Abstract
Nucleotide excision repair (NER) plays a central role in maintaining genomic integrity by detecting and repairing a wide variety of DNA lesions. Xeroderma pigmentosum complementation group A protein (XPA) is an essential component of the repair machinery, and it is thought to be involved in the initial step as a DNA damage recognition and/or confirmation factor. Human replication protein A (RPA) and XPA have been reported to interact to form a DNA damage recognition complex with greater specificity for damaged DNA than XPA alone. The mechanism by which these two proteins recognize such a wide array of structures resulting from different types of DNA damage is not known. One possibility is that they recognize a common feature of the lesions, such as distortions of the helical backbone. We have tested this idea by determining whether human XPA and RPA proteins can recognize the helical distortions induced by a DNA triple helix, a noncanonical DNA structure that has been shown to induce DNA repair, mutagenesis, and recombination. We measured binding of XPA and RPA, together or separately, to substrates containing triplexes with three, two, or no strands covalently linked by psoralen conjugation and photoaddition. We found that RPA alone recognizes all covalent triplex structures, but also forms multivalent nonspecific DNA aggregates at higher concentrations. XPA by itself does not recognize the substrates, but it binds them in the presence of RPA. Addition of XPA decreases the nonspecific DNA aggregate formation. These results support the hypothesis that the NER machinery is targeted to helical distortions and demonstrate that RPA can recognize damaged DNA even without XPA.
Keywords: triple helix‖nucleotide excision repair
Because genome stability is critical for cell survival, extremely sensitive DNA repair mechanisms have evolved to protect the genome from both internal and external assaults. Defects in these repair mechanisms can lead to severe disorders such as xeroderma pigmentosum, ataxia telangiectasia, Fanconi anemia, and Bloom syndrome (reviewed in refs. 1–5). One of the major consequences of these defects is an enhanced predisposition to cancer. It has been reported that 80–90% of human cancers are the result of DNA damage (6).
Many types of DNA damage, spontaneous and induced, develop from both endogenous and exogenous sources. Exogenous damages, both chemical and physical, arise from exposure to UV and ionizing radiation and from natural and synthetic chemicals. Cellular metabolic processes lead to internal sources of DNA damage; for example, it is estimated that loss of a purine base (depurination) occurs at a rate of nearly 20,000 events per cell per day (7).
In humans, different types of DNA damage are thought to be repaired by one of several mechanisms, including nucleotide excision repair (NER; reviewed in refs. 8 and 9). NER removes covalent DNA lesions and is the only known mechanism for removing bulky DNA adducts in humans. The damaged base is removed by endonucleolytic cleavage of the phosphodiester bonds several bases away from the lesion on either side, resulting in the release of an oligonucleotide containing the damage, followed by repair synthesis and ligation (10).
Although much is known about NER, very little is known about the first and rate-limiting step, DNA damage recognition. Several damage-recognition proteins have been identified by their preference for binding to damaged DNA, including the xeroderma pigmentosum complementation group A protein (XPA) and replication protein A (RPA) complex (11–15) and the xeroderma pigmentosum complementation group C protein (XPC) and human Rad23B protein (hHR23B) complex (16, 17). Although the exact mechanisms by which these proteins recognize DNA damage are not understood, damage recognition signals have been proposed including defects in the DNA helical structure and modification of the DNA chemistry. Hess et al. (18) describe a “bipartite” model of DNA damage recognition that requires both structural distortions and chemical modifications to the DNA for a lesion to become a substrate for NER. We hypothesize that damaged DNA is recognized by conformational alterations in the helical structure at the site of damage. One approach to test this hypothesis is to use DNA substrates whose structures differ substantially from B-form DNA in protein–DNA interaction studies.
One class of such distorted substrates in which a wide range of structural variations can be readily introduced is triplex DNA. Triplex-forming oligonucleotides (TFOs) recognize and bind to specific sites in duplex DNA, forming a triple-stranded DNA helix. Studies of triplex structure by using circular dichroism, NMR, and x-ray crystallography (19–25) have revealed that binding of the third strand induces substantial structural distortions in the underlying duplex, so that the helical geometry more closely resembles A-form DNA, while maintaining the normal pattern of Watson–Crick hydrogen bonding.
In addition to their advantages as probes of the structural basis of damage recognition, the recognition and repair of triplex structures is of interest because of the growing use of TFOs to manipulate gene structure and function in vitro and in vivo (reviewed in ref. 26). Recently, we have found that systemically administered TFOs can induce mutagenesis of a targeted gene in somatic cells of mice (27). In addition, intermolecular triplex formation has been found to stimulate recombination in mammalian cells and cell-free extracts (28, 29). Evidence suggests that the ability of triplexes to induce mutagenesis and recombination depends on the capacity of triplex structures to provoke DNA repair (28–30). Moreover, it has been suggested that intramolecular triplex DNA structures may exist transiently in vivo and play a role in gene expression and genomic instability (31, 32).
We report here results of experiments designed to determine the roles of XPA and RPA, separately and together, in the recognition of several structurally distinct DNA substrates, all containing triple helices. These experiments have revealed that RPA recognizes all covalent triplex structures tested, whereas XPA alone does not bind the substrates under our experimental conditions. RPA alone displays limited specificity in triplex recognition, but at high concentrations forms nonspecific aggregates with DNA, whose formation is inhibited by XPA. Thus, RPA and XPA work together as a complex to recognize the structural distortions common to all these triplex structures and to distinguish them from undamaged DNA, thus targeting them as substrates for NER.
Materials and Methods
Oligonucleotides.
The sequences of the TFOs and duplex targets used in this study are shown in Fig. 1. Psoralen was incorporated on the 5′ end by using the derivative 2-[4′-(hydroxymethyl)-4,5′,8-trimethylpsoralen]-hexyl-1-O-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite (HMT), from Glen Research (Sterling, VA) as described (33, 34). The concentration of DNA was determined by UV absorbance at 260 nm.
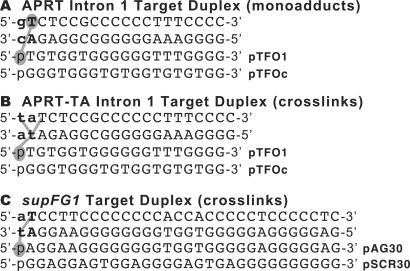
Figure 1.
Sequences of the APRT and supFG1 triplex target site duplexes and TFOs. (A) Nucleotide sequences of the APRT intron 1 target site duplex with the expected location and orientation of TFO binding. PTFO1 is the specific 19-base oligonucleotide that binds the APRT intron 1 site with high affinity. PTFOc is a 19-base control oligonucleotide that contains the same base composition as pTFO1, but a scrambled sequence and is not capable of binding the APRT intron 1 site. (B) The sequence of the modified APRT target site duplex, APRT-TA, containing a 5′ TA at the triplex-duplex junction for efficient psoralen crosslink formation. (C) The sequences of the supFG1 target site duplex and TFO. PAG30 binds the supFG1 site with high affinity, and the control oligonucleotide (pSCR30) does not. The psoralen intercalation site at the triplex-duplex junction is listed in bold for all duplex substrates. Psoralen is shaded and the base capable of monoadduct formation only. Lines indicate potential covalent interaction sites with psoralen after UVA irradiation.
DNA Substrates.
A 188-bp EcoRI to SacI fragment containing the supFG1 triplex target site was isolated from plasmid, pSupFG1 (35). Oligonucleotides (57 nucleotides) corresponding to the supFG1 sequence were annealed in a 1:1 molar ratio to form the synthetic target duplex at a final concentration of 5 × 10−6 M (Fig. 1C). For the adenine phosphoribosyltransferase (APRT) site, a 196 EcoRV to PvuII plasmid fragment containing the APRT intron 1 target site was isolated from plasmid, pGS37 (36). A 38-bp synthetic duplex was formed by annealing complementary strands in a 1:1 molar ratio. The duplexes were 5′-end-labeled with T4 polynucleotide kinase and [γ-32P]ATP, gel purified, and quantified by UV absorbance at 260 nm.
Psoralen-TFO Photoadduct Formation.
Triplex structures were formed by incubating radiolabeled duplexes (5 × 10−7 M) with TFOs (5 × 10−7 M) in a buffer containing 10 mM Tris⋅HCl, pH 7.6, 10 mM MgCl2, and 10% (vol/vol) glycerol at 37°C for 16 h. Psoralen-TFOs were then UVA irradiated (1.8 J/cm2, unless indicated otherwise) to form monoadducts or crosslinks at the triplex-duplex junction. Similarly, radiolabeled duplex DNA was incubated with free HMT before UVA treatment to form psoralen monoadducts and crosslinks in the absence of triplex formation. The resulting substrates were then either used in protein-binding assays as described below, or were subjected to denaturing gel electrophoresis (15% polyacrylamide gels, TBE, 7 M urea) to separate the psoralen-adducted substrates from free duplex or triplex DNA. To determine efficiency of photoadduct formation, gels were dried and radioactivity in each band measured by using a PhosphorImager (Molecular Dynamics).
Protein Synthesis and Purification.
The XPA-maltose-binding protein fusion protein was expressed in Escherichia coli PR745 from pMAL constructs (37). Protein expression was induced with isopropyl-β-D-thiogalactoside, cells were lysed by sonication, and the fusion protein purified by affinity chromatography on amylose resin according to manufacturer's instructions (New England Biolabs). The three-subunit histidine-tagged RPA complex (38) was expressed by coinfection of Sf9 cells at a multiplicity of infection of 5 for His-RPA1 and RPA2 baculoviruses and a multiplicity of infection of 10 for the RPA3 baculovirus. The infected cells were harvested, and the expressed complex was purified by Ni2+-chelate chromatography as described (38) and further purified by salt gradient elution from a Mono-Q FPLC column. Recombinant XPA and RPA were approximately 95% pure as judged by SDS-polyacrylamide gel electrophoresis and Coomassie brilliant blue staining.
Binding Assays.
Duplex and triplex substrates were prepared as described above, with or without photoadduct formation. Proteins were preincubated in a buffer containing 25 mM Hepes, pH 7.0, 50 mM KCl, 5 mM MgCl2, 0.7 μg BSA, 10% (vol/vol) glycerol, 0.5 mM DTT, 0.01% Nonidet P-40 in a 20-μl reaction volume at 30°C for 10 min. XPA was added at 100 ng and RPA at 10 ng for all reactions unless indicated otherwise. Radiolabeled triplex or duplex DNA substrates (at 5 × 10−8 M) were then added to the proteins in the same buffer and incubated for the indicated times at 30°C. The samples were then subjected to native polyacrylamide gel electrophoresis on 4% or 6% gels (29:1 acrylamide:bis) buffered in TBE and 4% glycerol. Gels were run at 4°C for ≈2 h at 25 mA, dried, and visualized by autoradiography.
Supershift Analyses.
Samples were incubated for 30 min as described above, and then antibodies against XPA (described in ref. 29) or RPA (NeoMarkers, Fremont, CA) were added to the appropriate samples and further incubated for 20 min at 30°C. The samples were then subjected to gel electrophoresis as described above, dried, and visualized by autoradiography.
Protein–Protein Interaction Assays.
Protein–protein interactions were analyzed by an ELISA. Ninety-six-well plates were incubated overnight at 4°C with 1.5 μg of purified protein in PBS as indicated in the legend (see Fig. 7C), washed in PBS, blocked overnight in PBS containing 3% BSA, and washed in PBS. The coated plates were incubated with increasing amounts of purified XPA for 1 h at 4°C in PBS containing 0.1% BSA followed by washes with PBS. Bound XPA was detected by incubation for 45 min with a 1:1000 dilution of an anti-XPA antibody (described in ref. 29). Bound antibody was detected by a horseradish peroxidase-coupled goat anti-rabbit IgG conjugate according to the manufacturer's instructions (Dako, Glostrup, Denmark). Optical densities were read at 490 nm.
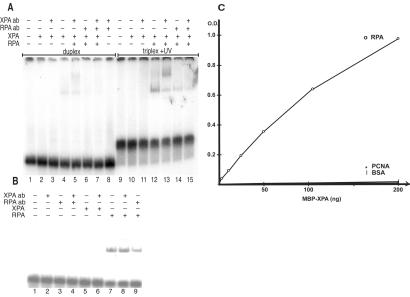
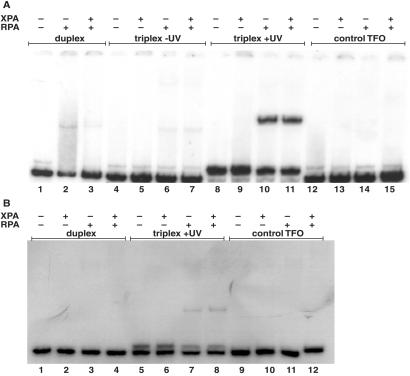
Figure 7.
Antibody supershifting of XPA–RPA–DNA complexes and XPA-RPA binding interactions. (A) The psoralen-modified TFOs were incubated with a radiolabeled 57-bp duplex substrate to form a psoralen-crosslinked triplex of 30 base triplets at a concentration of 5 × 10−8 M. Triplex + UV lanes indicate a psoralen-crosslinked 57-bp supFG1 triplex substrate incubated with XPA and RPA proteins. Antibodies against XPA (XPA ab) or RPA (RPA ab) were added to the indicated lanes. Duplex lanes contain the 57-bp duplex only. Samples were analyzed by EMSA after incubation with proteins and subjected to autoradiography. (B) Control samples were treated as described above with proteins and antibodies incubated with the psoralen-crosslinked 57-bp triplex substrate only. (C) ELISA analysis of XPA and RPA binding. Purified recombinant proteins [RPA, proliferating cell nuclear antigen (PCNA), or BSA] were coated on 96-well plates and then incubated with increasing amounts of XPA protein. Specific binding interactions were detected as OD units after incubation with anti-XPA primary antibody and a horseradish peroxidase-conjugated secondary antibody.
Results
Recognition of Triplex Structures by Human RPA and XPA Proteins.
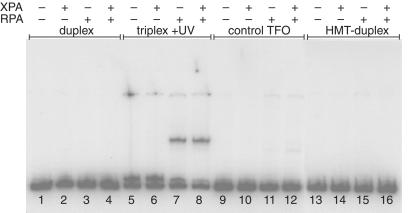
To determine whether triple helical structures are substrates for NER, 32P-labeled DNA triplex substrates of 196 bp were incubated with recombinant RPA and XPA proteins. We chose to study protein binding to the high-affinity triplex-forming site in intron 1 of the hamster APRT gene (Fig. 1A) that we had characterized (33, 36). A psoralen-modified TFO was used to form covalent psoralen-monoadducted triplex structures after treatment with UVA irradiation. In this structure, the psoralen is conjugated to the 5′ end of the TFO and forms a psoralen monoadduct with a thymidine on one strand of the target duplex (see Fig. 1A). As shown in Fig. 2, RPA (at 4 × 10−9 M) binds psoralen-monoadducted triplex DNA specifically and with high affinity, and the shifted band seems to run similarly in the presence of both RPA (at 4 × 10−9 M) and XPA (at 6 × 10−8 M). In contrast, binding of RPA or both RPA and XPA to the duplex DNA substrate or the psoralen-adducted duplex structure is not detectable under the conditions of the assay (see Fig. 2, lanes 1–4 and 13–16, respectively). XPA alone does not shift the mobility of either duplex or triplex. The third strand is clearly an important element of the RPA-DNA recognition because simply crosslinking the duplex with free psoralen (HMT) in the absence of a TFO, does not lead to the highly specific binding observed with the triple helix (Fig. 2, compare lanes 5–8 with lanes 13–16), despite the formation of both monoadducts and crosslinks on the target duplex DNA after incubation with HMT and irradiation with UVA, as confirmed by denaturing gel electrophoresis (data not shown). The 196-bp duplex fragment containing the APRT triplex target site contains seven potential psoralen-crosslinking sites including the triplex-targeted site at the triplex-duplex junction, yet the HMT-duplex DNA substrate is not bound by XPA or RPA in the absence of triplex formation. The presence of a control oligonucleotide incapable of forming a triplex structure with this duplex yields a result essentially identical to that obtained with duplex alone (Fig. 2, compare lanes 1–4 with 9–12).
Figure 2.
Binding of human XPA and RPA proteins to psoralen-monoadducted triplex DNA. XPA (100 ng) and RPA (10 ng) proteins were coincubated with 196 bp DNA duplexes or triplexes containing the APRT intron 1 triplex target site at a concentration of 5 × 10−8 M. Protein–DNA interactions were assessed by electrophoretic mobility-shift assay (EMSA) on a 4% polyacrylamide gel buffered with TBE and 4% glycerol. Duplex sample lanes indicate the 196-bp DNA substrate alone, triplex + UV lanes indicate a 19-base psoralen-monoadducted triplex within the 196-bp duplex, control TFO lanes are samples containing the control oligonucleotide which does not form triplex, and HMT-duplex lanes indicate the 196-bp duplex treated with the free psoralen derivative (HMT) plus UVA irradiation in the absence of the TFO.
Concentration Dependence of RPA Binding to Triplex DNA.
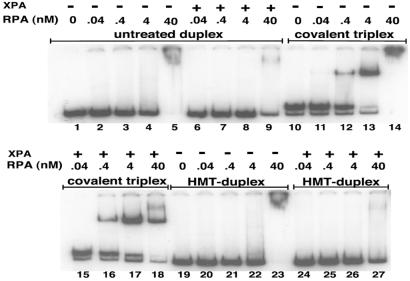
To determine the strength of RPA recognition for triplex substrates, we varied RPA concentrations across a range spanning three orders of magnitude (Fig. 3), from 40 pM to 40 nM. Binding to the covalent triplex structure was detected at RPA concentrations as low as 0.4 nM, and was half-maximal at concentrations between 0.4 nM and 4 nM. In the absence of XPA, RPA (at 4 × 10−8 M) formed large DNA aggregates that failed to enter the gel, characteristic of nonspecific DNA binding, but this binding is reduced in the presence of XPA (at 6 × 10−8 M). In contrast, binding of RPA to a psoralen-monoadducted triplex structure is not diminished by XPA, so that the net effect of XPA is an enhancement of the formation of the lower-mobility-specific RPA-triplex complex (Fig. 3, compare lanes 14 and 18) which, as discussed below, likely also includes bound XPA. These results underscore the ability of RPA to recognize a triple-helical substrate with high affinity, and the ability of XPA to affect complex formation, likely through binding to RPA in a way that inhibits nonspecific aggregate formation.
Figure 3.
Concentration-dependent binding of RPA to covalent triplexes. RPA was incubated with the APRT 196-bp DNA substrate (5 × 10−8 M) at the concentrations indicated in the presence or absence of XPA (100 ng). The covalent triplex substrate indicates a psoralen-monoadducted triplex DNA structure. After substrate formation and incubation with XPA and RPA, the protein-DNA interactions were analyzed by EMSA. The HMT-duplex substrate is the 196-bp APRT substrate after incubation with HMT and UVA irradiation to induce psoralen photoadducts in the DNA duplex.
To better understand the effect of XPA on preventing the DNA-dependent aggregation of RPA at high concentrations, we performed order-of-addition experiments by either preincubating RPA with XPA before addition of DNA substrate, or by allowing RPA to interact with DNA first, and then adding XPA. The results seemed similar regardless of when XPA was added (data not shown), suggesting that the DNA-RPA aggregation effect is both preventable and reversible by addition of XPA, consistent with results shown in Fig. 3.
Recognition of Psoralen-Crosslinked Triplex Structures by XPA and RPA.
Although RPA clearly recognizes psoralen-monoadducted triplex structures, we were interested in determining its potential to bind a more helix-distorting lesion, a psoralen-crosslinked triplex. In this structure, the psoralen is conjugated to the TFO and also forms photoadducts on both strands of the duplex, thereby crosslinking them, so that all three strands of the triplex are covalently linked (see Fig. 1B). Because the 196-bp substrate contains a 5′ TG at the triplex-duplex junction that has been shown to form predominantly psoralen monoadducts (33), we used a modified plasmid fragment containing a 5′ TA (APRT-TA, Fig. 1B) at the triplex-duplex junction that is capable of forming psoralen crosslinks with high efficiency (33). Again, RPA bound the triplex DNA substrate with high affinity (Fig. 4A) and specificity. By itself, XPA does not bind substantially to the substrate. Just less than half of the psoralen-crosslinked triplex was bound by RPA at a concentration of 4 × 10−9 M, indicating a similar affinity for a psoralen-crosslinked triplex as compared with the psoralen-monoadducted triplex substrate. At this low concentration of RPA, there is relatively little nonspecific binding to duplex, and those complexes that form seem to dissociate during electrophoresis, resulting in a smear of radioactivity (Fig. 4A, lane 2). Nonetheless, this weak nonspecific binding is again diminished by addition of XPA (Fig. 4A, lane 3). Binding to noncovalent triplex was not observed for either RPA alone, or the XPA–RPA complex, although the triplex likely dissociates under the conditions of the gel shift assay. Because the triplex + UV substrate was formed with high efficiency, we wanted to determine the specificity of RPA binding to covalent triplex DNA substrate when both triplex and duplex DNA substrates were present. Thus, in Fig. 4B, we subjected the preformed triplex to a lower dose of UVA irradiation (0.18 J/cm2) so that less than 50% of the duplex was psoralen-TFO conjugated. As shown in Fig. 4B, lanes 7 and 8, RPA alone and the XPA–RPA complex seemed to specifically shift DNA substrate from the triplex substrate (the upper band in the radiolabeled substrate), demonstrating a triplex-specific interaction with the DNA repair proteins.
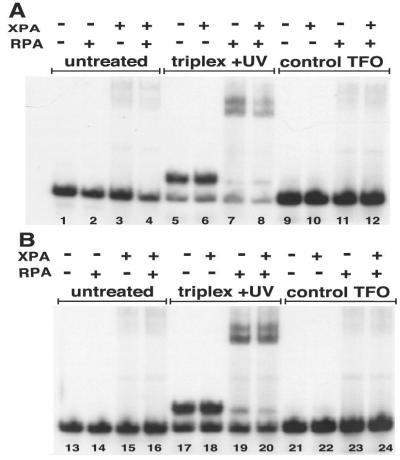
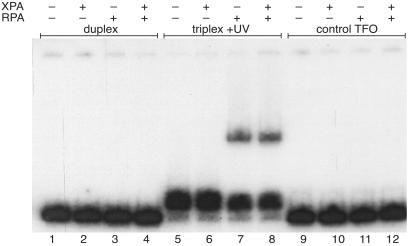
Figure 4.
Recognition of a psoralen-crosslinked triplex by XPA and RPA. (A) The 196-bp modified APRT site (APRT-TA) containing a 5′ TA at the triplex-duplex junction was incubated with the specific psoralen-modified TFO, pTFO1, in the presence or absence of UVA irradiation to form a psoralen-crosslinked triple helix substrate (listed as triplex + UV) or a noncovalent triplex substrate (triplex − UV). Duplex APRT-TA also was incubated with the specificity control psoralen-modified oligonucleotide, pTFOc, (control TFO lanes) and substrates were incubated with RPA and XPA. The samples were subjected to EMSA analyses. All DNA substrates were at a concentration of 5 × 10−8 M. (B) The reactions were performed as in A, only the UVA irradiation dose was decreased to 0.18 J/cm2 so that crosslinked-triplex DNA substrate constituted less than half of the available DNA substrate in the triplex + UV lanes.
Recognition of Short Triplexes by XPA and RPA.
To determine whether binding of XPA and RPA to triplex structures is limited by substrate length, we tested their ability to bind the same APRT triplex-binding site, but with a short (37-bp) synthetic target duplex. We assessed protein binding to the triplex substrates by using radiolabeled 37-bp duplexes containing the APRT triplex target site with a 5′ TG or a 5′ TA at the triplex-duplex junction (see Fig. 1 A and B) after the formation of psoralen monoadducts or crosslinks, respectively. As shown in Fig. 5A, the psoralen-monoadducted triplex was recognized by XPA and RPA specifically, and with high affinity, similar to their affinity for the longer 196-bp DNA substrate. Again, the psoralen-crosslinked triplex was recognized by the proteins with a similar affinity to that of the monoadducted triplex (Fig. 5B).
Figure 5.
Binding of XPA and RPA proteins to short covalent triplex substrates. (A) The psoralen-modified TFOs were incubated with a radiolabeled 37-bp duplex substrate containing the APRT intron 1 site to form a triplex of 19 base triplets (5 × 10−8 M). Triplex + UV lanes indicate a psoralen-monoadducted triplex substrate incubated with XPA and RPA. Control TFO samples were incubated with the specificity control oligonucleotide (pTFOc). Samples were analyzed by EMSA after incubation with proteins. (B) The modified APRT site (APRT-TA) incorporated into a 37-bp synthetic duplex was incubated with pTFO1 to form a psoralen-crosslinked triplex substrate (triplex + UV lanes). Reactions with XPA and RPA were performed as described in A. Control TFO lanes indicate the 37-bp duplex incubated with the specificity control oligonucleotide, pTFOc that does not bind the triplex target site.
Binding of a Triplex in the supFG1 Target Site by XPA and RPA.
Although RPA clearly recognizes the APRT triplex structure on both long and short substrates, we considered the possibility that these effects were unique to that particular triplex site. To address this question we tested a triplex formed on a different TFO-recognition site, the supFG1 triplex site, which has been well characterized (27, 30, 34, 35). Triplexes formed at this site are thought to be substrates for NER (28–30). We used a radiolabeled 188-bp plasmid fragment containing a 5′ AT psoralen crosslinking site at the triplex-duplex junction (Fig. 1C) as the test substrate under the same binding conditions as those used for the APRT triplex substrates. The results shown in Fig. 6 indicate that for this DNA substrate RPA, and the XPA–RPA complex (see below) bind with high affinity and specificity to the psoralen-crosslinked triplex only, consistent with our observations with the APRT triplex substrate (see Fig. 4).
Figure 6.
XPA and RPA recognize the supFG1 covalent triplex. A 188-bp plasmid fragment containing a 30-bp triplex target site in the supFG1 gene was incubated with a psoralen-modified TFO, pAG30, to form a triplex. Selected samples were irradiated with UVA (triplex + UV) as indicated to form psoralen-crosslinked triplex substrates (at 5 × 10−8 M) and then incubated with XPA and RPA as indicated by the (+) and (−) signs. The control TFO lanes contained radiolabeled duplex incubated with the control oligonucleotide, pSCR30. Protein binding was assessed by EMSA on a 4% polyacrylamide gel.
Presence of XPA in the Mobility-Shifted DNA–RPA Complex.
To test both DNA length limitations on XPA and RPA binding to the supFG1 triplex and to confirm the presence of the proteins in the shifted DNA complexes, we used a short synthetic duplex as the target substrate for electrophoretic mobility shift assays (EMSA) and antibody-supershifting experiments. The results of XPA and RPA binding to the 57-bp duplex target in the presence of the 30 base TFO with psoralen crosslinks are shown in Fig. 7A. Again, the short length of the substrate does not appear to hinder binding by the proteins to the triplex structures, consistent with the results from the APRT triplexes. Addition of XPA-specific antibodies gives rise to a distinct supershift of the XPA–RPA–DNA complex, as shown in Fig. 7. Although XPA binding to the RPA–DNA complex does not by itself induce a further shift in mobility, likely because the mass-to-charge ratio of the complex is unchanged upon binding of the negatively charged XPA, the XPA-dependent supershifting effect of XPA antibodies implies that XPA must be present in the protein DNA complex. RPA antibodies, in contrast, did not induce supershifts (Fig. 7 A and B). Rather, they diminished slightly the amount of shifted RPA–DNA complexes, indicating that antibody binding may interfere with the damage recognition site on RPA.
XPA and RPA Binding in the Absence of DNA.
To test whether our preparations of recombinant proteins interact as expected, we performed ELISA analysis with the purified proteins used in the studies described in this work and the XPA-specific antibody. As demonstrated in Fig. 7C, XPA binds tightly to immobilized RPA but not to the control proteins BSA and proliferating cell nuclear antigen (PCNA). This result is consistent with our conclusion that in the presence of XPA, RPA binds triplex DNA as an XPA–RPA complex.
Discussion
Triplexes and NER.
Previous work has provided indirect evidence for the recognition of triplex DNA structures by the NER machinery. For example, we have demonstrated triplex-induced mutagenesis in the supFG1 reporter gene on plasmids, on chromosomal targets in cells, and in transgenic mice (27, 34, 35), but when TFOs were targeted to a plasmid substrate in NER-deficient XPA cells, no induction of mutagenesis was detected (30). This result suggested a requirement for NER in the triplex-directed mutagenic pathway. However, the mechanisms for triplex recognition are not known, and no direct evidence for triplex binding by NER proteins exists. In this study, we have obtained direct evidence for recognition of triplexes by the human XPA–RPA repair complex and uncovered a distinct role for each of these repair proteins. RPA binds to triplexes with high affinity, with or without XPA, whereas XPA binds only in conjunction with RPA, but contributes to specificity by selectively diminishing nonspecific binding and/or aggregation of RPA to undamaged DNA.
Binding of XPA and RPA to Psoralen Monoadducts and Crosslinks.
Psoralen monoadducts and crosslinks are, at least in part, recognized and repaired by NER in human cells (39–46). Psoralen monoadducts are somewhat helix-stabilizing, whereas crosslinks are helix-destabilizing (47, 48). If XPA and RPA recognize helical distortions, then we would not expect a psoralen monoadduct to be recognized with the same efficiency as a psoralen crosslink. However, we found that neither a psoralen monoadduct nor crosslink are recognized by XPA or RPA under the conditions of our assay in the absence of the third strand TFO, which suggests that the triplex structure itself provides a strong damaged DNA signal to the NER machinery. Because the gel-mobility shift assay did not detect binding of XPA or RPA to the noncovalent triplex DNA alone, the covalent attachment of the third strand to the underlying duplex via psoralen may be necessary to maintain the integrity of the triplex structure in the presence of the proteins and the binding buffer used in these studies. For example, high levels of potassium are known to inhibit triplex formation (36, 49), and in our studies inclusion of potassium in the buffer was necessary for stable protein-DNA-binding interactions. Alternatively, the addition of the psoralen monoadduct or crosslink at the triplex-duplex junction may induce a greater distortion to the DNA helix than either the triplex or psoralen photoadducts alone, thus providing a stronger signal to the DNA damage recognition proteins.
Substrate Length Independence of Binding by RPA and the XPA–RPA Complex.
It has been suggested that a minimum length of DNA for efficient cleavage by the excision nuclease of the NER machinery is near 100 bp (50). To determine whether the size and structure of the DNA substrate would pose a limit to triplex recognition by the XPA–RPA complex, we measured the binding of XPA and RPA to two different triplexes, formed on targets presented by short synthetic duplexes (37 bp or 57 bp) or on longer (188 bp or 196 bp) plasmid fragments. We found no substantial length-dependent differences in the binding affinities of RPA to the triplex structures in this range. Several differences occur between the triplex sites, including the length and sequence of the triplex itself and the sequence context surrounding the triplex target site. Because the APRT and supFG1 triplexes were recognized with near equal affinities by XPA and RPA, perhaps it is the overall distortion in the helix or the reverse Hoogsteen hydrogen-bonding pattern that is being recognized by NER, regardless of the length or sequence of the individual triplex.
RPA Binds Triple-Stranded DNA Structures.
In humans, the RPA protein is involved in many aspects of DNA metabolism including DNA repair, recombination, and replication (reviewed in ref. 51). RPA has a strong preference for binding to single-stranded DNA, but also has an affinity for double-stranded DNA. We have now demonstrated that RPA can also recognize and bind to triple-stranded DNA structures, which may be of utility in the cell where H-DNA (intramolecular triplexes) structures are thought to form transiently during DNA metabolic processes such as transcription and may play a role in certain human hereditary disorders (32).
DNA Damage Recognition.
We have demonstrated the recognition of triplex structures by the NER damage recognition complex, XPA–RPA. This finding expands our understanding of both the recognition of distorted DNA structures by NER, and the processing of triplexes by NER that may contribute to the triplex-induced mutagenesis reported by our group and others (27, 30, 34, 35, 52–58). In addition, both in vitro and in vivo studies have revealed a role for XPA in the induction of recombination by triple helix formation (28, 29). The results presented here support the proposed model in which the triplex is recognized by XPA-RPA, leading to DNA repair activity that generates recombination intermediates.
Stability of the triplex structure is afforded by Hoogsteen (or reverse Hoogsteen) hydrogen bond formation through the major groove by the nucleotides on the TFO pairing with the purine-rich strand of the underlying duplex. Thus, in addition to the helical distortions produced upon triplex formation, the hydrogen-bonding pattern is altered as well (the Watson–Crick hydrogen bonds in the underlying duplex are not disrupted). Our results indicate that the XPA–RPA complex may recruit the NER machinery to sites of triplex-induced helical distortions, and the Hoogsteen hydrogen bonds, as well as the third strand itself, may play a role in the recognition process. This finding is consistent with a “bipartite” recognition model (18) in that a triplex structure produces both a distortion of the DNA structure and a modification of the DNA chemistry. Further studies are required to determine the physical and chemical details of the recognition signal(s) in human NER.
The mechanism involved in the initial damage recognition step is currently being studied (reviewed in ref. 59), and much discussion still goes on as to which protein or protein complex is the first to recognize and bind to damaged DNA. The kinetic interactions of the XPA–RPA complex and the XPC–hHR23B complex with damaged DNA and with each other have yet to be clearly elucidated. For example, a previous report has suggested, on the basis of order-of-addition experiments, that XPC may be the initial damage recognition protein, with XPA and RPA acting as damage verification proteins subsequent to XPC binding (17). Additional studies will be needed to determine the contribution of XPC to recognition of unusual DNA structures, including triplex structures.
Acknowledgments
We thank T. G. Wensel for critical review of the manuscript, R. Black for technical assistance, and M. Gardiner for manuscript preparation. This work was supported by National Institutes of Health Grants CA93729 (to K.M.V.), AI26109 (to J.C.), CA64186 (to P.M.G.), the Leukemia and Lymphoma Society (to P.M.G.), and the Anna Fuller Fund and National Research Service Award CA75723 (to K.M.V.).
Abbreviations
- NER
nucleotide excision repair
- RPA
replication protein A
- TFO
triplex-forming oligonucleotide
- XPA
xeroderma pigmentosum complementation group A protein
- APRT
adenine phosphoribosyltransferase
- HMT
2-[4′-(hydroxymethyl)-4,5′,8trimethylpsoralen]-hexyl-1-O-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Auerbach A D, Verlander P C. Curr Opin Pediatr. 1997;9:600–616. doi: 10.1097/00008480-199712000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Berneburg M, Lehmann A R. Adv Genet. 2001;43:71–102. doi: 10.1016/s0065-2660(01)43004-5. [DOI] [PubMed] [Google Scholar]
- 3.Cleaver J E, Thompson L H, Richardson A S, States J C. Hum Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 4.de Boer J, Hoeijmakers J H. Carcinogenesis. 2000;21:453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- 5.Woods C G. Arch Dis Child. 1998;78:178–184. doi: 10.1136/adc.78.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doll R, Peto R. J Natl Cancer Inst. 1981;66:1191–1308. [PubMed] [Google Scholar]
- 7.Helene C. Adv Biophys. 1985;20:177–186. doi: 10.1016/0065-227x(85)90036-x. [DOI] [PubMed] [Google Scholar]
- 8.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 9.Wood R D. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 10.Friedberg E C, Walker G C, Siede W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 11.Asahina H, Kuraoka I, Shirakawa M, Morita E H, Miura N, Miyamoto I, Ohtsuka E, Okada Y, Tanaka K. Mutat Res. 1994;315:229–237. doi: 10.1016/0921-8777(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 12.Buschta-Hedayat N, Buterin T, Hess M T, Missura M, Naegeli H. Proc Natl Acad Sci USA. 1999;96:6090–6095. doi: 10.1073/pnas.96.11.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones C J, Wood R D. Biochemistry. 1993;32:12096–12104. doi: 10.1021/bi00096a021. [DOI] [PubMed] [Google Scholar]
- 14.Robins P, Jones C J, Biggerstaff M, Lindahl T, Wood R D. EMBO J. 1991;10:3913–3921. doi: 10.1002/j.1460-2075.1991.tb04961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wakasugi M, Sancar A. J Biol Chem. 1999;274:18759–18768. doi: 10.1074/jbc.274.26.18759. [DOI] [PubMed] [Google Scholar]
- 16.Reardon J T, Mu D, Sancar A. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 17.Sugasawa K, Ng J M, Masutani C, Iwai S, van der Spek P J, Eker A P, Hanaoka F, Bootsma D, Hoeijmakers J H. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 18.Hess M T, Schwitter U, Petretta M, Giese B, Naegeli H. Proc Natl Acad Sci USA. 1997;94:6664–6669. doi: 10.1073/pnas.94.13.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asensio J L, Dosanjh H S, Jenkins T C, Lane A N. Biochemistry. 1998;37:15188–15198. doi: 10.1021/bi980057m. [DOI] [PubMed] [Google Scholar]
- 20.Han Z J, Rhee S, Liu K, Miles H T, Davies D R. Acta Crystallogr D. 2000;56:104–105. doi: 10.1107/s0907444999012895. [DOI] [PubMed] [Google Scholar]
- 21.He Y, Scaria P V, Shafer R H. Biopolymers. 1997;41:431–441. doi: 10.1002/(SICI)1097-0282(19970405)41:4<431::AID-BIP7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 22.Johnson K H, Durland R H, Hogan M E. Nucleic Acids Res. 1992;20:3859–3864. doi: 10.1093/nar/20.15.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kan L S, Callahan D E, Trapane T L, Miller P S, Ts'o P O, Huang D H. J Biomol Struct Dyn. 1991;8:911–933. doi: 10.1080/07391102.1991.10507857. [DOI] [PubMed] [Google Scholar]
- 24.Rhee S, Han Z, Liu K, Miles H T, Davies D R. Biochemistry. 1999;38:16810–16815. doi: 10.1021/bi991811m. [DOI] [PubMed] [Google Scholar]
- 25.Sun X G, Cao E H, He Y J, Qin J F. J Biomol Struct Dyn. 1999;16:863–872. doi: 10.1080/07391102.1999.10508298. [DOI] [PubMed] [Google Scholar]
- 26.Vasquez K M, Wilson J H. Trends Biochem Sci. 1998;23:4–9. doi: 10.1016/s0968-0004(97)01158-4. [DOI] [PubMed] [Google Scholar]
- 27.Vasquez K M, Narayanan L, Glazer P M. Science. 2000;290:530–533. doi: 10.1126/science.290.5491.530. [DOI] [PubMed] [Google Scholar]
- 28.Faruqi A F, Datta H J, Carroll D, Seidman M M, Glazer P M. Mol Cell Biol. 2001;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta H J, Chan P P, Vasquez K M, Gupta R C, Glazer P M. J Biol Chem. 2001;270:18018–18023. doi: 10.1074/jbc.M011646200. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Seidman M M, Glazer P M. Science. 1996;271:802–805. doi: 10.1126/science.271.5250.802. [DOI] [PubMed] [Google Scholar]
- 31.Wells R D. J Biol Chem. 1988;263:1095–1098. [PubMed] [Google Scholar]
- 32.Bowater R P, Wells R D. Nucleic Acids Res. 2001;66:159–202. doi: 10.1016/s0079-6603(00)66029-4. [DOI] [PubMed] [Google Scholar]
- 33.Vasquez K M, Wensel T G, Hogan M E, Wilson J H. Biochemistry. 1996;35:10712–10719. doi: 10.1021/bi960881f. [DOI] [PubMed] [Google Scholar]
- 34.Vasquez K M, Wang G, Havre P A, Glazer P M. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang G, Levy D D, Seidman M M, Glazer P M. Mol Cell Biol. 1995;15:1759–1768. doi: 10.1128/mcb.15.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vasquez K M, Wensel T G, Hogan M E, Wilson J H. Biochemistry. 1995;34:7243–7251. doi: 10.1021/bi00021a040. [DOI] [PubMed] [Google Scholar]
- 37.Li L, Peterson C A, Lu X, Legerski R J. Mol Cell Biol. 1995;15:1993–1998. doi: 10.1128/mcb.15.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen J, Cotmore S F, Tattersall P. J Virol. 1995;69:5422–5430. doi: 10.1128/jvi.69.9.5422-5430.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Averbeck D, Papadopoulo D, Moustacchi E. Cancer Res. 1988;48:2015–2020. [PubMed] [Google Scholar]
- 40.Averbeck D, Dardalhon M, Magana-Schwencke N. J Photochem Photobiol. 1990;6:221–236. doi: 10.1016/1011-1344(90)85092-b. [DOI] [PubMed] [Google Scholar]
- 41.Calsou P, Sage E, Moustacchi E, Salles B. Biochemistry. 1996;35:14963–14969. doi: 10.1021/bi9607261. [DOI] [PubMed] [Google Scholar]
- 42.Cheng S, Sancar A, Hearst J E. Nucleic Acids Res. 1991;19:657–663. doi: 10.1093/nar/19.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reardon J T, Spielmann P, Huang J C, Sastry S, Sancar A, Hearst J E. Nucleic Acids Res. 1991;19:4623–4629. doi: 10.1093/nar/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sancar A, Franklin K A, Sancar G, Tang M S. J Mol Biol. 1985;184:725–734. doi: 10.1016/0022-2836(85)90316-x. [DOI] [PubMed] [Google Scholar]
- 45.Sibghatullah, Husain I, Carlton W, Sancar A. Nucleic Acids Res. 1989;17:4471–4484. doi: 10.1093/nar/17.12.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sladek F M, Melian A, Howard-Flanders P. Proc Natl Acad Sci USA. 1989;86:3982–3986. doi: 10.1073/pnas.86.11.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunz D, Hess M T, Naegeli H. J Biol Chem. 1996;271:25089–25098. doi: 10.1074/jbc.271.41.25089. [DOI] [PubMed] [Google Scholar]
- 48.Shi Y B, Griffith J, Gamper H, Hearst J E. Nucleic Acids Res. 1988;16:8945–8952. doi: 10.1093/nar/16.18.8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng A J, Van Dyke M W. Nucleic Acids Res. 1993;21:5630–5635. doi: 10.1093/nar/21.24.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang J C, Sancar A. J Biol Chem. 1994;269:19034–19040. [PubMed] [Google Scholar]
- 51.Wold M S. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 52.Barre F X, Ait-Si-Ali S, Giovannangeli C, Luis R, Robin P, Pritchard L L, Helene C, Harel-Bellan A. Proc Natl Acad Sci USA. 2000;97:3084–3088. doi: 10.1073/pnas.050368997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Havre P A, Glazer P M. J Virol. 1993;67:7324–7331. doi: 10.1128/jvi.67.12.7324-7331.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Havre P A, Gunther E J, Gasparro F P, Glazer P M. Proc Natl Acad Sci USA. 1993;90:7879–7883. doi: 10.1073/pnas.90.16.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majumdar A, Khorlin A, Dyatkina N, Lin F-L M, Powell J, Liu J, Fei Z, Khripine Y, Watanabe K A, George J, et al. Nat Genet. 1998;20:212–214. doi: 10.1038/2530. [DOI] [PubMed] [Google Scholar]
- 56.Panyutin I G, Neumann R D. Nucleic Acids Res. 1994;22:4979–4982. doi: 10.1093/nar/22.23.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandor Z, Bredberg A. Nucleic Acids Res. 1994;22:2051–2056. doi: 10.1093/nar/22.11.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sandor Z, Bredberg A. FEBS Lett. 1995;374:287–291. doi: 10.1016/0014-5793(95)01133-y. [DOI] [PubMed] [Google Scholar]
- 59.Wood R D. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]