Abstract
Chloroplast DNA of the green alga Chlamydomonas reinhardtii is maternally inherited. Methylation mapping directly revealed that, before mating, chloroplast DNA of maternal (mating type plus; mt+) gametes is heavily methylated whereas that of paternal (mating type minus; mt−) gametes is not. Indirect immunofluorescence analyses with anti-5-methylcytosine mAbs visually showed methylation to occur exclusively in chloroplast DNA of mt+ gametes, and not in mt− gametes or nuclear DNA of either mt. To clarify the relationship between methylation and maternal inheritance of chloroplast DNA, we have isolated and characterized a cDNA encoding a DNA methyltransferase. The deduced protein, CrMET1, consists of 1,344 aa and contains a conserved catalytic domain at the C terminal and a nonconserved N-terminal region. The predicted N-terminal region has an arginine-rich domain, suggesting CrMET1 is transferred to chloroplasts. This finding could be directly shown by green fluorescent protein epifluorescence microscopy analyses. CrMET1 transcripts were found to be absent in both mt+ and mt− vegetative cells. Upon gametogenesis, however, transcript levels clearly increased in mt+ but not mt− cells. These experiments suggest that the CrMET1 protein is located in chloroplasts and that it specifically methylates cytosine residues of chloroplast DNA in mt+ gametes. This conclusion was further strengthened by the observation that, during gametogenesis, CrMET1 is expressed in a mt− mutant, mat-1, whose chloroplast DNA is heavily methylated in gametes and paternally inherited. The results provide evidence that cytosine methylation plays a critical role in maternal inheritance of chloroplast genes in C. reinhardtii.
The soil organism Chlamydomonas reinhardtii is a member of a large genus including soil, fresh water, and marine forms. It is a unicellular sexual microorganism with a simple sexual life cycle, showing segregation by the mating type (mt), denoted by plus (mt+) and minus (mt−) (1). C. reinhardtii in liquid culture grow exponentially at rates of 1–4 doublings in 24 h, depending on the culture medium and conditions. Exponentially growing cultures do not mate. Upon nitrogen starvation, however, they differentiate into gametes capable of mating. After a clumping reaction, pairs of cells of opposite mt fuse to form zygotes, which are the only diploid stage during the usual life cycle. After a period of maturation for several days, zygotes germinate with the release of four zoospores, the four products of meiosis (1).
The inheritance of chloroplast genes is maternal: chloroplast genes from the mt+ parent are transmitted to all progeny, whereas the corresponding alleles from the mt− parent are permanently lost. It was earlier proposed that this maternal pattern is regulated by a methylation and restriction system analogous to that in bacteria (2). Indeed, southwestern hybridization using antibodies against 5-methylcytosine (m5C) directly demonstrated that chloroplast DNA from mt+ gametes is heavily methylated, in contrast to that from mt− gametes or vegetative cells of either mt (3). A recent study using 5-azacytidine, a powerful inhibitor of DNA methylation in vivo, further demonstrated that hypomethylation of chloroplast DNA of mt+ gametes in fact reduces the frequency of maternally inherited progeny (4). Although these experimental data indicate a strong correlation between maternal inheritance of chloroplast DNA and its hypermethylation, the available evidence is indirect. The present work was thus initiated to determine the cause-result relationship between the two phenomena.
Materials and Methods
Cell Strains and Culture Conditions.
Wild-type strains, CC125 (mt+) and CC124 (mt−), and mutant strains of chloroplast DNA methylation, CC1312 (mat-1, mt−), CC1154 (me-1, mt+), and CC1155 (me-1, mt−), were accessed from the Chlamydomonas Genetics Center, Duke University, Durham, NC. A cell wall-less mutant (cw-15, mt+) used for transformation was a generous gift from K. Shimogawara, Teikyo University, Tokyo. Vegetative cells were cultured in Tris/acetate/phosphate (TAP) liquid medium (5) at 25°C under continuous light, and gametes were prepared by incubating the vegetative cells on 1/5 N TAP solid medium at 28oC for 4 days, with a regimen of 16 h of light followed by 8 h of darkness.
Methylation Mapping Assay.
Total DNA was extracted (6) and linearized with restriction enzymes (EcoRI, PstI, and StyI). One microgram of the linearized DNA and 4 μg of pGEM plasmid as carrier were diluted to 50 μl and denatured at room temperature by addition of 5 μl of 2 M NaOH for 10 min. A 1 ml of bisulfite solution (2.5 M sodium metabisulfite, 100 mM hydroquinone, pH 5.0 adjusted with NaOH) was then added to the template mixture and incubated at 50°C for 4 h (7). After three times of ethanol precipitation to remove bisulfite, 10 ng of the bisulfite-modified DNA was amplified by 30 cycles of PCR (94°C 15 sec, 50°C 15 sec, 72°C 1 min). Primers used were as follows: for the A strand (the sense strand), pRBCLA1, 5′-CTAAGAGCTCTAATTTAAATTTAATTATACAACC-3′, pRBCLA2, 5′-ATTGGTCGACATAAAGTTTATTATATTGGAGAGG-3′; for the B strand (the antisense strand), pRBCLB1, 5′-TAAAGTCGACAACACTACCTCTAATAAAATCTAC-3′, pRBCLB2, 5′-TTTTGAGCTCAAAGATTTAATTTAGGTTTGATTG-3′ (SacI and SalI sites used for subcloning are indicated by underlines). They were used to amplify an 850-bp segment of the ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit gene (rbcL, GenBank accession no. J01399) on the chloroplast DNA. The resulting fragments were cloned and sequences were determined (7).
Indirect Immunofluorescence Staining.
Cells were fixed on a slide glass with 4% paraformaldehyde in PBS solution, denatured with 70% formamide in 2× SSC, and immediately chilled in 70%, 95%, and 99% ethanol for 2 min each and air-dried. Staining were essentially performed as described (8) with an anti-m5C mAb (9). For DNA identification, slides were mounted in 1 μg/ml 4′,6 diamidino-2-phenylindole (DAPI)/Vectashield solution (Vector Laboratories). Fluorescent images of DAPI and FITC were captured by using UV- and B-excitation filters and merged with the assistance of photoshop (version 5, Adobe).
Full-Length cDNA Isolation of CrMET1.
To isolate C. reinhardtii DNA methyltransferase, a set of degenerate primers was designed in accordance with the conserved regions of the Cyanobacteria methyltransferase gene as follows: pCyano1F, 5′-CTSTTCGCSGGYTGCGGYGG-3′; pCyano4R, 5′-AARCCCTGGCASGGSGGRCC-3′. A 359-bp cDNA fragment encoding the catalytic domain of CrMET1 (domain I-IV) was obtained from the initial screening of a cDNA pool of me-1 mt+ gametes. A genomic nucleotide sequence including the 5′ regulatory region was obtained and determined by inverse PCR with genomic DNA from me-1 mt+ vegetative cells. The full-length cDNA was finally obtained by reverse transcription (RT)-PCR, 5′ rapid amplification of cDNA ends (RACE) (5′-Full RACE Core Kit, Takara, Kyoto) and 3′ RACE with the first cDNA of me-1 mt+ gametes by referring to the genomic DNA sequence.
Green Fluorescent Protein (GFP) Epifluorescence Microscopy Analysis.
Vector construction was carried out essentially as described (10, 11) (see Fig. 3a). Transformed cells were selected by plating cell suspensions after electroporation onto Tris/acetate/phosphate (TAP) solid medium containing 10 mg/l of zeocin (Invitrogen), and observed under a fluorescence microscope equipped with UV- and B-excitation filters (Ax70, Olympus, Tokyo). Fluorescent images were captured separately by using a cooled charge-coupled device camera (CoolSNAP-HQ, Photometrics, Tucson, AZ).
Figure 3.
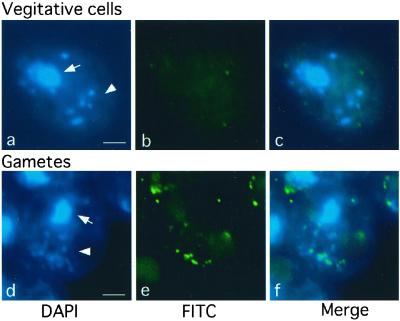
Methylation of chloroplast DNA visualized by immunocytochemistry labeling with anti-m5C antibodies. After fixation on slide glasses, me-1 cells were stained with DAPI to show fluorescence images (a and d). Note that up to 10 chloroplast nucleoids containing chloroplast DNA are visible. Simultaneously, samples were treated with monoclonal anti-m5C antibodies followed by signal amplification by the second antibodies to show FITC signal images (b and e). DAPI and FITC signal images were merged (c and f), showing chloroplast DNA is exclusively methylated in gamete cells (f). (a–c) Vegetative cells; (d–f) gamete cells. Arrows and arrow heads indicate nuclei and chloroplast DNAs, respectively. (Bars = 2 μm.)
RNA and DNA Blot Analyses.
Total RNA was extracted from vegetative cells and gametes (12). Poly(A)+ RNA was purified by using an mRNA Purification Kit (Amersham Pharmacia), and 1.5-μg aliquots were used for RNA blot analyses. The probe was a 1.7-kb cDNA fragment of CrMET1. For the DNA blot analysis, 10 μg of total genomic DNA from vegetative cells was digested with BamHI or SphI and subjected to hybridization with CrMET1 as a probe.
RT-PCR.
The initial cDNA pool was synthesized from 10 μg of total RNA from vegetative cells and gametes by using an oligo(dT) primer. A 0.9-kb fragment of ATP synthase subunit C (atpC1) gene was used as a quantitative standard. A 25-cycle PCR (98°C 10 sec, 68°C 1 min) was performed with primers specific for the atpC1 gene, (pATPC1F: CGCTGACGACGAGATCTTCAAGC, and pATPC1R: ACAGGTCCGTTGTCTTCACCTCC), and a 1-kb cDNA fragment of CrMET1 was amplified from the initial cDNA pool by a 30-cycle PCR with primers (pCrMETF6: CCATGAGCGCCTTCCAGTGG, pCrMETR10: CTGATGCTGGCACTGCATACC). The PCR products were subjected to electrophoresis and stained with ethidium bromide.
Results
Identification of Methylated DNA During Gametogenesis.
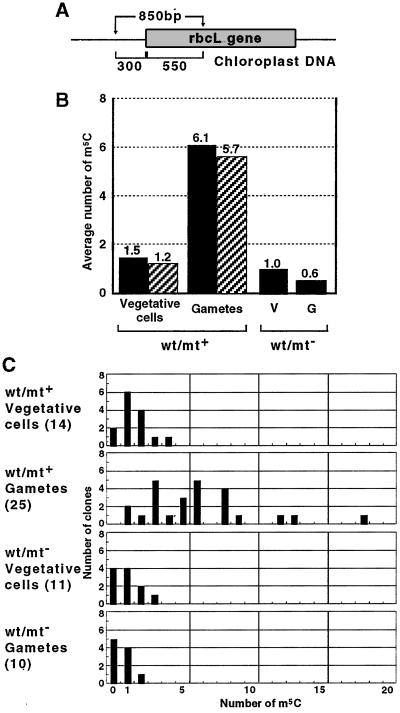
Our previous analyses with southwestern hybridization and HPLC revealed selective methylation of chloroplast DNA in mt+ gamete cells (3, 13). This finding was further confirmed by direct methylation mapping (see Fig. 7, which is published as supporting information on the PNAS web site, www.pnas.org), using the bisulfite method, which determines positions of m5C in the rbcL locus of chloroplast DNA. The sequence examined covers 850 bp containing the 5′ upstream promoter region and part of the coding region (Fig. 1A). The average numbers of m5C in both A and B strands were estimated in each clone derived from vegetative and gamete cells of both mts. The results showed that the average number of m5C in mt+ vegetative cells was one or two, whereas in mt+ gametes this was increased to six (Fig. 1B). In mt− vegetative and gamete cells, however, the number of m5C was about one or slightly less (Fig. 1B). Among methylated sites in mt+ gametes, CpG was predominant, accounting for 36.8% of the total m5Cs, followed by CpNpG (27.6%). The other sites (35.5%) were apparently randomly distributed among CpA, CpT, and CpC. Methylation occurred at equal frequency in both promoter and coding regions. The number of m5C in a single clone was always less than four in vegetative cells of both mts and in mt− gametes. In contrast, more than half of the clones of mt+ gametes showed more than six m5Cs, with an extreme example of 18 m5Cs (Fig. 1C). Because a single cell of C. reinhardtii contains ≈80 copies of chloroplast DNA, the above observation suggests that, even in mt+ gametes, methylation does not occur uniformly in the entire chloroplast DNA population. Some molecules are heavily methylated whereas others are not.
Figure 1.
Methylation mapping of the rbcL gene in wild-type (wt) cells. (A) Analyzed region of the rbcL locus in chloroplast DNA. The 850 bp contains 300 bp and 550 bp of promoter and coding regions, respectively. The total number of cytosine residues is 149 in the A strand (sense strand) and 148 in the B strand (antisense strand). (B) Average numbers of m5C. After 30 cycles of PCR (94°C 15 sec, 50°C 15 sec, 72°C 1 min) using 10 ng of the bisulfite-modified DNA, the products were digested by SalI and SacI, gel-purified, and cloned into pBluescript KS−. Individual clones were sequenced and sites of methylated cytosines were identified. The numbers of sequenced clones were 14 and 11 for mt+ and mt− vegetative cells, respectively, and 25 and 10 for mt+ and mt− gametes, respectively. Average numbers of m5C in A and B strands are indicated by solid and hatched bars, respectively. (C) Numbers of m5C in individual clones. The value for a given clone is indicated on the horizontal scale, and the number of clones containing the indicated m5C number is shown on the vertical scale. The sequence specificity of the m5C is not shown, but 36.8% of the total m5C appeared in CpG, 27.6% in CpNpG, and 35.5% in other sequences, apparently at random.
Preferential Methylation of Chloroplast DNA.
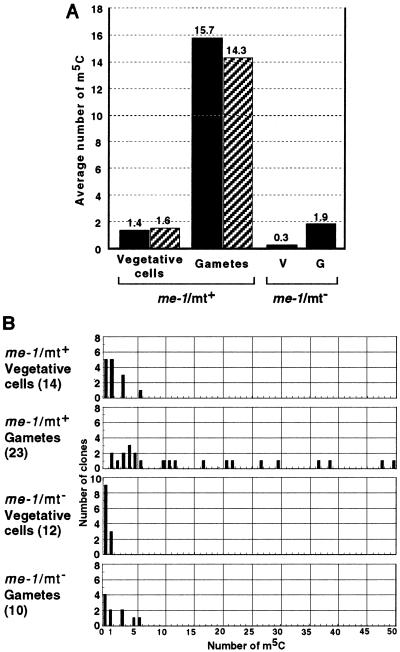
The me-1 mutant was originally described to obey normal maternal inheritance for chloroplast genes, despite the hypermethylation of chloroplast DNA in vegetative and gamete cells of both mts (14). On direct methylation mapping, however, the overall methylation status of both mts at the rbcL locus was similar to that of wild type: hypomethylated in vegetative cells, and heavily methylated only in mt+ gemetes, although the level was more than 2.5-fold higher than in the wild type (Fig. 2). Taking advantage of this hypermethylation, we visualized the methylation status of chloroplast and nuclear DNAs by indirect immunofluorescence staining with anti-m5C antibodies. With this technique, the chloroplast nucleoid structure, which consists of chloroplast DNA and proteins, is clearly visible. Whereas mt+ vegetative cells did not show detectable methylation of either chloroplast or nuclear DNAs, mt+ gemete cells showed exclusive methylation of chloroplast DNA (Fig. 3). It appeared, however, that not all chloroplast nucleoids were equally methylated: some were heavily methylated whereas others were not. This finding is consistent with the results of direct methylation mapping, showing a diverse population in terms of methylation (Fig. 2). Chloroplast and nuclear DNAs from mt− vegetative and gamete cells remained unmethylated, showing similar images to those of mt+ vegetative cells (data not shown). Note that nuclear DNA was apparently not methylated at any stage in either mt.
Figure 2.
Methylation mapping of the rbcL gene in me-1 mutant cells. (A) Average numbers of m5C. Experiments were performed as described in the legend for Fig. 1. The numbers of sequenced clones were 14 and 12 for mt+ and mt− vegetative cells, respectively, and 23 and 10 for mt+ and mt− gametes, respectively. Average numbers of m5C in A and B strands are indicated by solid and hatched bars, respectively. (B) Numbers of m5C in individual clones. See the legend for Fig. 1. Of the total m5C, 38.4% appeared in CpG, 14.1% in CpNpG, and 47.5% in other sequences.
Isolation of a Gene Encoding a DNA Methyltransferase.
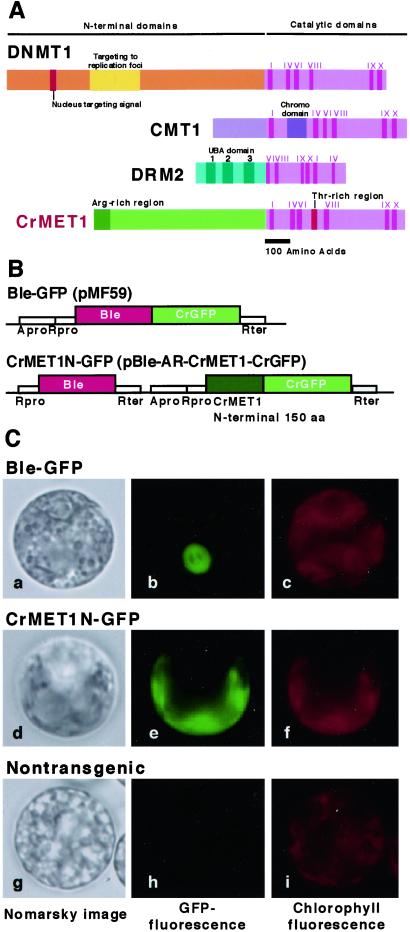
To examine the molecular mechanisms of chloroplast DNA methylation, we attempted to isolate a gene for a DNA methyltransferase, which is considered responsible for methylation of chloroplast DNA. Initial trials with PCR using a set of primers designed from plant methyltransferase sequences were unsuccessful, but we finally obtained a 359-bp fragment that was amplified with primers designed after Cyanobacteria methyltransferase. Subsequently, a full-length cDNA clone of 4.6 kb encoding a protein with 1,344 aa and a calculated molecular mass of 142 kDa was isolated (Fig. 4A). The deduced polypeptide contained well-conserved domains of many DNA methyltransferases at the C terminus and a nonconserved N-terminal region with an arginine-rich domain in the extreme N-terminal region. The gene was therefore designated as CrMET1 (C. reinhardtii DNA methyltransferase 1). Sequence comparison of the catalytic domain with known methyltransferases indicated CrMET1 to resemble ZmMET1 from maize (15). To estimate its enzymatic activity, a 2-kb fragment of CrMET1 encoding the C-terminal region with a 68-kDa polypeptide was expressed in insect cells by using a baculo-system, in which the maximal size of proteins to be expressed is around 120 kDa. The purified protein, however, showed no methylation activity (data not shown). The presence of the arginine-rich region, which may serve as the signal peptide, suggested CrMET1 to be translocated in the organelles. Subsequent examination of a GFP fusion protein with the N-terminal 150 aa directly demonstrated it to be exclusively present in chloroplasts (Fig. 4 B and C).
Figure 4.
Structure and localization of CrMET1. (A) Schematic representation of CrMET1 and other methyltransferases. Several functional domains based on homology with other known sequences could be identified: note the I-X catalytic motifs and the arginine-rich region at the N terminus. (B) Plasmid constructs used for GFP assay. pMF59 and pCrGFP (Entelechon, Regensburg, Germany) were used for construction of pBle-AR-CrMET1-CrGFP to express the CrMET1N-GFP fusion protein in C. reinhardtii. A 450-bp cDNA fragment of the N-terminal coding region of the CrMET1 gene (150 aa) was introduced into pBle-AR-CrMET1-CrGFP adapted to the cgfp gene in-frame. The pMF59 and pBle-AR-CrMET1-CrGFP were transformed into cw-15 vegetative cells (23). (C) Localization of CrMET1-GFP fusion proteins. A GFP fusion protein with the N-terminal 150 aa of CrMET1 was introduced into the C. reinhardtii cw15 strain and assayed for Nomarsky images (Left), GFP fluorescence (Center), and chlorophyll autofluorescence (Right). Samples are positive control cells transformed with a vector carrying Ble-GFP, which migrates into the nucleus (Top), cells transformed with a vector carrying CrMET1N-GFP (Middle), and nontransgenic cells as a negative control (Bottom).
Genomic Organization and Expression of CrMET1.
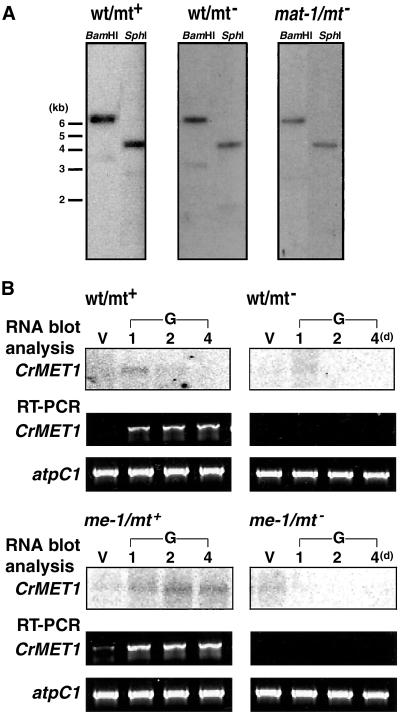
Southern hybridization indicated CrMET1 to be a single copy gene in the C. reinhardtii genome (Fig. 5A). There was no difference in restriction patterns between mt+ and mt− in either coding or promoter regions (Fig. 5A), suggesting separation from the hypervariable mating loci, which show a high polymorphism among mts (16). Northern hybridization with poly(A)+ RNA as well as RT-PCR showed CrMET1 transcripts to accumulate only during mt+ gametogenesis (Fig. 5B). The high levels of CrMET1 transcripts observed in the me-1 gamete is consistent with the findings for m5C (Fig. 2). In contrast, they were totally absent in vegetative cells of either mt and in mt− gametes from both wild-type and me-1 cells (Fig. 5B).
Figure 5.
Genomic organization and expression of CrMET1. (A) Southern blot analysis. Genomic DNA (10 μg per lane) was digested with BamHI or SphI, separated on an agarose gel, blotted, and probed with a radioactively labeled CrMET1 cDNA fragment. DNA samples were from mt+ (Left) and mt− (Center) wild-type (wt) cells, and from mat-1 cells (Right). (B) Northern blot analysis and RT-PCR. RNA blot analysis were carried out with 1.5 μg of poly(A)+ RNA extracted from vegetative cells (V) and gametes at 1, 2, and 4 days after gametogenesis induction (G, 1, 2, 4). Hybridization was performed with a radioactively labeled CrMET1 cDNA fragment. As a control, atpC1 (24) was amplified under the same conditions as for CrMET1. Assayed samples were mt+ (Upper Left) and mt− (Upper Right) wild-type cells, and mt+ (Lower Left) and mt− (Lower Right) me-1 mutant cells.
Relationship between DNA Methylation and Maternal Inheritance of Chloroplast DNA.
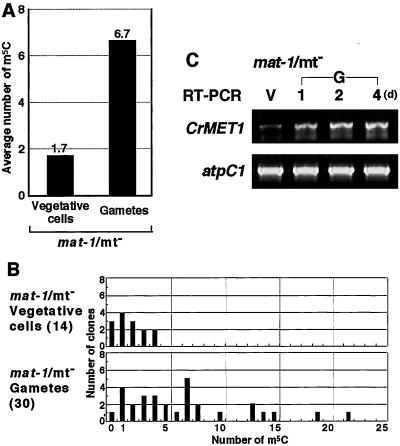
The mat-1 is an mt− mutant, which stably transmits chloroplast DNA to progeny, resulting in the paternal inheritance (17). Upon gametogenesis, chloroplast DNA was previously shown to be hypermethylated (18). We confirmed this observation by direct methylation mapping, showing an increased methylation level in gametes and similar methylation sequences, equivalent to those in wild-type mt+ gametes (Figs. 1 and 6 A and B). Transcripts of CrMET1 increased during gamete formation of this mutant, strongly suggesting that CrMET1 is responsible for the observed specific methylation (Fig. 6C). However, the genomic organization was the same as that in the wild type, indicating that the CrMET1 locus itself is not mutated (Fig. 5A). The results are in agreement with the hypothesis that chloroplast DNA methylation is indeed catalyzed by CrMET1, and consequently support the idea that methylation of chloroplast DNA is responsible for its persistence in progeny.
Figure 6.
Methylation mapping and expression of CrMET1 in mat-1 mutant cells. (A) Average numbers of m5C. Experiments were performed as described in the legend for Fig. 1. The numbers of sequenced clones was 14 and 30 for mt− vegetative and gamete cells, respectively. The average number of m5C in the A strand is indicated by solid bars. (B) Numbers of m5C in individual clones. Data are expressed as described in the legend for Fig. 1. Of the total m5C, 33.7% appeared in CpG, 23.3% in CpNpG, and 43.0% in other sequences. (C) Transcript accumulation. RT-PCR was carried out by using total RNA prepared from vegetative cells (V) and gametes at 1, 2, and 4 days after gametogenesis induction (G, 1, 2, 4). As a control, atpC1 was amplified under the same conditions as for CrMET1.
Discussion
Since Sager and Lane first proposed the restriction-modification system to account for the molecular basis of maternal inheritance of Chlamydomonas chloroplast DNA in 1972 (2), considerable experimental data have accumulated in support of their hypothesis (see ref. 19). For example, selective methylation of chloroplast DNA has been consistently observed in mt+ gametes with buoyant density centrifugation (13), restriction enzyme cleavage patterns (6), and by southwestern hybridization with anti-m5C antibodies (3). A gamete- and zygote-specific DNA methyltransferase with a high molecular mass also was identified (20). However, the evidence was indirect, and critical experiments showing a direct association of methylation with persistence of mt+ chloroplast DNA during mating have hitherto not been performed to our knowledge.
At the same time, the hypothesis has been challenged in two independent reports. One showed maintenance of maternal inheritance in mt+ cells, in which methylation was reduced by treatment with 5-azacytidine, a drug that inhibits in vivo methylation (21). The other showed that a hypermethylation mutant (me-1) of both mts still obeys normal maternal inheritance (14). Consequently, it has remained controversial whether DNA methylation functions in maternal inheritance of chloroplast DNA. Curiously, however, no attempts to address this question have been presented since the mid-80s, until the recent report describing methylation to be only necessary for differential replication of chloroplast DNA in germinating zygotes (4). When mt+ gametes were treated with 5-azadeoxycitidine and successively crossed to mt− gametes, the number of exceptional progeny carrying the mt− chloroplast DNA marker increased (4). However, hypomethylated chloroplast DNA of mt+ cells could persist during zygote maturation, and it was proposed that methylation functions not for protection against nucleases, but for promotion of DNA replication by, for example, properly packaging chloroplast DNA into a protective nucleoid conformation (4).
In the present study, we confirmed selective methylation in mt+ gamete chloroplast DNA by direct mapping. Among the identified m5C residues, two-thirds were located at CpG and CpNpG sequences, and one-third occurred in other sites such as CpA and CpT. This result is in clear contrast to findings with m5Cs in most eukaryotic organisms, restricted to CpG and CpNpG. It appears that, in C. reinhardtii, it is not the site of methylation that is important but rather the methylation itself of chloroplast DNA. Among ≈80 copies of chloroplast DNA per cell, methylation was found to occur heterogenously. Whereas some were heavily methylated, ≈1/3 remained unmethylated. Because chemical conversion was complete under our experimental conditions, and because more than 2/3 cells underwent gametogenesis (unpublished observation), the results indicate that only a limited number of copies are preferentially methylated. This finding is consistent with unequal immunofluorescence staining of chloroplast nucleoids. Judging from the site-independent methylation patterns and biased methylation among the chloroplast DNA population, it can be speculated that only DNA molecules that undergo high methylation can be transmitted to progeny. The unmethylated population may be excluded, like those of mt− origin. The biological significance of such a biased methylation remains to be clarified.
In our previous study, we identified a mt+ gamete- and zygote-specific DNA methyltransferase of ≈200 kDa, which methylates both unmethylated and hemimethylated chloroplast DNA (20). Its identity may in fact be CrMET1, given the molecular mass of ≈150 kDa, and the mt+ gamete specificity. Structurally, conserved domains in the C terminus resemble those of type I methyltransferases from other plants. For example, the F-X-G-X-G in motif I, P-C in motif IV, E-N-V in motif VI, and Q-X-R-X-R in motif VIII are well conserved (22). However, other parts of the catalytic domain show little homology with other methyltransferases. In addition, a threonine-rich region between motif VI and VIII is distinct only in CrMET1. Furthermore we could not find any similarity of the N terminus to known proteins, except for a signal peptide for chloroplast translocation at the extreme N terminal. In this sense, CrMET1 constitutes a member of a novel family of DNA methyltransferases with specific features like being chloroplast-resident. Whether or not this type of DNA methyltransferase is common among plants is not clear. Among 10 genes for putative DNA methyltransferases so far identified in the Arabidopsis genome, none has a transit peptide, which implies CrMET1 specifically evolved in Chlamydomonas.
The mt locus covering a 1.1-Mb region was previously isolated and shown to be highly polymorphic because of chromosomal rearrangement among mt+ and mt− cells (16). The rearranged region contains several mt-specific genes, one of which could be mt+ specific (16). To determine whether or not CrMET1 is one such gene, restriction analysis of the coding region and direct sequencing of the promoter region were here performed (Fig. 5 and unpublished observation). Because no difference was apparent in the structure of the CrMET1 locus between mt+ and mt− and considering that the CrMET1 expression is mt+ gamete specific, either CrMET1 resides on unidentified mating locus or its transcription factors are under the control of mt locus products. Currently we cannot determine which is the case, but favor the latter, judging from fact that the mt− mutant, mat-1, in which CrMET1 is highly expressed in gametes, has a restriction profile the same as that of the wild type. The behavior of the me-1 mutant provides further support: the original report described that, despite heavy methylation of chloroplast DNA from vegetative cells of either mt, the maternal inheritance of chloroplast DNA was normally conducted as in the wild type (14). In the present study, we could not confirm this finding, but found methylation to occur solely in mt+ gametes. Expression of CrMET1 during mt+ gametogenesis was also higher than in the wild type. The simplest explanation is that transcription was enhanced because of some alteration in transcriptional machinery.
Taking into account all of the available information, we conclude that CrMET1 is indeed responsible for mt+ gamete-specific chloroplast DNA methylation, and that this may be critical for maintenance of maternal chloroplast genes. The question thus arises as to the responsible mechanism. A simple restriction-modification system may not be the case, because (i) no restriction enzyme has so far been identified in Chlamydomonas, and (ii) hypomethylated chloroplast DNA persists throughout the zygote maturation period (4). It is conceivable that m5C serves as the marker for some proteins to assemble on DNA, thereby forming complexes that may affect replication.
Supplementary Material
Acknowledgments
This paper is dedicated to the late Professor Ruth Sager, who first proposed that DNA methylation might drive the maternal inheritance of chloroplast DNA in Chlamydomonas. We thank Drs. K. Shimogawara and M. Fuhrmann (University of Regensburg, Germany) for generous gifts of the cw-15 strain and the pMF59 plasmid, respectively. We also thank Dr. M. Moore (Intermal, Nagoya) for critical reading of the manuscript. This work was supported by grants for the Core Research for Evolutional Science and Technology (CREST) from the Japan Science and Technology Corporation (JST), and for the Research for the Future Program (JSPS-RFTF 00L01604) from the Japan Society for the Promotion of Science. R.N. was a recipient of a Core Research for Evolutional Science and Technology research fellowship during the performance of this work.
Abbreviations
- mt
mating type
- m5C
5-methylcytosine
- rbcL
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit
- DAPI
4′,6 diamidino-2-phenylindole
- RT
reverse transcription
- GFP
green fluorescent protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB073989).
References
- 1.Sager R. Cytoplasmic Genes and Organelles. New York: Academic; 1972. [Google Scholar]
- 2.Sager R, Lane D. Proc Natl Acad Sci USA. 1972;69:2410–2413. doi: 10.1073/pnas.69.9.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sano H, Royer H D, Sager R. Proc Natl Acad Sci USA. 1980;77:3581–3585. doi: 10.1073/pnas.77.6.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Umen J G, Goodenough U W. Genes Dev. 2001;15:2585–2597. doi: 10.1101/gad.906701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorman D S, Levine R P. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Royer H D, Sager R. Proc Natl Acad Sci USA. 1979;76:5794–5798. doi: 10.1073/pnas.76.11.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raizis A M, Schmitt F, Jost J P. Anal Biochem. 1995;226:161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- 8.Ohmido N, Fukui K. Plant Mol Biol. 1997;35:963–968. doi: 10.1023/a:1005822504690. [DOI] [PubMed] [Google Scholar]
- 9.Sano H, Imokawa M, Sager R. Biochem Biophys Acta. 1988;951:158–165. doi: 10.1016/0167-4781(88)90036-x. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrmann M, Oertel W, Hegemann P. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- 11.Schroda M, Blocker D, Beck C F. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- 12.Kirk M M, Kirk D L. Cell. 1985;41:419–428. doi: 10.1016/s0092-8674(85)80015-5. [DOI] [PubMed] [Google Scholar]
- 13.Burton W G, Grabowy C T, Sager R. Proc Natl Acad Sci USA. 1979;76:1390–1394. doi: 10.1073/pnas.76.3.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolen P L, Grant D M, Swinton D, Boynton J E, Gillham N W. Cell. 1982;28:335–343. doi: 10.1016/0092-8674(82)90351-8. [DOI] [PubMed] [Google Scholar]
- 15.Steward N, Kusano T, Sano H. Nucleic Acids Res. 2000;28:3250–3259. doi: 10.1093/nar/28.17.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferris P J, Goodenough U W. Cell. 1994;76:1135–1145. doi: 10.1016/0092-8674(94)90389-1. [DOI] [PubMed] [Google Scholar]
- 17.Sager R, Ramanis Z. Proc Natl Acad Sci USA. 1974;71:4698–4702. doi: 10.1073/pnas.71.12.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sager R, Grabowy C, Sano H. Cell. 1981;24:41–47. doi: 10.1016/0092-8674(81)90499-2. [DOI] [PubMed] [Google Scholar]
- 19.Sager R, Sano H, Grabowy C T. Curr Top Microbiol Immunol. 1984;108:157–172. doi: 10.1007/978-3-642-69370-0_11. [DOI] [PubMed] [Google Scholar]
- 20.Sano H, Grabowy C, Sager R. Proc Natl Acad Sci USA. 1981;78:3118–3122. doi: 10.1073/pnas.78.5.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng T Y, Chiang K S. Proc Natl Acad Sci USA. 1984;81:3438–3442. doi: 10.1073/pnas.81.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vertino P M. In: S-Adenosylmethionine-Dependent Methyltransferases: Structures and Functions. Cheng X, Blumenthal R M, editors. Singapore: World Scientific; 1999. pp. 341–372. [Google Scholar]
- 23.Shimogawara K, Fujiwara S, Grossman A, Usuda H. Genetics. 1998;148:1821–1828. doi: 10.1093/genetics/148.4.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu L M, Selman B R. J Biol Chem. 1988;263:19342–19345. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.