT and SMRT-ID1 and -ID2 domains. Deletion of the RXR AF-2 helix at amino acid 443 (RXR443) strongly enhances interaction with cSMRT and SMRT-ID2. (B) A GST-pull-down assay showing binding of 35S-RXR443 with GST-cSMRT and its point mutants, mID1 (V2142A/I2143A), mID2 (I2345A/I2346A), and mID1/2. The two unrelated mutations, m3 (S2285E/K2286E/K2287E) and m4 (L2421A/I2422A), show wild-type binding. The mID1 has no effect on RXR443 binding, whereas mID2 and mID1/2 abolish the interactions to background levels. (C) Deletion of the AF-2 helix converts RXR into a potent transcriptional repressor in the absence of ligand. HEK293 cells were transfected with Gal4 DBD or Gal4 DBD fusions of either wild-type RXR or RXR443. The RXR443 has an ≈4-fold repression activity compared with Gal4 DBD alone. (D) A schematic representation of SMRT mutants and their interactions with RXR443. The amino acid residues in SMRT are according to the human SMRTe sequence (GenBank accession no. AF125672). Δ indicates the lack of 46 aa between residues 2353 and 2398 in this SMRT isoform.
To confirm the preferential binding of SMRT-ID2 to RXR, we mutated each corepressor motif in the context of cSMRT (Fig. 1B). As expected, mutation of the ID1 motif (mID1) has no effect on RXR443 binding. In contrast, mutation of the ID2 motif (mID2) abolishes RXR443 binding. Similarly, double mutation of ID1 and ID2 motifs (mID1/2) also abolishes the interaction. In contrast, two unrelated mutations, m3 and m4, which are outside of the corepressor motifs, show no effect on RXR443 binding. We also correlate the SMRT binding with the transcriptional repression activity of RXR in a reporter gene assay (Fig. 1C). As expected, the wild-type RXR does not repress basal transcription; however, deletion of the AF-2 helix converts RXR into a potent repressor. A schematic representation of the SMRT mutants and their interactions with RXR443 are summarized in Fig. 1D. Together, these data indicate that RXR443 interacts preferentially with SMRT-ID2, and this interaction correlates with its transcriptional repression activity.
Determinants Within the AF-2 Helix That Inhibit SMRT-ID2 Binding.
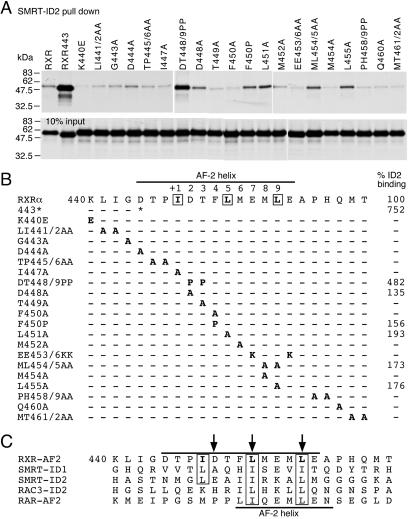
The data above indicate that the AF-2 helix masks RXR's repression activity by inhibiting SMRT-ID2 binding. To determine the key AF-2 residues that are responsible for inhibition of SMRT binding, we systematically mutated each amino acid within the AF-2 helix of RXR (Fig. 2). These mutant proteins were tested by GST-pull-down assay for binding with purified SMRT-ID2 protein (Fig. 2 A and B). Most of these mutations have no effect on SMRT-ID2 binding, except for mutations DT448/9PP, D448A, F450P, L451A, ML454/5AA, and L455A that increase SMRT-ID2 binding. The proline mutations, DT448/9PP and F450P, probably disrupt the helical structure because alanine substitutions at these positions have no effect on SMRT-ID2 binding. These results suggest that the AF-2 helical structure is an essential determinant for inhibition of corepressor binding. More importantly, the enhancement of SMRT-ID2 binding by single-point mutations D448A, L451A, and L455A suggests that the amino acid sequence of the AF-2 helix is also a critical determinant in regulating corepressor binding.
Figure 2.
Determinants of the RXR AF-2 helix that inhibit SMRT binding. (A) GST-pull-down assays showing bindings of RXR AF-2 helix mutants to SMRT-ID2. The in vitro translated probes of the AF-2 helix mutants are shown at the bottom. (B) Summary of the RXR AF-2 helix mutants and their relative bindings to SMRT-ID2. Only those mutants that consistently enhance SMRT-ID2 binding are shown. (C) Sequence comparison of the RXR AF-2 helix (upper line) with SMRT-ID1, -ID2, RAC3-ID2 LxxLL motif, and the RAR AF-2 helix (lower line). The three amino acid residues in RXR AF-2 helix that are most critical for inhibition of SMRT binding are indicated by arrows.
Sequence comparison reveals a similarity of the RXR AF-2 helix with the corepressor motif (Fig. 2C). In contrast, the RAR AF-2 helix is more related to the coactivator LxxLL motif because of the presence of two proline residues preceding the canonical LxxLL sequence. The sequence similarity of the RXR AF-2 helix with the corepressor motif is further supported by the RXR tetramer structure, where the AF-2 helix contains a three-turn α-helix, analogous to the proposed structure of the corepressor motif. These data suggest that the ability of the AF-2 helix to inhibit RXR binding to SMRT is determined by both the primary amino acid sequences and the secondary structure of the helix.
RXR Helix 11 May Not Be Involved in SMRT-ID2 Binding.
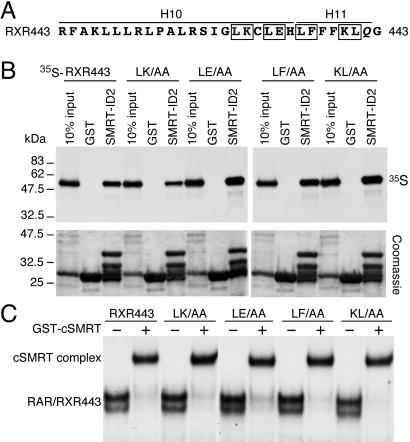
On the basis of deletion and domain-swapping experiments, it has been suggested recently that the NCoR-ID2 corepressor motif binds to the helix 11 region of RXR (29). These data are puzzling considering that the corepressor motif sequence is similar to the RXR AF-2 helix, which does not contact helix 11 but binds to the coactivator-binding site of its adjacent monomer in the tetramer structure (16). Therefore, we further investigated the involvement of RXR helix 11 in SMRT-ID2 binding. Several alanine-substituted mutations were created across the helix 10 and helix 11 in the context of RXR443 (Fig. 3A) including LK430/1AA, LE433/4AA, LF436/7AA, and KL440/1AA. None of these mutations affect SMRT-ID2 binding in GST-pull-down and gel-mobility-shift assays (Fig. 3 B and C). These observations suggest that the helix 11 of RXR may not be involved in direct binding with the SMRT-ID2.
Figure 3.
RXR helix-11 is not involved in SMRT-ID2 binding. (A) Amino acid sequence of the helix-10 (H10) and helix-11 (H11) regions of RXR. The boxed residues were mutated into alanines to create double mutants, L430A/K431A (LK/AA), L433A/E434A (LE/AA), L436A/F437A (LF/AA), and K440A/L441A (KL/AA) in the context of RXR443. In the RXR443 mutant, residue 442 was mutated from an isoleucine (I) to a glutamate (Q) during plasmid construction. (B) GST-pull-down assay showing the interactions of SMRT-ID2 with RXR443 and the helix 10/11 mutants. Upper, the results of the GST-pull-down assay; Lower, the Coomassie blue-stained protein gel of the recovered GST and SMRT-ID2 proteins. Note that the purified GST-SMRT-ID2 contains multiple bands and the major upper band at about 35 kDa represents the full-length fusion protein. (C) Gel mobility-shift assay showing the binding of GST-cSMRT to RAR/RXR443 heterodimer on DR5 elements. Only the shifted RAR/RXR443 DNA complexes and the cSMRT-supershifted DNA complexes are shown.
SMRT-ID2 Binds to an Overlapped Coactivator Pocket on RXR.
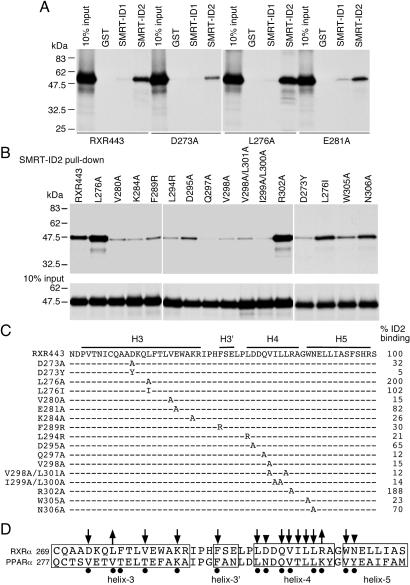
The results above prompted us to study further the RXR surface responsible for SMRT-ID2 binding. On the basis of the binding model of the RXR AF-2 helix into the coactivator-binding site of its neighboring monomer, and the sequence similarity between the corepressor motif and the RXR AF-2 helix, we hypothesize that RXR may use overlapped pockets for interactions with the coactivator and the corepressor motifs. To test this possibility and to pinpoint the exact amino acids involved in this interaction, we created a series of site-directed mutations in the coactivator pocket of RXR in the context of RXR443, and tested their effects on SMRT-ID2 binding (Fig. 4). The residues were chosen on the basis of their involvement in interactions with the LxxLL motif of the SRC-1 coactivator and the recent crystal structure of the PPARα/SMRT-ID2 complex (33). As expected, most of these residues are essential for interaction with the SMRT-ID2. The following mutations result in reduction to less than 35% of SMRT-ID2 binding: D273A, D273Y, V280A, K284A, F289R, L294R, Q297A, V298A, V298A/L301A, I299A/L300A, and W305A. The E281A, D295A, and N306A mutations have more modest effects (65–82% of wild-type binding). Mutations L276A and R302A enhance SMRT-ID2 binding, in contrast to their essential role in PPARα for SMRT-ID2 binding (Fig. 4D). These results demonstrate that SMRT-ID2 binds to an overlapped coactivator pocket on RXR and suggest a potentially conserved mechanism of SMRT-ID2 interactions with different NRs.
Figure 4.
SMRT-ID2 binds to the coactivator pocket on RXR. (A) GST-pull-down assays showing interactions of three RXR coactivator pocket mutants, D273A, L276A, and E281A, with SMRT-ID1 and -ID2. (B) GST-pull-down assays showing the interactions of SMRT-ID2 with other coactivator pocket mutants. The 10% input of the wild-type and mutant proteins is shown at the bottom. (C) Summary of the RXR coactivator pocket mutations and their relative bindings to SMRT-ID2. The positions of helix 3 (H3), helix 3′ (H3′), helix 4 (H4), and helix 5 (H5) are indicated. (D) Summary of amino acid residues in RXRα and PPARα involved in SMRT-ID2 binding. Downward arrows indicate decreases in SMRT-ID2 binding, whereas upward arrows indicate increases in SMRT-ID2 binding. Arrowheads indicate that the effects were less than 50%. Black dots show PPARα residues involved in SMRT-ID2 binding as revealed by the PPARα/SMRT-ID2 crystal structure (33).
A Structure Model of the RXRα/SMRT-ID2 Complex.
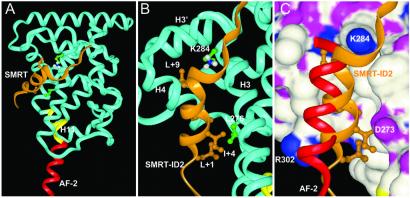
To understand better the molecular interactions between SMRT-ID2 and RXR, we built a structure model based on the crystal structure of the PPARα/SMRT-ID2 complex (33). Because the 14 PPARα residues that contact the SMRT-ID2 motif in the crystal structure are highly conserved in RXR, we would expect a similar binding mode of SMRT-ID2 to RXR. In this model, the SMRT-ID2 corepressor motif also adopts a three-turn helix that docks into the coactivator pocket comprising helices 3, 4, and 5 (Fig. 5A). The C terminus of the corepressor helix is capped by the conserved K284 from the end of helix 3, which also caps the coactivator helix (Fig. 5B). Compared with the binding of coactivator helix to RXR (15), the corepressor helix is angled 15° closer into the coactivator-binding groove (Fig. 5C). In this binding mode, the residues of L+1, I+5, and L+9 of the corepressor motif form the core hydrophobic interface with RXR, the same as observed in the PPARα/SMRT complex.
Figure 5.
A structural model for the RXRα/SMRT-ID2 complex. (A) An overview of the RXRα/SMRT-ID2 complex. The SMRT is shown in gold and RXR is in blue except the AF-2 helix in red and helix -11 in yellow. (B) A close view of the RXRα/SMRT-ID2 interface. The key residues of SMRT-ID2 and RXR are noted. (C) A superposition of the RXR AF-2 helix (red) from its tetramer structure with the SMRT corepressor helix (gold) in the RXR coactivator-binding site (shown in a surface representation colored with atom types).
The molecular interactions between SMRT and the RXR observed in this model provide an explanation of our mutagenesis data. In the RXR model, the same 14 residues as observed in the PPARα structure made up the surface for SMRT binding. As resulted, the mutations in these residues, except for L276 and R302, decrease binding of SMRT to RXR (Fig. 4). Especially, the D273Y mutation only retains 5% of wild-type SMRT binding and this same mutation has been identified in ERα from the tamoxifen-resistant breast cancers (36). In the structure model, the D273Y mutation creates a steric hindrance to SMRT binding by the large tyrosine side chain. Furthermore, the structural model also helps explain the only two mutations of 14 residues that increase SMRT binding. As mentioned above, because of the 15° rotation of the SMRT helix around its C terminus, the N-terminal portion of the SMRT helix would fit more closely to the receptor surface. However, the large side chains of L276 and R302 would prevent such a rotation; thus, mutations to a small side chain like alanine would improve the fitting between SMRT and RXR.
Discussion
We have investigated the molecular interactions that determine the assembly of the RXRα/SMRT-ID2 complex and revealed the molecular mechanism of AF-2 helix-mediated inhibition of SMRT binding. A series of amino acid residues within the RXR AF-2 helix and coactivator-binding site were identified as being critical for regulating SMRT binding. We propose a structure model to understand the interaction of SMRT-ID2 with RXR. Our data provide new insights into the regulation of RXR transcriptional activity by the corepressor SMRT.
Consistent with a positive role in transcriptional activation, the AF-2 helix is thought to play a negative role in repression by inhibiting corepressor binding. Deletion of the AF-2 helix from RXR significantly increases SMRT binding, which is mediated exclusively through the SMRT-ID2 domain (Fig. 1A). Our results demonstrate that full-length RXR binds preferentially to SMRT-ID2 (Fig. 1A). The strong SMRT-ID2 preference of RXR is intriguing, because the two corepressor motifs are similar in sequence and the ID preferences for RAR and thyroid hormone receptor are not as prominent (23, 24). A major difference between ID1 and ID2 is at the +9 position, where ID1 contains an isoleucine and ID2 contains a leucine (Fig. 2C). In the model, this leucine corresponds to the last leucine in the coactivator LxxLL motif in the RXR/SRC-1 structure (15). An isoleucine at this position in ID1 may compromise the exact docking of the ID1 into the coactivator pocket as the ID2 would. Furthermore, the ID2 contains a pair of charged residues (E+2 and R+6) that form direct hydrogen bond interactions with the receptors. This pair of charged residues is missing in ID1. These differences may collectively contribute to the SMRT-ID2 preference of RXR.
The interaction of SMRT-ID2 with RXR increases dramatically in the absence of AF-2 helix (Fig. 1A). We hypothesize that the RXR AF-2 helix may mimic SMRT-ID2 corepressor motif and compete with SMRT-ID2 binding to the coactivator pocket on RXR. This hypothesis is supported by the following observations. First, the primary amino acid sequence of RXR AF-2 helix correlates well with the consensus of the corepressor motif. This good correlation is in contrast with the AF-2 helix of RAR (Fig. 2C), which resembles the shorter coactivator motif because of the presence of two proline residues at the +2/+3 positions, immediately N-terminal to the LxxLL core sequence (13, 37). Second, in the absence of ligand, the RXR AF-2 helix contains three α-helical turns similar in length to the corepressor motif (16, 20). Third, the crystal structure of the inactive RXR tetramer (16) shows a reciprocal intermolecular interaction of the RXR AF-2 helix with the coactivator pocket of a neighboring monomer. This structure information is consistent with the idea that the AF-2 helix competes with SMRT-ID2 for binding to the same site, and that the binding of the AF-2 helix and SMRT-ID2 could be mutually exclusive.
Our scanning mutagenesis analysis of the RXR AF-2 helix reveals three amino acid residues that are critical for the function of the AF-2 helix in inhibition of SMRT binding, namely D448, L451, and L455 (Fig. 2). These three amino acids correspond precisely to the key residues that are important for the binding of the AF-2 helix to the coactivator pocket of the adjacent monomer (16). The hydrophobic side chains of L451 and L455 fit directly into the hydrophobic groove of the coactivator pocket. The carbonyl group of L455 forms a hydrogen bond with the side chain of K284 from helix 3, and the D448 forms another hydrogen bond with R302. Therefore, we expect that removal of side chains from these three amino acids would decrease the binding of AF-2 helix to the coactivator pocket, thus increasing the binding of corepressor. In addition to the specific amino acid sequence, proline mutations at residues 448, 449, and 450 also illustrate the essential role of the helical structure of the AF-2 helix on inhibition of SMRT binding (Fig. 2). Our results therefore establish that the primary amino acid sequence and the secondary structure of the AF-2 helix are both critical for the function of AF-2 helix in inhibition of corepressor binding. Paradoxically, a previous study reported that polyalanine substitution is sufficient to restore the function of RXR AF-2 helix on inhibition of transcriptional repression and N-CoR binding (28). Although the current study uses a different corepressor with a different assay, it raises an intriguing possibility that, in addition to the length of the helix, specific amino acids within the helix also play an important role in regulating corepressor binding.
The NCoR-ID2 motif has been reported to bind to the helix 11 region of the RXR LBD (29). In contrast, we have mapped the SMRT-ID2-binding site to an overlapped surface of the coactivator pocket in RXR LBD. Several point mutations with RXR helix 11 suggest that this region may not be critical for SMRT-ID2 binding (Fig. 3). Similarly, it has been reported that the helix 11 of thyroid hormone receptor does not seem to be essential for N-CoR recruitment (25, 38). These results are consistent with our structural model of the RXRα/SMRT-ID2 complex, where the RXR helix 11 is too far from the corepressor-binding site to make direct contacts with the corepressor motif (Fig. 5A). It is possible that the previous domain-swapping mutations might have affected the conformation of coactivator pocket indirectly. However, our current study does not necessarily contradict the previous work (29), because we used GST fusion of the SMRT-ID2 domain containing 97 aa in GST-pull-down assay, whereas the previous work used the Gal4 DBD fusion of N-CoR core motif of 14 aa in a mammalian two-hybrid assay. Neither can our current data rule out the possibility that the RXR helices 9–11 region may somehow stabilize binding of SMRT-ID2 motif to the coactivator pocket, nor the possibility that the SMRT-ID2 motif might interact with RXR differently than the NCoR-ID2 motif.
Further investigation of the SMRT-ID2 binding on RXR reveals 12-aa residues within the coactivator pocket that are essential for SMRT-ID2 binding. The crystal structure of PPARα/SMRT-ID2 complex also supports this conclusion (33). Comparison of the SMRT-ID2 contacting residues between PPARα and RXRα reveals a conservative mode of interaction, suggesting that the mechanism of SMRT-ID2 binding to NRs may be conserved. Two striking differences between ID2 interactions with PPARα and RXRα are observed. First, the L276A mutation in RXRα seems to enhance, rather than reduce, SMRT-ID2 binding. The corresponding residue in PPARα is a valine, which contributes to the hydrophobic interaction with the isoleucine at the +4 position of the SMRT-ID2. An alanine substitution at the corresponding position in thyroid hormone receptor significantly reduces SMRT-ID2 binding (33). Another striking difference is the R302A mutation, which enhances RXR binding to SMRT-ID2 as well. This R302 residue forms an intermolecular hydrogen bond with D448 in the AF-2 helix of a neighboring monomer in the symmetry dimer, which is expected to stabilize tetramer formation (16). Because the R302A mutation would disrupt this hydrogen bonding, it could reduce the binding affinity of the AF-2 helix to the coactivator site and disrupt the tetramer formation. However, compared with the AF-2 helix, the N terminus of the SMRT helix docks much closer to the receptor (Fig. 5C), and the R302A mutation may improve this close fit by removing the large side chain of R302. In PPARs and other class I receptors, the corresponding residue has a conserved lysine, which side chain is more flexible than an arginine to accommodate the corepressor helix. Together, the differences in residues 276 and 302 may contribute to subtle changes of SMRT binding to RXRα from PPARα.
Overall, we have provided a potential molecular mechanism of interactions that determine the assembly of the RXRα/SMRT-ID2 complex. Our data reveal the structural basis of the inhibition of SMRT-ID2 binding by the RXR AF-2 helix. We also demonstrate that SMRT-ID2 binds to an overlapped site at the coactivator pocket on RXR. Therefore, it seems that a common underlying mechanism may exist for NR interactions with the coactivator motif, corepressor motif, and the AF-2 helix. Thus, these studies provide insights into the molecular interactions between RXR and the SMRT corepressor, which may have important implications for understanding the regulation of RXR's broad biological activities by corepressors.
Acknowledgments
We thank Amy Chen for excellent technical support, David Johnson for critical reading of the manuscript, and members of the Chen Laboratory for helpful discussions throughout the course of this work. J.D.C. is a Research Scholar funded by the Leukemia and Lymphoma Society. This work was made possible by Grants DK52542 and DK52888 from the National Institutes of Health (to J.D.C.) and RPG-98–085-01LBC from the American Cancer Society. Its contents are solely the responsibility of the authors and do not necessarily represent the funding agencies.
Abbreviations
- RXR
retinoid X receptor
- RAR
retinoic acid receptors
- NR
nuclear receptors
- PPAR
peroxisome proliferator-activated receptor
- DBD
DNA-binding domain
- LBD
ligand-binding domain
- SMRT
silencing mediator for retinoid and thyroid hormone receptors
- GST
glutathione S-transferase
- ID
interacting domain
- m
mutation, N-CoR, nuclear receptor corepressor
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 2.Willy P J, Umesono K, Ong E S, Evans R M, Heyman R A, Mangelsdorf D J. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- 3.Moore J T, Kliewer S A. Toxicology. 2000;153:1–10. doi: 10.1016/s0300-483x(00)00300-0. [DOI] [PubMed] [Google Scholar]
- 4.Honkakoski P, Zelko I, Sueyoshi T, Negishi M. Mol Cell Biol. 1998;18:5652–5658. doi: 10.1128/mcb.18.10.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangelsdorf D J, Borgmeyer U, Heyman R A, Zhou J Y, Ong E S, Oro A E, Kakizuka A, Evans R M. Genes Dev. 1992;6:329–344. doi: 10.1101/gad.6.3.329. [DOI] [PubMed] [Google Scholar]
- 6.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leo C, Chen J D. Gene. 2000;245:1–11. doi: 10.1016/s0378-1119(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 8.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, et al. Nature (London) 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 9.Chen J D, Evans R M. Nature (London) 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 10.Park E J, Schroen D J, Yang M, Li H, Li L, Chen J D. Proc Natl Acad Sci USA. 1999;96:3519–3524. doi: 10.1073/pnas.96.7.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ordentlich P, Downes M, Xie W, Genin A, Spinner N B, Evans R M. Proc Natl Acad Sci USA. 1999;96:2639–2644. doi: 10.1073/pnas.96.6.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna N J, O'Malley B W. J Steroid Biochem Mol Biol. 2000;74:351–356. doi: 10.1016/s0960-0760(00)00112-6. [DOI] [PubMed] [Google Scholar]
- 13.Bourguet W, Vivat V, Wurtz J M, Chambon P, Gronemeyer H, Moras D. Mol Cell. 2000;5:289–298. doi: 10.1016/s1097-2765(00)80424-4. [DOI] [PubMed] [Google Scholar]
- 14.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gampe R T, Jr, Montana V G, Lambert M H, Miller A B, Bledsoe R K, Milburn M V, Kliewer S A, Willson T M, Xu H E. Mol Cell. 2000;5:545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 16.Gampe R T, Jr, Montana V G, Lambert M H, Wisely G B, Milburn M V, Xu H E. Genes Dev. 2000;14:2229–2241. doi: 10.1101/gad.802300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Nature (London) 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 18.Shiau A K, Barstad D, Loria P M, Cheng L, Kushner P J, Agard D A, Greene G L. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 19.Xu H E, Lambert M H, Montana V G, Parks D J, Blanchard S G, Brown P J, Sternbach D D, Lehmann J M, Wisely G B, Willson T M, et al. Mol Cell. 1999;3:397–403. doi: 10.1016/s1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 20.Bourguet W, Ruff M, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;375:377–382. doi: 10.1038/375377a0. [DOI] [PubMed] [Google Scholar]
- 21.Egea P F, Mitschler A, Rochel N, Ruff M, Chambon P, Moras D. EMBO J. 2000;19:2592–2601. doi: 10.1093/emboj/19.11.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Leo C, Schroen D J, Chen J D. Mol Endocrinol. 1997;11:2025–2037. doi: 10.1210/mend.11.13.0028. [DOI] [PubMed] [Google Scholar]
- 23.Hu X, Lazar M A. Nature (London) 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 24.Nagy L, Kao H Y, Love J D, Li C, Banayo E, Gooch J T, Krishna V, Chatterjee K, Evans R M, Schwabe J W. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perissi V, Staszewski L M, McInerney E M, Kurokawa R, Krones A, Rose D W, Lambert M H, Milburn M V, Glass C K, Rosenfeld M G. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cohen R N, Wondisford F E, Hollenberg A N. Mol Endocrinol. 1998;12:1567–1581. doi: 10.1210/mend.12.10.0188. [DOI] [PubMed] [Google Scholar]
- 27.Zamir I, Zhang J, Lazar M A. Genes Dev. 1997;11:835–846. doi: 10.1101/gad.11.7.835. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Hu X, Lazar M A. Mol Cell Biol. 1999;19:6448–6457. doi: 10.1128/mcb.19.9.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu X, Li Y, Lazar M A. Mol Cell Biol. 2001;21:1747–1758. doi: 10.1128/MCB.21.5.1747-1758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangelsdorf D J, Ong E S, Dyck J A, Evans R M. Nature (London) 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 31.Umesono K, Murakami K K, Thompson C C, Evans R M. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frangioni J V, Neel B G. Anal Biochem. 1993;210:179–187. doi: 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
- 33.Xu H E, Stanley T B, Montana V G, Lambert M H, Shearer B G, Cobb J E, McKee D D, Galardi C M, Plunket K D, Nolte R T, et al. Nature (London) 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 34.Lambert M H. In: Practical Application of Computer-Aided Drug Design. Charifson P S, editor. New York: Dekker; 1997. pp. 243–303. [Google Scholar]
- 35.Chen J D, Umesono K, Evans R M. Proc Natl Acad Sci USA. 1996;93:7567–7571. doi: 10.1073/pnas.93.15.7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamamoto Y, Wada O, Suzawa M, Yogiashi Y, Yano T, Kato S, Yanagisawa J. J Biol Chem. 2001;276:42684–42691. doi: 10.1074/jbc.M107844200. [DOI] [PubMed] [Google Scholar]
- 37.Renaud J P, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Nature (London) 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 38.Marimuthu A, Feng W, Tagami T, Nguyen H, Jameson J L, Fletterick R J, Baxter J D, West B L. Mol Endocrinol. 2002;16:271–286. doi: 10.1210/mend.16.2.0777. [DOI] [PubMed] [Google Scholar]