Abstract
The general transcription factor TFIID facilitates recruitment of the transcription machinery to gene promoters and regulates initiation of transcription by RNA polymerase II. hTAFII130, a component of TFIID, interacts with and serves as a coactivator for multiple transcriptional regulatory proteins, including Sp1 and CREB. A yeast two-hybrid screen has identified an interaction between hTAFII130 and heterochromatin protein 1 (HP1), a chromatin-associated protein whose function has been implicated in gene silencing. We find that hTAFII130 associates with HP1 in an isoform-specific manner: HP1α and HP1γ bind to hTAFII130, but not HP1β. In addition, we show that endogenous hTAFII130 and components of TFIID in HeLa nuclear extracts associate with glutathione S-transferase-HP1α and -HP1γ. hTAFII130 possesses a pentapeptide HP1-binding motif, and mutation of the hTAFII130 HP1 box compromises the interaction of hTAFII130 with HP1. We demonstrate that Gal4-HP1 proteins interfere with hTAFII130-mediated activation of transcription. Our results suggest that HP1α and HP1γ associate with hTAFII130 to mediate repression of transcription, supporting a new model of transcriptional repression involving a specific interaction between a component of TFIID and chromatin.
The general transcription factor TFIID, through the activities of its composite TATA-binding protein (TBP) and TBP-associated factors (TAFIIs), plays a central role in the regulation of transcription by RNA polymerase II (reviewed in refs. 1–4). TAFIIs participate in transcription by serving as molecular integrators of signals mediated by site-specific transcription factors, assisting in promoter recognition, enzymatically modifying target proteins, and facilitating the nucleation of the preinitiation complex formation. A plethora of biochemical and genetic data support the notion that TAFIIs function at the interface between gene-specific transcriptional regulators and general transcription machinery.
Human TFIID, composed of TBP and 13 associated TAFIIs, is required for activator-dependent transcription in vitro. hTAFII130 and its Drosophila homologue dTAFII110 directly contact the glutamine-rich activation domains of Sp1 and function as Sp1's coactivator (5–7). dTAFII110 and hTAFII130 also interact with the Q2 activation domain of the cAMP-responsive transcription factor CREB and mediates its activation function (7–9). In addition, hTAFII130 increases transcriptional activation by the retinoic acid, vitamin D3, and thyroid hormone receptors without directly contacting their activation domains (10).
We have mapped the domains of hTAFII130 that interact with Sp1 and CREB to the central glutamine-rich regions (refs. 11 and 12, Fig. 1A). hTAFII130 shares two highly conserved regions, CI and CII, with dTAFII110, Caenorhabditis elegans TAF-5 (13), and hTAFII105, a human TAFII first identified in B cells and recently shown to be essential for ovarian development (refs. 14 and 15). The CII is involved in interactions with other TAFIIs as well as TFIIA and is required for incorporation of hTAFII130 into the TFIID complex (16). hTAFII130, through a histone-like motif in CII, heterodimerizes with hTAFII20 to form a histone-like pair in TFIID (17). The histone-fold motifs found in many TAFIIs are thought to mediate subunit interactions in the TFIID complex (reviewed in ref. 18). Studies to date have focused largely on the coactivator function of hTAFII130; few reports have pointed to a role of hTAFII130 in supporting transcriptional repression.
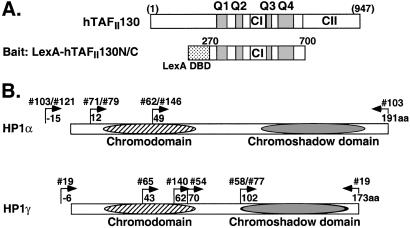
Figure 1.
Multiple clones of human HP1α and HP1γ are isolated in a yeast two-hybrid screen by using the central domain of hTAFII130 as bait. (A) A schematic representation of the bait protein LexA-hTAFII130N/C composed of the DNA-binding domain (DBD) of LexA fused to residues 270–700 of hTAFII130 (numbering according to ref. 7). Conserved region I (CI), conserved region II (CII), and glutamine-rich regions (Q1 to Q4) are indicated. (B) A schematic representation of HP1α and HP1γ clones isolated from the yeast screen. Arrows demarcate the sequence boundaries for each clone. The 3′ end of all isolated HP1 clones included the complete C-terminal coding sequence of HP1. The clone numbers and their amino acid positions are indicated for each isolate. The smallest clone contained only the chromoshadow domain of HP1γ. Protein interaction was not detectable between HP1 and LexA DBD alone or with other transcription factors. The expression of HA-tagged HP1 proteins was confirmed in yeast cell lysates by immunoblotting with α-HA antibody.
Modulation of chromatin structure plays a fundamental role in gene expression because transcription factors must contend with the nucleosomes, which are generally inhibitory to transcription. Genetic and biochemical approaches have led to the discovery of multiple transcription factor complexes that are thought to activate or repress transcription by targeting histones or nucleosomes (reviewed in refs. 19–21). A diverse array of posttranslational modifications is made to the core histone tails that are thought to bring about distinct events affecting gene expression (reviewed in refs. 22 and 23). It has been proposed that histone N-terminal modifications serve as a code to recruit specific proteins to the chromatin template and regulate gene expression (24). Recent findings have implicated a functional link between TFIID and chromatin components. Several TAFIIs in TFIID contain histone-like motifs (18, 25), and the largest TAFII250 subunit possesses acetyltransferase (26) and ubiquitin-conjugating activities (27) that target histones and likely contribute to alterations in chromatin structure and facilitation of transcription. Furthermore, the bromodomains of TAFII250 have been shown to bind to the acetylated tails of histone H4 (28). Here, we report that hTAFII130 interacts with certain isoforms of HP1, a chromatin-associated protein whose function has been implicated in gene silencing. We have examined the interaction of HP1 with hTAFII130 in the context of TFIID and analyzed the effect of HP1 on transcriptional activation mediated by hTAFII130. Our results support the notion that hTAFII130 mediates transcriptional repression through an interaction with HP1.
Methods
Yeast Two-Hybrid Methods.
The Brent yeast two-hybrid system (29) was used to screen a HeLa cell cDNA library in pJG4–5 (a gift from M. Garabedian, New York University School of Medicine) with hTAFII130N/C in pEG202 as bait (12) in the yeast strain EGY188 carrying the reporter plasmid pJK103. High-efficiency transformation of the library plasmids using TRAFO protocol (30) yielded 7.5 × 106 colonies that were plated on selective media and plates containing 5-bromo-4-chloro-3-indolyl β-d-galactoside. Blue colonies were picked and their pJG4–5 plasmids were isolated by yeast DNA miniprep protocol and electroporated into the KC8 (Trp−) bacterial strain, as described (31). Restriction digests were performed to confirm the presence of library inserts in pJG4–5, and positive clones were sequenced.
Plasmids.
Glutathione S-transferase (GST)-HP1 fusions were generated by PCR amplification of HP1 coding sequence and ligation into the vector pGEX-4T-1 by using EcoRI and XhoI. DNA templates used for amplification of HP1α and HP1γ were pJG4–5 HeLa cDNA library plasmids isolated from the yeast two-hybrid screen. An expressed sequence tag clone containing the coding sequence of HP1β (GenBank accession no. BE315541) was used as template. HP1γΔN lacking the N-terminal 70 amino acids and HP1γΔC lacking the C-terminal 25 amino acids were constructed by PCR amplification of corresponding DNA fragments. All DNA constructs generated by PCR were sequenced. The same HP1 DNA fragments were subcloned into pcDNA3.1-HA-Gal4 (1–94) for expression in mammalian cells. CMV-LexA-hTAFII130N/C was constructed by subcloning a DNA fragment encoding hTAFII130N/C into pCS2+ expression vector containing LexA DBD (ref. 32; a gift from K. Struhl, Harvard Medical School, Boston). For in vitro translation, hTAFII130 (1–947) and derivatives were subcloned into the vector pTβSTOP (33). hTAFII130N/C-DE was generated by using QuikChange Site-Directed Mutagenesis Kit (Stratagene) with the following oligonucleotides harboring the mutation (underlined). 5′-GTTGACGCAGACACCTATGGACGCCGAGCGGCAGCCTCACAAC-3′ and 5′-GTTGTGAGGCTGCCGCTCGGCGTCCATAGGTGTCTGCGTCAAC-3′. Mutant clones were identified by the loss of NcoI cleavage site.
GST Pull-Down Assays.
All GST fusion proteins were made in the low protease Escherichia coli strain SG1117 (a gift from H. Samuels, New York University School of Medicine). E. coli cultures were grown to an OD600 of ≈0.6, induced with isopropyl thio-β-d-galactoside for 45 min at 30°C and resuspended in HEMG buffer [25 mM Hepes-KOH, pH 7.9/0.1 mM EDTA/12.5 mM MgCl2/20% (vol/vol) glycerol] containing 0.1M KCl, 0.1% Nonidet P-40, and protease inhibitors (Roche Molecular Biochemicals). Recombinant proteins were purified following incubation with glutathione Sepharose 4B (Amersham Pharmacia Biotech). In vitro-translated [35S]methionine-labeled proteins were synthesized by using the TNT-coupled reticulocyte lysate system (Promega). Radiolabeled lysate (5–10 μl) was added to each binding reaction containing 1–2 μg of purified GST protein in 200 μl of YE buffer [150 mM NaCl/20 mM Hepes, pH 7.4/10% (vol/vol) glycerol/0.05% BSA/0.05% Nonidet P-40] (34). The reactions were carried out at 4°C for 2–3 h with nutation. After five washings with YE buffer, bound proteins were separated by SDS/PAGE and analyzed by autoradiography. Experiments to identify endogenous TFIID components associating with GST-HP1 resins used mininuclear extracts (35) prepared from HeLa cells stably transfected with a control plasmid or a plasmid expressing inducible HA-hTAFII130 (1–947) (a gift from S. Giannakopoulos, New York University School of Medicine). One milligram of nuclear extract prepared from induced (removal of doxycycline) or uninduced (growth in 30 ng/ml doxycycline) cells was incubated with each resin for 3 h with nutation at 4°C. After five washings in YE buffer, bound proteins were separated by SDS/PAGE, transferred onto nitrocellulose membrane, and sequentially probed with the following antibodies: α-HA (12CA5), α-hTAFII130, α-hTAFII250, α-hTAFII100 (α-hTAFII antibodies were gifts from E. Wang, University of Washington, Seattle). α-hTBP (SL39, a gift from N. Hernandez, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) was used in a separate experiment. Immunoreactive bands were detected with an Enhanced ChemiLuminescence kit (Amersham Pharmacia).
Transfection Assays.
Transient transfections were performed by using the calcium phosphate precipitation method in 35-mm tissue culture dishes of subconfluent HeLa cells that had been passaged 1 day before transfection. The total DNA per transfection was 3 μg. The mammalian expression plasmids used were: pcDNA3.1-HA-Gal4 (1–94) fused to HP1α, HP1β, HP1γ, or HP1γΔC, and CMV-hTAFII130N/C (12) or CMV-LexA-hTAFII130N/C. 0.5 μg of the 2XGal/2XLex-E1bTATA-luciferase reporter plasmid (32) (a gift from K. Struhl, Harvard Medical School, Boston) was cotransfected with 50 ng of LexA-hTAFII130N/C and 50 ng of the Gal4-HP1 expression plasmids. Comparable levels of protein expression were confirmed by immunoblotting. Luciferase assays were performed as described (12). Data shown are from a representative experiment carried out a minimum of three times.
Results
Identification of an Interaction Between hTAFII130 and HP1.
To gain further insight into the functions of hTAFII130, we set out to identify hTAFII130-interacting proteins by using a yeast two-hybrid screen. The bait was composed of the central domain of hTAFII130 including four glutamine-rich regions and CI fused to the LexA DNA-binding domain (LexA-hTAFII130N/C, Fig. 1A). We knew from previous studies that hTAFII130N/C fragment interacted positively with the activation domains of Sp1 and CREB, which served as positive controls (11, 12). Approximately 7.5 × 106 transformants were screened from a HeLa cell cDNA library and, unexpectedly, clones encoding human HP1α or HP1γ were each isolated six times (Fig. 1B).
The HP1 family, composed of three isoforms (α, β, and γ) in mammals, are chromatin-associated factors, whose Drosophila homologue has a well established function in epigenetic silencing (36). The three mammalian isoforms of HP1 exhibit distinct localization in the nucleus: HP1α and β localize predominantly to heterochromatin, and HP1γ localizes to both heterochromatin and euchromatin (ref. 37 and references therein). Although earlier studies have implicated HP1 in the regulation of chromatin structure through interactions with proteins in heterochromatin, HP1 also has been found to associate with euchromatic regions where it may play a more dynamic role in the regulation of gene expression (reviewed in refs. 38 and 39).
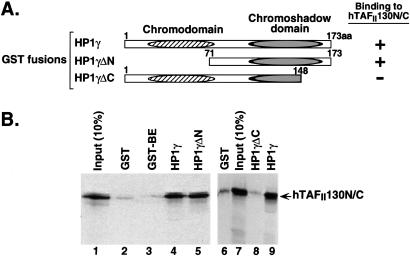
The multiple isolates of HP1α (three unique clones isolated two times each) and HP1γ (five unique clones) shared a common region corresponding to the “chromoshadow domain” of HP1. The chromoshadow domain shares sequence homology with the chromodomain that lies near the N terminus of HP1. Chromodomains have been identified in many factors that affect gene expression and chromatin structure (39), whereas the chromoshadow domain is unique to HP1 (40). To define further the region of HP1γ required for interaction with hTAFII130, we performed binding studies by using an in vitro-translated and -radiolabeled hTAFII130 polypeptide fragment that had been used as bait in the yeast two-hybrid screen (Fig. 2). hTAFII130N/C was retained on GST-HP1γ resin but not on GST alone (Fig. 2B, lanes 2, 4, 6, and 9). Deletion of the C-terminal 25 residues of HP1γ (HP1γΔC) that truncated the chromoshadow domain abrogated the interaction (lanes 8 and 9). In contrast, deletion of the N-terminal 70 residues encompassing the entire chromodomain (HP1γΔN) had no effect on binding of hTAFII130N/C to HP1γ (lanes 4 and 5). Comparable amounts of GST proteins were used in all experiments (data not shown). These results indicate that the chromoshadow domain is essential for the interaction with hTAFII130, consistent with the findings from the yeast two-hybrid screen.
Figure 2.
The HP1 C-terminal domain is required for its association with hTAFII130 in vitro. (A) A schematic representation of the GST-HP1 fusion proteins used in the study. (B) In vitro-translated hTAFII 130N/C was incubated with comparable amounts of the indicated GST fusion proteins. The result (summarized in A at right) suggests that an intact chromoshadow domain of HP1γ is required for hTAFII130 interaction. GST-BE is a control fusion protein containing a small fragment of hTAFII130 similar in size to HP1γ.
A Pentapeptide Motif in hTAFII130 Is Important for Interaction with HP1γ.
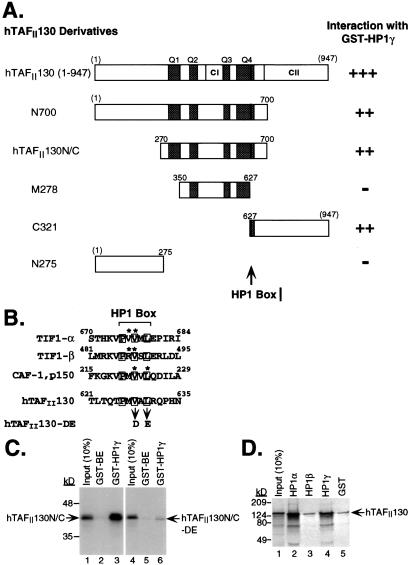
By using immobilized GST-HP1γ and in vitro-translated derivatives of hTAFII130, we set out to identify the region of hTAFII130 that is important for interaction with GST-HP1γ (Fig. 3A). Significantly, a derivative of hTAFII130, slightly smaller than hTAFII130N/C, corresponding to the central 278 residues (M278) failed to interact with GST-HP1γ. By contrast, derivative C321 containing the C-terminal 321 residues efficiently bound to HP1γ. Based on the retention patterns of hTAFII130N/C, M278 and C321, we identified a region (residues 627–700) near the C terminus of hTAFII130N/C that was essential for interaction with GST-HP1γ (Fig. 3A). In addition, we found that CII was dispensable for the interaction, but its presence increased the binding of hTAFII130 to the GST-HP1γ up to five-fold. The N terminus of hTAFII130 did not contribute to the interaction with HP1γ, and the N-terminal 275 residues (N275) did not interact with HP1γ.
Figure 3.
hTAFII130 binds to HP1 through an HP1 box present in hTAFII130. (A) hTAFII130 derivatives were translated in vitro and incubated with GST-HP1γ. Relative amounts of hTAFII130 derivative bound to GST-HP1γ are indicated qualitatively at right. The thick vertical bar represents newly identified HP1 interaction motif (HP1 box) present in all hTAFII130 constructs that associate with HP1. (B) An alignment of HP1-associated protein sequences containing an HP1 box (43). Conserved residues (consensus PXVXL where X is any amino acid) are boxed. *, residues that, when mutated, compromised the binding of each protein to HP1 in vitro. hTAFII130-DE is a mutant with alterations of the two conserved amino acids in the HP1 box. (C) Point mutations in the HP1 box compromised hTAFII130 binding to HP1γ. In vitro-translated wild type and the DE mutant of hTAFII130N/C was incubated with GST-HP1γ. A comparable amount of GST-BE was used as a control for nonspecific binding. (D) hTAFII130 bound to HP1α and HP1γ but not to HP1β. GST fusions of HP1α, HP1β, and HP1γ were incubated with in vitro-translated hTAFII130 (1–947). The bound fractions were separated by SDS/PAGE and visualized by autoradiography.
Studies have shown that HP1 interacts with the transcriptional corepressor TIF-1β/KAP-1 and the p150 subunit of the chromatin assembly factor-1 (CAF-1) through a pentapeptide motif termed an “HP1 box” (41, 42). Inspection of the amino acid sequence of the C-terminal region of hTAFII130N/C overlapping with the derivative C321 identified a pentapeptide sequence (PMVAL) resembling the HP1 box (consensus PXVXL, where X is any amino acid). We compared known HP1-interacting proteins and their HP1 boxes with the potential hTAFII130 HP1 box (ref. 43, Fig. 3B). Mutations that changed the conserved residues within the HP1 box from hydrophobic to charged residues have been shown to compromise their binding to HP1 (44). To address directly whether the potential hTAFII130 HP1 box is important for interaction with HP1γ, we mutated the pentapeptide motif from the wild-type sequence of PMVAL to PMDAE (Fig. 3B). Mutation of the hTAFII130 HP1 box in the context of hTAFII130N/C (to create hTAFII130N/C-DE) dramatically reduced its ability to bind to GST-HP1γ, indicating that the HP1 box is critical for the interaction between hTAFII130N/C and HP1γ (Fig. 3C).
hTAFII130 Exhibits Differential Binding to the Three HP1 Isoforms.
Intrigued by our isolation of multiple clones of HP1α and HP1γ but not HP1β in the yeast two-hybrid screen, we sought to determine if hTAFII130 exhibits preferences for interacting with the HP1 isoforms. We generated GST fusions of full-length HP1α, HP1β, and HP1γ and examined their ability to bind to in vitro-translated hTAFII130 (Fig. 3D). hTAFII130 (1–947) was efficiently retained by GST-HP1α and GST-HP1γ but not by GST-HP1β (lanes 2–4). hTAFII105, a human TAFII closely related to hTAFII130 (14), did not bind to the three GST-HP1 resins (data not shown), consistent with the lack of an apparent HP1 box in hTAFII105.
Also, we have detected the association of BRG1, an ATPase subunit of the mammalian SWI/SNF complex reported to bind to HP1α (45), with all three isoforms of HP1 as determined by immunoblotting (data not shown). Mass spectrometric analysis of HeLa nuclear proteins retained by the GST-HP1 resins but not by GST-HP1ΔC identified TIF-1β/KAP-1 (44, 45) as well as AHNAK, a 700-kDa cell cycle-regulated protein (46) as HP1-associated proteins (data not shown). Because both BRG1 and TIF-1β were found to associate with all three isoforms of HP1, the inability of hTAFII130 to bind to HP1β is unlikely because of misfolding of HP1β. A reciprocal binding experiment carried out with immobilized GST-hTAFII130 (residues 410–947) demonstrated that purified recombinant HP1α bound specifically to hTAFII130 (410–947) as well as to all three HP1 isoforms (data not shown), confirming HP1's ability to self-associate (47). In summary, the in vitro binding experiments demonstrated isoform-specific interactions between hTAFII130 and HP1.
HP1 Associates with Endogenous hTAFII130 in the Context of TFIID in a Mammalian Nuclear Extract.
Having demonstrated specific interactions between hTAFII130 and HP1α and HP1γ in vitro, we sought to examine these interactions by using endogenous material from mammalian cells. We had previously generated a HeLa cell line that stably expressed an inducible HA-tagged hTAFII130 (1–947, HA-hTAFII130; S. Giannakopoulos and N.T., unpublished work). Nuclear extracts prepared from a control cell line or the HA-hTAFII130 cell line under induced and uninduced conditions were incubated with GST-HP1γ resin. Proteins that remained bound after extensive washings were resolved by SDS/PAGE and visualized by immunoblotting (Fig. 4A). In the extract obtained from cells induced to overexpress HA-hTAFII130 (Input, lane 2), HA-hTAFII130 was retained by GST-HP1γ (lane 8). There was no retention of HA-hTAFII130 on the GST control resin (lane 5). To determine whether endogenous hTAFII130 was retained by GST-HP1γ under these conditions, we reprobed the nitrocellulose membrane with a monoclonal antibody specific for hTAFII130 (Fig. 4A, second panel). A signal that corresponded to the endogenous hTAFII130 (slightly larger than recombinant HA-hTAFII130) was detected in all three extracts bound to GST-HP1γ (lanes 7–9). As expected, GST alone did not retain any hTAFII130 (lanes 4–6).
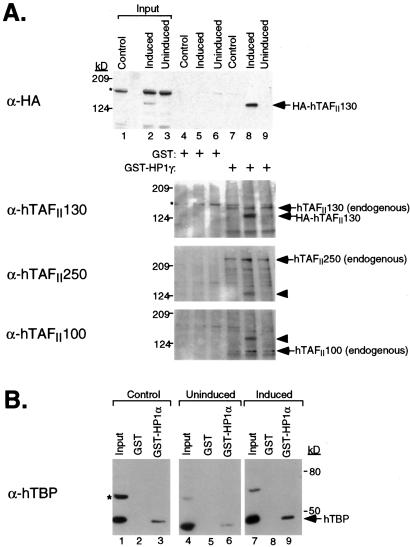
Figure 4.
Endogenous TAFIIs and TBP from HeLa cells bind to GST-HP1. (A) Nuclear extracts were prepared from control HeLa cells (lanes 1, 4, and 7), and a HeLa cell line induced (lanes 2, 5, and 8) or uninduced (lanes 3, 6, and 9) for HA-hTAFII130 expression. The extracts were incubated with GST (lanes 4–6) or GST-HP1γ (lanes 7–9); bound proteins were analyzed by sequential immunoblotting with α-HA, α-hTAFII130, α-hTAFII250, and α-hTAFII100 antibody. Endogenous hTAFII130 is larger in size than the recombinant HA-hTAFII130, which lacks the extreme N-terminal sequence. The recombinant HA-hTAFII130 in the induced cell extract is detected in the fraction bound to HP1γ, whereas endogenous hTAFII130, hTAFII250, and hTAFII100 were detected in all three nuclear extracts bound to HP1γ. The arrowheads in the third and fourth panels correspond to HA-hTAFII130, whose signal remained after sequential immunoblotting with the monoclonal antibodies. *, nonspecific background bands. (B) The same nuclear extracts used in A were incubated with GST or GST-HP1α, and the bound fractions were analyzed for the presence of hTBP with α-hTBP antibody.
To determine whether the other components of endogenous TFIID complex associated with GST-HP1γ, we sequentially probed the same membrane with antibodies to hTAFII250 and hTAFII100. We detected the recovery of endogenous hTAFII250 and hTAFII100 on the GST-HP1γ resin but not on the control GST resin (Fig. 4A, third and fourth panels). Interestingly, we observed increased recovery of endogenous TAFIIs associating with GST-HP1γ in the extracts induced for HA-hTAFII130 expression compared with noninduced extract, suggesting that endogenous hTAFII130 might be limiting in HeLa cells. In addition to the three TAFII components of endogenous TFIID, we also asked whether TBP could be recovered by using GST-HP1. As shown in Fig. 4B, endogenous hTBP was found to associate with GST-HP1α but not with GST alone. As before, the induction of HA-hTAFII130 increased the level of hTBP associating with GST-HP1α (compare lanes 6 and 9). Because we did not detect direct binding of individually in vitro-translated hTAFII250, hTAFII100, and hTBP to GST-HP1 (data not shown), we think that the components of TFIID, through interaction with hTAFII130, associate with GST-HP1. The observations that several endogenous TAFIIs as well as TBP indirectly associate with GST-HP1 support a model in which HP1 targets hTAFII130 in the context of TFIID.
Gal4-HP1 Fusions Repress Transcriptional Activation by hTAFII130.
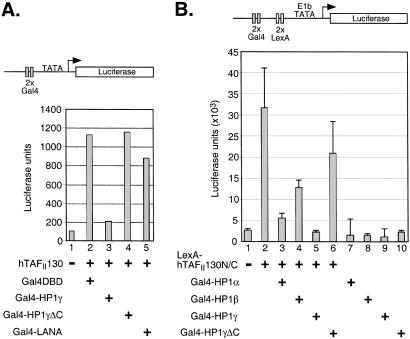
To determine the functional significance of the newly discovered hTAFII130-HP1 interaction, we performed transient transfection assays in HeLa cells. We initially examined the effects of transfected HP1 on reporter activity and found that HP1 repressed hTAFII130-mediated transcription by two-fold (data not shown). We think the modest effect is caused by the low expression levels of recombinant HP1 compared with endogenous HP1 that are abundant in cells. It is also possible that the transfected HP1 becomes associated with the endogenous protein, and only a small fraction of HP1 may be involved in transcriptional regulation at euchromatic gene promoters. Consistent with this possibility, we were unable to copurify endogenous or transfected HP1 with hTAFII130. However, when fused to a heterologous DBD, HP1 proteins function as repressors of transcription (48, 49). We examined whether Gal4-HP1γ can affect hTAFII130-mediated transcriptional activation. Transcription from the UAS-Luc reporter bearing two Gal4-binding sites was enhanced when transfected with hTAFII130 expression plasmid in the presence of Gal4 DBD (Fig. 5A, lanes 1 and 2). Significantly, increased transcription mediated by hTAFII130 was dramatically reduced upon cotransfection of Gal4-HP1γ (lane 3). This repression depended on the C-terminal domain of HP1γ that is required for the interaction with hTAFII130 because HP1γΔC truncated for this domain failed to repress hTAFII130-mediated activation (lane 4). Gal4 DBD fused to an unrelated repressor protein LANA (50) also failed to repress transcription by hTAFII130 (lane 5). None of the Gal4 fusion proteins affected basal transcription in the absence of hTAFII130 (data not shown), indicating that the observed effect is specific to activation mediated by hTAFII130.
Figure 5.
HP1 interferes with hTAFII130-mediated stimulation of transcription. (A) HeLa cells transfected with a plasmid-expressing hTAFII130 stimulated UAS-Luc reporter containing two Gal4-binding sites upstream of the minimal angiotensinogen promoter (lane 2). This activation was inhibited by the coexpression of Gal4-HP1γ (lane 3) but not by Gal4-HP1γΔC (lane 4). An unrelated repressor protein Gal4-LANA also did not block activation by hTAFII130 (lane 5). (B) HeLa cells were cotransfected with 2XGal/2XLex-E1bTATA-luciferase reporter plasmid and plasmids expressing LexA-hTAFII130N/C and/or Gal4-HP1, as indicated. Gal4-HP1α and Gal4-HP1γ more potently inhibited LexA-hTAFII130N/C-mediated activation compared with Gal4-HP1β.
The central domain of hTAFII130 activates transcription when fused to a heterologous DNA-binding domain (11). A LexA-hTAFII130N/C fusion, containing the same hTAFII130 subdomain used in the yeast two-hybrid screen and in vitro binding studies activated transcription from a reporter plasmid bearing two Gal4- and two LexA-binding sites (Fig. 5B, lane 2). To examine the effects of Gal4-HP1 proteins on transcriptional activation by LexA-hTAFII130N/C, we cotransfected plasmids expressing Gal4 DBD fused to the three isoforms of HP1. Gal4-HP1α and Gal4-HP1γ potently inhibited LexA-hTAFII130N/C-mediated activation, whereas Gal4-HP1β was not as effective in inhibiting transcription (lanes 3–5). Partial repression detected by Gal4-HP1β is likely caused by other mechanisms of repression mediated by HP1 involving self-association and/or association with HDAC activity (39, 49). Furthermore, preferential repression of hTAFII130 activity by HP1α and HP1γ compared with HP1β was lost when the reporter was activated by LexA-hTAFII130N/C-DE carrying the mutations that abolished the binding of hTAFII130N/C to HP1 (data not shown). Therefore, the ability of HP1 isoforms to repress transcription by LexA-hTAFII130N/C correlates with their ability to interact with hTAFII130 in vitro. Significantly, Gal4-HP1γΔC lacking the hTAFII130 interaction domain did not inhibit LexA-hTAFII130N/C-mediated activation. All Gal4-HP1 fusions had similar modest effects on basal transcription, indicating that the isoform-dependent inhibitory effect is specific to activation by hTAFII130. These data are consistent with the idea that hTAFII130-HP1 interaction plays a role in transcriptional regulation mediated by hTAFII130.
Discussion
HP1 Isoform-Specific Interactions with hTAFII130.
By using a yeast two-hybrid screen, we have isolated α and γ isoforms of HP1 as hTAFII130-interacting proteins. Several proteins have been described to bind to HP1; however, many bind to HP1 without any preference for certain isoforms. Our finding that hTAFII130 interacts with HP1α and HP1γ but not HP1β makes hTAFII130 unique in its ability to discriminate among closely related HP1 proteins. We propose that HP1α and HP1γ interact with hTAFII130 to regulate transcriptional repression of target genes. The lack of demonstrable association of hTAFII130 with HP1β may be indicative of HP1β's predominant association with heterochromatin, a highly condensed, gene-sparse region of the chromatin that is likely to require little TFIID activity.
Functional Implications of hTAFII130-HP1 Interaction.
HP1 proteins are abundant nonhistone chromatin-associated factors that participate in heterochromatin formation and gene silencing (reviewed in refs. 38 and 39); however, the precise mechanisms of HP1 repression are not well understood. HP1 has been shown to play a role not only in heterochromatic silencing but also in normal repression of genes in euchromatin (51). In mammalian cells and Schizosaccharomyces pombe, it has been reported that HP1/Swi6 chromodomain binds to histone H3 methylated by the methyltransferase SUV39H1/Clr4 at Lys-9 (52–54). Interestingly, Rb was shown to target SUV39H1 to histone H3 and to recruit HP1, leading to transcriptional repression of the endogenous cyclin E promoter (55). In vitro transcription assays reconstituted with chromatin and nuclear extracts have demonstrated HP1β-dependent repression of transcription from a template methylated by SUV39H1 (56). In this study, the authors failed to detect repression by HP1β in a highly reconstituted transcription system and speculated that other factors in the nuclear extract might be necessary to establish full repression. Because we find preferential association between hTAFII130 and HP1α and HP1γ, it is possible that HP1β is insufficient to establish repression in the reconstituted transcription system.
The interaction of hTAFII130 and HP1 may result in transcriptional repression by blocking the association of activators with components of TFIID. Alternatively, promoter-bound TFIID may be held in a repressed state by hTAFII130-HP1 interaction. Indeed, there is increasing evidence suggesting that the general transcription factors (GTFs) are bound at promoters repressed by heterochromatin or by similar repressive protein complexes (reviewed in refs. 57 and 58). In Drosophila, chromatin-immunoprecipitation analysis of promoter regions bound by the Polycomb group (PcG) of proteins showed that repressed promoters are bound by GTFs (59). Moreover, PcG proteins interact with GTFs in vitro, suggesting that PcG complexes may silence target gene expression by inhibiting the activation function of GTFs. Significantly, purification of a major PcG complex from Drosophila embryos has identified multiple TAFIIs associating with known PcG proteins (60). It is possible that hTAFII130 may interact with HP1α and HP1γ to facilitate the retention of TFIID in a promoter-specific manner forming repressed “preloaded” TFIID poised for rapid activation. The binding of site-specific activators upstream of the core promoter may compete for a surface on hTAFII130, causing dissociation of HP1 and relieving its repressive activity.
Another intriguing possibility is that hTAFII130-HP1 association may play a direct role in the activation of certain gene promoters by facilitating recruitment of TFIID to histones specifically modified by acetylation and methylation and bound by HP1. Structural studies have indicated that hTAFII250 double bromodomain binds to multiple acetylated tails of histone H4 (28). It is tempting to speculate that TFIID may be recruited to promoters bound by specifically modified histones through contacts with hTAFII130 and hTAFII250, an idea that is consistent with the histone code hypothesis (24).
Acknowledgments
We thank Michael Garabedian, Herb Samuels, Edith Wang, Jerry Crabtree, Nouria Hernandez, Kevin Struhl, Bob Tjian, and Angus Wilson for generously providing the reagents used in this study. We thank Michael Garabedian, Herb Samuels, Angus Wilson, Keith Blackwell, and David Levy for their valuable suggestions throughout the project and critical reading of the manuscript. We acknowledge Adam Hittelman for his instrumental help with the yeast two-hybrid screen, Stavros Giannakopoulos for the nuclear extracts from stable cell lines expressing TAFIIs, Tom Neubert and New York University Protein Analysis Facility for mass spectrometric analysis, and Stuart Brown of New York University Research Computing Resources for help with molecular biology computing. This work was supported by National Institutes of Health Grant GM51314 and by an award from the Lauri Strauss Leukemia Foundation. We thank the National Science Foundation for its support of the Computing Resources through Grant BIR-9318128. M.F.V. was supported in part by National Institutes of Health Grant T32 AI07810. N.T. was supported in part by The Irma T. Hirschl Trust.
Abbreviations
- GST
glutathione S-transferase
- TBP
TATA-binding protein
- TAFIIs
TBP-associated factors
- DBD
DNA-binding domain
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Albright S R, Tjian R. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Bell B, Tora L. Exp Cell Res. 1999;246:11–19. doi: 10.1006/excr.1998.4294. [DOI] [PubMed] [Google Scholar]
- 3.Struhl K, Moqtaderi Z. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- 4.Green M R. Trends Biochem Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- 5.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 6.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 7.Tanese N, Saluja D, Vassallo M F, Chen J L, Admon A. Proc Natl Acad Sci USA. 1996;93:13611–13616. doi: 10.1073/pnas.93.24.13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferreri K, Gill G, Montminy M. Proc Natl Acad Sci USA. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakajima T, Uchida C, Anderson S F, Parvin J D, Montminy M. Genes Dev. 1997;11:738–747. doi: 10.1101/gad.11.6.738. [DOI] [PubMed] [Google Scholar]
- 10.Mengus G, May M, Jacq X, Staub A, Tora L, Chambon P, Davidson I. EMBO J. 1995;14:1520–1531. doi: 10.1002/j.1460-2075.1995.tb07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojo-Niersbach E, Furukawa T, Tanese N. J Biol Chem. 1999;274:33778–33784. doi: 10.1074/jbc.274.47.33778. [DOI] [PubMed] [Google Scholar]
- 12.Saluja D, Vassallo M F, Tanese N. Mol Cell Biol. 1998;18:5734–5743. doi: 10.1128/mcb.18.10.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walker A K, Rothman J H, Shi Y, Blackwell T K. EMBO J. 2001;20:5269–5279. doi: 10.1093/emboj/20.18.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dikstein R, Zhou S, Tjian R. Cell. 1996;87:137–146. doi: 10.1016/s0092-8674(00)81330-6. [DOI] [PubMed] [Google Scholar]
- 15.Freiman R N, Albright S R, Zheng S, Sha W C, Hammer R E, Tjian R. Science. 2001;293:2084–2087. doi: 10.1126/science.1061935. [DOI] [PubMed] [Google Scholar]
- 16.Furukawa T, Tanese N. J Biol Chem. 2000;275:29847–29856. doi: 10.1074/jbc.M002989200. [DOI] [PubMed] [Google Scholar]
- 17.Gangloff Y G, Werten S, Romier C, Carre L, Poch O, Moras D, Davidson I. Mol Cell Biol. 2000;20:340–351. doi: 10.1128/mcb.20.1.340-351.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gangloff Y G, Romier C, Thuault S, Werten S, Davidson I. Trends Biochem Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 19.Workman J L, Kingston R E. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 20.Pollard K J, Peterson C L. BioEssays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 21.Wolffe A P, Guschin D. J Struct Biol. 2000;129:102–122. doi: 10.1006/jsbi.2000.4217. [DOI] [PubMed] [Google Scholar]
- 22.Marmorstein R. Nat Rev Mol Cell Biol. 2001;2:422–432. doi: 10.1038/35073047. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Reinberg D. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 24.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 25.Hoffmann A, Oelgeschlager T, Roeder R G. Proc Natl Acad Sci USA. 1997;94:8928–8935. doi: 10.1073/pnas.94.17.8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizzen C A, Yang X-J, Kokubo T, Brownell J E, Bannister A J, Owen-Hughes T, Workman J, Wang L, Berger S L, Kouzarides T, et al. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 27.Pham A D, Sauer F. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 28.Jacobson R H, Ladurner A G, King D S, Tjian R. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 29.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 30.Gietz R D, Schiestl R H, Willems A R, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 31.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. Vol. 2. New York: Wiley; 1988. [Google Scholar]
- 32.Dorris D R, Struhl K. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jantzen H M, Chow A M, King D S, Tjian R. Genes Dev. 1992;6:1950–1963. doi: 10.1101/gad.6.10.1950. [DOI] [PubMed] [Google Scholar]
- 34.Ye Q, Callebaut I, Pezhman A, Courvalin J C, Worman H J. J Biol Chem. 1997;272:14983–14989. doi: 10.1074/jbc.272.23.14983. [DOI] [PubMed] [Google Scholar]
- 35.Lee K A, Bindereif A, Green M R. Gene Anal Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 36.Elgin S C. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 37.Minc E, Courvalin J C, Buendia B. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- 38.Eissenberg J C, Elgin S C. Curr Opin Genet Dev. 2000;10:204–210. doi: 10.1016/s0959-437x(00)00058-7. [DOI] [PubMed] [Google Scholar]
- 39.Jones D O, Cowell I G, Singh P B. BioEssays. 2000;22:124–137. doi: 10.1002/(SICI)1521-1878(200002)22:2<124::AID-BIES4>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 40.Aasland R, Stewart A F. Nucleic Acids Res. 1995;23:3168–3174. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lechner M S, Begg G E, Speicher D W, Rauscher F J., III Mol Cell Biol. 2000;20:6449–6465. doi: 10.1128/mcb.20.17.6449-6465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murzina N, Verreault A, Laue E, Stillman B. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 43.Smothers J F, Henikoff S. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 44.Ryan R F, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Douarin B, Nielsen A L, Garnier J M, Ichinose H, Jeanmougin F, Losson R, Chambon P. EMBO J. 1996;15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- 46.Shtivelman E, Bishop J M. J Cell Biol. 1993;120:625–630. doi: 10.1083/jcb.120.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brasher S V, Smith B O, Fogh R H, Nietlispach D, Thiru A, Nielsen P R, Broadhurst R W, Ball L J, Murzina N V, Laue E D. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lehming N, Le Saux A, Schuller J, Ptashne M. Proc Natl Acad Sci USA. 1998;95:7322–7326. doi: 10.1073/pnas.95.13.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwam D R, Luciano R L, Mahajan S S, Wong L, Wilson A C. J Virol. 2000;74:8532–8540. doi: 10.1128/jvi.74.18.8532-8540.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hwang K K, Eissenberg J C, Worman H J. Proc Natl Acad Sci USA. 2001;98:11423–11427. doi: 10.1073/pnas.211303598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bannister A J, Zegerman P, Partridge J F, Miska E A, Thomas J O, Allshire R C, Kouzarides T. Nature (London) 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 53.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Nature (London) 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 54.Nakayama J, Rice J C, Strahl B D, Allis C D, Grewal S I. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen S J, Schneider R, Bauer U M, Bannister A J, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera R E, Kouzarides T. Nature (London) 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 56.Loyola A, LeRoy G, Wang Y H, Reinberg D. Genes Dev. 2001;15:2837–2851. doi: 10.1101/gad.937401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eissenberg J C. BioEssays. 2001;23:767–771. doi: 10.1002/bies.1111. [DOI] [PubMed] [Google Scholar]
- 58.Gross D S. Trends Biochem Sci. 2001;26:685–686. doi: 10.1016/s0968-0004(01)01985-5. [DOI] [PubMed] [Google Scholar]
- 59.Breiling A, Turner B M, Bianchi M E, Orlando V. Nature (London) 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- 60.Saurin A J, Shao Z, Erdjument-Bromage H, Tempst P, Kingston R E. Nature (London) 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]