Abstract
In response to IFN-γ, the latent cytoplasmic protein signal transducers and activators of transcription 1 (Stat1) becomes phosphorylated on Y701, dimerizes, and accumulates in the nucleus to activate transcription of IFN-γ-responsive genes. For maximal gene activation, S727 in the transcription activation domain of Stat1 also is inducibly phosphorylated by IFN-γ. We previously purified a group of nuclear proteins that interact specifically with the Stat1 transcription activation domain. In this report, we identified one of them as the multifunctional Ca2+/calmodulin-dependent kinase (CaMK) II. We demonstrate that IFN-γ mobilizes a Ca2+ flux in cells and activates CaMKII. CaMKII can interact directly with Stat1 and phosphorylate Stat1 on S727 in vitro. Inhibition of Ca2+ flux or CaMKII results in a lack of S727 phosphorylation and Stat1-dependent gene activation, suggesting in vivo phosphorylation of Stat1 S727 by CaMKII. Thus two different cellular signaling events, IFN-γ receptor occupation and Ca2+ flux, are required for Stat1 to achieve maximal transcriptional activation through regulation of phosphorylation.
After ligand binding to cell surface cytokine receptors, the latent cytosolic STAT proteins are phosphorylated on a single tyrosine residue by a receptor-associated Janus kinase, dimerize, and enter the nucleus where they bind to specific DNA sequences and activate transcription (1, 2). The seven mammalian STAT proteins, each performing a specific physiological role, share common structural features such as the DNA binding domain and SH2 domain (3, 4). The carboxyl-terminal region of STATs serves as a transcription activation domain (TAD) for recruitment of transcription coactivators such as CBP/p300 (5–10). Although there is little sequence homology between the TADs, several TADs have a conserved serine residue at or near position 727 that is phosphorylated in response to ligands and are required for maximal transcription activity (8, 11). The interaction between Stat1 TAD and the DNA helicase MCM5 was shown to depend on the phosphorylated S727 in the Stat1 TAD (8). Phosphorylation of S727 in Stat1 can be induced by a variety of stimuli such as IFN-γ, lipopolysaccharide, and UV, and p38 mitogen-activated protein (MAP) kinase has been implicated in lipopolysaccharide- or UV-induced phosphorylation of S727 in Stat1 (12, 13). However, for IFN-γ-induced S727 phosphorylation in Stat1, the involvement of p38 MAP kinase is controversial (13, 14). The phosphatidylinositol 3-kinase/Akt pathway also could be involved in the IFN-γ-induced phosphorylation of Stat1 S727 (15).
Calcium (Ca2+) is a secondary messenger involved in a wide variety of cellular processes such as synaptic communication between neurons, muscle contraction, immune response, cell proliferation, and gene transcription (16, 17). The frequency and magnitude of Ca2+ flux elicited by various signaling ligands confers specificity to the many different signaling pathways by activating different Ca2+-dependent enzymes (18, 19). One of the main target kinases of Ca2+ is the multifunctional, multimeric Ca2+/calmodulin-dependent kinase (CaMK) II, a family of serine/threonine kinases involved in many cellular processes (20–22). The CaMKII family includes the neuronal-specific α and β genes, and the ubiquitous γ and δ genes (23). There are many isoforms for each CaMKII gene because of alternative splicing with some isoforms containing a nuclear localization signal to function in the nucleus (24). In vivo, the CaMKII enzyme functions as a 12-unit holoenzyme, and autophosphorylation occurs after activation, rendering the enzyme Ca2+-independent (21). Many proteins have been shown to be CaMKII substrates; among them are transcription factors such as CREB, ATF, and CEBPβ, and their transcription activities are regulated through phosphorylation on specific serine residues (25, 26).
IFN-γ has been shown to elicit a Ca2+ flux in certain cell types (27–29), suggesting a possible involvement of Ca2+ flux in IFN-γ signaling. However, its physiological consequence was not clear. In this report, we describe a role of Ca2+ in IFN-γ-induced phosphorylation of S727 of Stat1. Furthermore, we identified CaMKII as the downstream serine kinase that phosphorylates S727 in Stat1 critical for IFN-γ-induced gene activation.
Materials and Methods
Cell Culture and Antibodies.
U3A cells (provided by G. Stark, Cleveland Clinic Foundation Research Institute, Cleveland, and I. Kerr, Imperial Cancer Research Foundation, London) were maintained in DMEM supplemented with 10% cosmic calf serum. NIH 3T3 cells (American Type Culture Collection) were maintained in DMEM supplemented with 10% bovine calf serum. Only NIH 3T3 cells with early-passage numbers were used for the various experiments. Bovine calf sera were from HyClone, and FBS was from Gemini Biological Products (Woodland, CA). Antibody against phosphoserine of Stat1 was from Upstate Biotechnology (Lake Placid, NY). Antibodies against phosphotyrosine of Stat1 and phospho-protein kinase C (PKC) were from Cell Signaling Technology (Beverly, MA). Antibodies against Stat1 and Stat3 were from Transduction Laboratories (Lexington, KY). Antibody against CaMKIIγ was from Santa Cruz Biotechnology. Anti-hemagglutinin (HA) antibody was from Roche Molecular Biochemicals. Recombinant mouse IFN-γ was from Roche Diagnostic Corporation (Indianapolis). Human IFN-γ was a gift from Amgen Biologicals. Cells were treated with human IFN-γ at 5 ng/ml or mouse IFN at 200 units/ml for lengths of time as indicated in each experiment. 1,2-Bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate (BAPTA)-acetoxymethyl ester (AM), KN93, bisindolylmaleimide, SB203580, calmodulin, and G418 were from Calbiochem. Fluo-3 AM was from Molecular Probes. CaMKIIα/β purified from rat brain was a gift from A. Nairn (Rockefeller University).
Glutathione S-Transferase (GST) Pull-Down Assays, Coimmunoprecipitation, and Western Blot Analysis.
GST-fusion proteins were purified from bacteria by using glutathione-Sepharose beads (Amersham Pharmacia). In vitro translation reactions were done by using the TNT T7 system (Promega). The preparation of nuclear extracts and GST pull-down assays were done as described (8). For the coimmunoprecipitation experiment, 0.5 mg of proteins from whole-cell extracts were dialyzed in buffer BC100 (50 mM Tris⋅HCl, pH 8.0/0.5 mM EDTA/100 mM NaCl/20% glycerol) and incubated overnight with 10–20 μg of antibodies in buffer BC100 (8). Immune complexes were brought down with protein A/G agarose beads (Santa Cruz Biotechnology), washed with the same buffer, and separated by SDS/PAGE. Western blot analyses were done by using chemiluminescence (Dupont/NEN).
Plasmid Constructions.
GST-Stat1 TAD was constructed as described (8). The cDNA of Stat1 was subcloned into pBluescript (Stratagene) to generate SK/Stat1. The HA epitope was placed at the amino terminus of Stat1 by PCR with the following overlapping oligonucleotides for the 5′ primer: 5′-AAGGAAAAAAGCGGCCGCACCATGGCATATCCATACGATGTGCCAGACTACGCG, 5′-TACGATGTGCCAGACTACGCGTCTCAGTGGTACGAACTTCAG, and 5′-GTCTGATTTCCATGGGAAAACTG for the 3′ primer to generate SK/HAStat1. The HA-tagged Stat1 cDNA then was subcloned into the RcCMV expression vector (Invitrogen). The CaMKIIγ wild type (WT) and the K43M dominant-negative (DN) mutant cDNA were gifts from H. Schulman (Stanford University, Stanford, CA). GSTCK-FL (residues 1–518), -RVA (273–518), -RcVA (291–518), -RN (273–290), -RC (291–315), and RcCMVCK-WT, -DN, and -A (386–518) constructs were generated by PCR subcloning of the various CaMKII fragments into pGEX-5x-1 (Amersham Pharmacia) and RcCMV (Invitrogen), respectively.
Transfection Experiments.
Transient transfections of NIH 3T3 or U3A cells were done with the Lipofectamine 2000 method (GIBCO) according to manufacturer instructions. All transfections were done with CMV-GFP cotransfected to monitor transfection efficiency. For Fig. 5A, 24 h after transfection cells were plated into medium containing 1 mg/ml G418 for 6 days to eliminate nontransfected cells, and G418-resistant cells were pooled for the experiments.
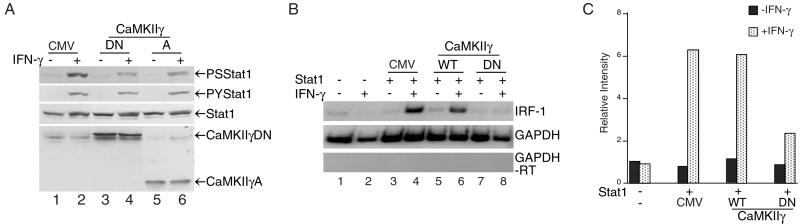
Figure 5.
DN forms of CaMKII inhibit Stat1 S727 phosphorylation and IFN-γ-induced transcription activation. (A) NIH 3T3 cells were transfected with the indicated mutant CaMKIIs and selected in G418-containing medium for 6 days. G418-resistant cells were pooled and treated with 200 units/ml mouse IFN-γ for 30 min, and whole-cell extracts were analyzed by Western blotting. CMV, RcCMV vector; (A) a fragment containing the association region of CaMKII; DN, CaMKII containing a K43M mutation. (B) U3A cells were transfected with Stat1 and the WT or DN CaMKII. Twenty-four hours after transfection, cells were treated with 5 ng/ml human IFN-γ for 3 h, and total RNA was analyzed by reverse transcription–PCR with [32P]dCTP and primer pairs for the indicated genes. (C) The gels of B were quantitated and analyzed by a PhosphorImager. The relative intensities were ratios of IRF-1/glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
RNA Analyses.
Total RNAs were prepared by using TRIzol (GIBCO). Northern blot and reverse transcription–PCR analyses were carried out as described (8, 30). Quantitation and analyses were performed on a Molecular Dynamics Storm PhosphorImager.
Measurement of [Ca2+]i.
NIH 3T3 cells were plated on a 96-well (2 × 104 cells per well), clear-bottom black plates (Falcon 356640 Biocoat). Cells were washed once with freshly prepared wash buffer (130 mM NaCl/2 mM CaCl2/5 mM KCl/10 mM glucose/0.45 mM KH2PO4/0.4 mM Na2HPO4/1.2 mM MgSO4/1.2 mM MgCl2/4.2 mM NaHCO3/20 mM Hepes, pH 7.4/2.5 mM probenecid/0.1% BSA) and loaded with 100 μl of dye-loading buffer (4 μM fluo-3/0.018% Pluronic/1% BSA in wash buffer). The plate was wrapped in aluminum foil and incubated for 45 min at room temperature and then 15 min at 37°C. The cells were washed three times with wash buffer on ice. Wash buffer (100 μl) was added to each well. The samples were incubated for 10 min at 37°C immediately before assay. Ligand was diluted to 3× concentration and added to an individual well via an injector mechanism supplied within the fluorospectrophotometer. The fluorescence of the cell population of an individual well was followed for 5 min after the addition of ligand and recorded for 20 msec at 1-sec intervals. The fluorescence of each well also was recorded first after incubating with 50 μl of 4% Triton X-100 for 10 min and then after quenching with 25 μl of 90 mM EGTA.
The data for calcium-flux experiments were collected on a Fluroskan Ascent FL fluorospectrophotometer (Labsystems, Chicago) and analyzed by ASCENT 2.4 software (Labsystems). The free cytoplasmic calcium concentration was calculated by using the following equation: [Ca2+] = Kdβ (R − Rmin)/(Rmax − R), where R is the ratio between fluorescence intensity at 485 and 538 nm, and Kdβ is a constant equal to 390 nM when using fluo-3 as the fluorescent calcium indicator. Rmin is the reading after EGTA, and Rmax is the reading after Triton X-100.
CaMK Assays.
A 50-μl CaMKII kinase assay reaction mixture contained 50 mM Pipes, pH 7.0, 20 mM MgCl2, 0.2 mg/ml BSA, 50 μM ATP, 5 μCi/ml (1 Ci = 37 GBq) of [γ-32P]ATP (3,000–6,000 Ci/mmol), 1 mM CaCl2, and 20 μg/ml calmodulin. Ca2+/calmodulin-independent activity was measured in the presence of 1 mM EGTA in the absence of exogenous Ca2+ and calmodulin. The concentrations of the various substrates were: 15 μM CaMKII-selective peptide substrate, autocamtide-3 (Figs. 2B and 4A), 50–100 ng/μl GST-fusion proteins (Fig. 4B), and 20 ng/μl histone H3. For Fig. 2B, NIH 3T3 cells with indicated treatment were lysed in whole-cell extract buffer (50 mM Tris, pH 8/280 mM NaCl/0.5% Nonidet P-40/0.2 mM EDTA/2 mM EGTA), and lysates were assayed for the Ca2+/calmodulin-independent activity. For Fig. 4, preparation of the CaMKIIγ/δ-containing eluate was as described (8). The eluate then was dialyzed in buffer BC100 (8) and concentrated with the Ultrafree-0.5 centrifugal device (Biomax-10) according to instructions from the manufacturer (Millipore). For Figs. 2B and 4A, the kinase reactions were initiated by the addition of 5 μl of freshly prepared ice-cold whole-cell extract (Fig. 2B) or the indicated kinases (Fig. 4A) to the mixture at 30°C. After 2 min, the reaction was terminated by the addition of 10 μl of cold 15% trichloroacetic acid solution and incubated on ice for 10 min. Each reaction was centrifuged at 13,000 rpm for 2 min, and 30 μl was spotted onto phosphocellulose paper (GIBCO). The paper was washed with 1% H3PO4 and dH2O twice. The [γ-32P]ATP incorporation was measured by using a scintillation counter (Beckman Coulter). The CaMKII activity was expressed as pmol of 32P/min/mg of protein. For Fig. 4 B and C, 250 ng of pCaMKIIα/β, ≈200 ng of pCaMKIIγ/δ from EPGSTS1C were used in the reactions. The reactions were stopped by the addition of SDS/PAGE sample loading buffer, and the whole reaction mixes were separated by SDS/PAGE followed by autoradiography.
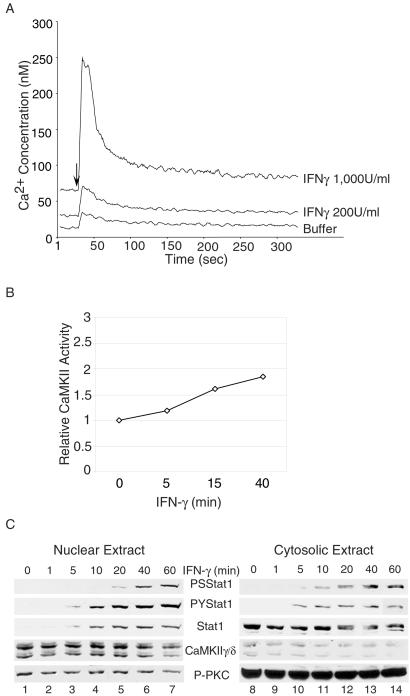
Figure 2.
IFN-γ induces Ca2+ transients and activates CaMKII. (A) NIH 3T3 cells were preloaded with fluo-3 AM, and the [Ca2+]i was measured at 1-sec intervals after IFN-γ treatment by a Fluroskan Ascent FL fluorospectrophotometer (Labsystems) and analyzed by the ASCENT 2.4 software (Labsystems). The arrow indicates the addition of IFN-γ to the cells. (B) NIH 3T3 cells were treated with 200 units/ml mouse IFN-γ for the times indicated, and cell lysates were analyzed in a CaMKII kinase assay with the CaMKII-selective autocamtide-3 as substrate. (C) Nuclear and cytoplasmic extracts were prepared from NIH 3T3 cells treated with 200 units/ml mouse IFN-γ for the times indicated and analyzed by Western blotting. PS, Phospho-S727; PY, Phospho-Y701; P-PKC, activated phospho-PKC.
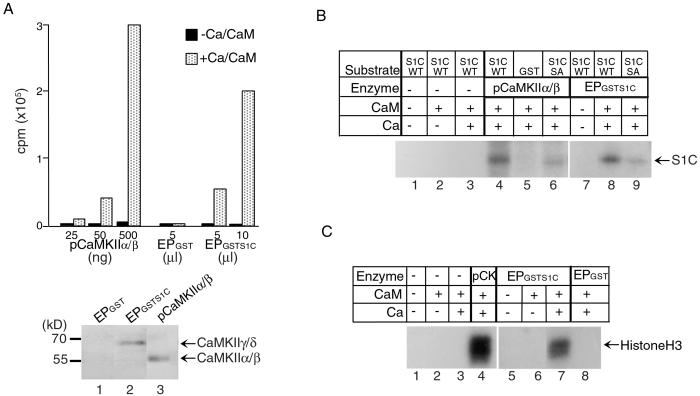
Figure 4.
CaMKII phosphorylates Stat1 S727 in vitro. (A Upper) Kinase assays were performed with purified rat brain CaMKIIα/β and a CaMKIIγ/δ-containing fraction purified from U3A nuclear extracts by using GST-Stat1 TAD affinity columns (EPGSTS1C) or eluates from a GST column (EPGST) as control. The incorporation of 32P in the CaMKII substrate, autocamtide-3, was measured by a scintillation counter. (Lower) Western blot analyses of 10 μl of eluates from a GST column (lane 1), from a GST-Stat1 TAD affinity column (lane 2), and 250 ng of pCaMKIIα/β (lane 3). S1C, Stat1 TAD; pCaMKII, purified CaMKIIα/β, Ca, calcium; CaM, calmodulin; EP, eluted protein. (B) GST-fusion proteins containing WT or S727A mutant Stat1 TAD were used as substrates for the indicated CaMKIIs. Approximately 250 ng of CaMKIIα/β, 200 ng of CaMKIIγ/δ, and 2.5 μg of various GST-fusion proteins were used in the kinase assays. The incorporation of 32P in the Stat1 TAD was visualized by autoradiography after SDS/PAGE. SA, S727A. (C) Purified histone H3 (1 μg) was used as a substrate for the indicated CaMKIIs, and the incorporation of 32P was visualized by autoradiography after SDS/PAGE. pCK, purified CaMKIIα/β.
Results
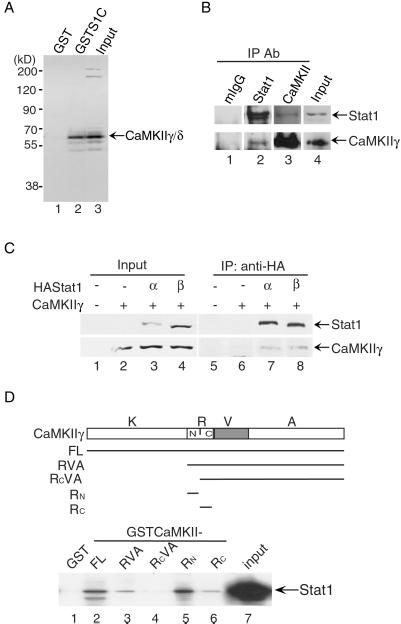
We previously purified a group of nuclear proteins that interact specifically with Stat1 TAD (8). Protein identification by mass spectrometry (31) confirmed that the p55 protein in this group of Stat1 TAD-interacting proteins was CaMKIIγ (data not shown). The GST pull-down assay showed that the Stat1 TAD (residues 710–750 including the S727 residue) could interact with CaMKIIγ/δ in nuclear extracts from U3A cells (Fig. 1A, lane 2). Furthermore, coimmunoprecipitation of whole-cell extracts from IFN-γ-treated NIH 3T3 cells showed that endogenous Stat1 and CaMKIIγ could interact in vivo (Fig. 1B, lanes 2 and 3). Coimmunoprecipitation of whole-cell extracts from U3A cells transfected with HA-tagged Stat1α or β and CaMKIIγ showed that both full-length Stat1α and β could interact with CaMKIIγ in vivo, suggesting that not only the Stat1 TAD (missing in Stat1β) but some other site in Stat1 could also interact with CaMKIIγ (Fig. 1C, lanes 7 and 8). To investigate which region in CaMKII can interact with Stat1, various regions of CaMKII were expressed as GST-fusion proteins according to their known structural and functional domains (ref. 21; Fig. 1D). The regulatory domain was divided further into the amino-terminal half and the carboxyl-terminal half (the calmodulin-binding domain; ref. 32). GST pull-down assays using these GST-fusion proteins showed that the amino-terminal half of the regulatory region (residues 273–290) of CaMKIIγ could interact with Stat1 directly (Fig. 1D, lane 5). Equivalent amounts of GST-fusion proteins were used in the assay (data not shown). These results indicate that there is a specific interaction between Stat1 and the ubiquitous members of the CaMKII family, i.e., CaMKIIγ/δ.
Figure 1.
Specific interactions between Stat1 and CaMKII in vivo and in vitro. (A) GST pull-down assays were performed with GST-fusion proteins containing the Stat1 TAD and nuclear extract of U3A cells. The bound proteins were analyzed by Western blotting. S1C, Stat1 TAD. (B) Coimmunoprecipitation (IP) analyses were performed with whole-cell extracts from NIH 3T3 cells. The endogenous Stat1 or CaMKII proteins in the immune complexes were analyzed by Western blotting. mIgG, control antibody from mouse serum. (C) Whole-cell extracts were prepared from U3A cells transfected with CaMKII-γ and HA-tagged Stat1-α or -β. The immune-precipitated complexes by anti-HA were analyzed by Western blotting. (D) GST pull-down assays were performed with GST-fusion proteins containing the various portions of CaMKIIγ and in vitro translated 35S-labeled Stat1. The bound proteins were separated by SDS/PAGE and visualized by autoradiography. K, kinase domain (residues 1–272); R, regulatory domain (273–315); V, variable region (316–385); A, association domain (386–518); Rn, amino-terminal half of the regulatory domain (273–290); Rc, carboxyl-terminal half of the regulatory domain (291–315); FL, full length (1–518).
To investigate any direct role CaMKII might play in the phosphorylation of S727 of Stat1, we first tested whether IFN-γ can activate CaMKII. To see whether IFN-γ can elicit a Ca2+ flux necessary for CaMKII activation in fibroblasts, NIH 3T3 cells were treated with IFN-γ, and the intracellular concentration of Ca2+ ([Ca2+]i) was measured. IFN-γ induced a rapid and sharp increase in [Ca2+]i in a dose-dependent manner (Fig. 2A), similar to results reported previously with other cell types (28, 29). Autophosphorylation of CaMKII occurs in situ accompanying enzymatic activation and renders CaMKII Ca2+-independent, and the Ca2+-independent activity is used to assess activation of CaMKII (21). CaMKIIγ/δ activities in whole-cell extracts from NIH 3T3 cells were assayed by direct substrate phosphorylation using the CaMKII-selective autocamtide-3 substrate (33). As shown in Fig. 2B, the Ca2+-independent activities of CaMKII were increased after treatment with IFN-γ. Cell fractionation showed that CaMKIIγ/δ was present both in the cytoplasm and the nucleus (Fig. 2C). The distribution patterns or amounts of CaMKII did not change significantly with IFN-γ treatment (Fig. 2C). As a control, the activation of the Ca2+-dependent PKC subfamily, PKCα/β/γ (20–22), was measured by Western blot with an antibody against the active form of phosphorylated PKCα/β/γ. PKC was not activated by IFN-γ treatment, but a constitutively autophosphorylated PKC was present predominantly in the cytoplasmic fraction in cells grown in serum (Fig. 2C) as reported previously (22, 24). Altogether, these results indicate that IFN-γ treatment induces a Ca2+ flux and activates CaMKII in NIH 3T3 cells.
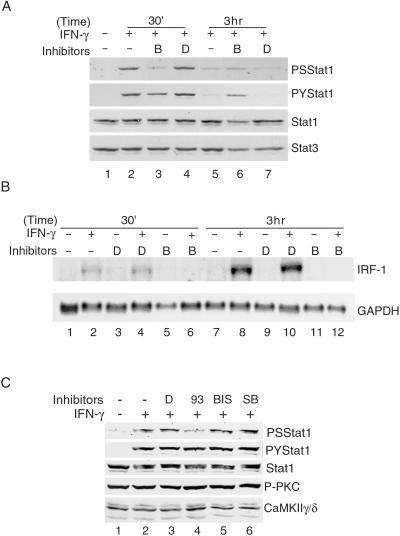
To determine whether a Ca2+ flux was required for the phosphorylation of Stat1 in response to IFN-γ, NIH 3T3 cells were pretreated with a Ca2+-chelating reagent, BAPTA-AM, which prevents an intracellular rise of [Ca2+]i caused by either influx of Ca2+ from the medium or release of Ca2+ from intracellular stores. Phosphorylation of S727 of Stat1 was inhibited substantially by BAPTA, whereas phosphorylation of Y701 was not inhibited significantly (Fig. 3A, compare lane 3 with lanes 2 and 4). The total amount of Stat1 protein was not affected by BAPTA treatment at 30 min (Fig. 3A, lane 3). As reported before (34), after 3 h of treatment with IFN-γ, the Stat1 protein level was increased because of an increase in the transcription of the Stat1 gene (Fig. 3A, compare lanes 5 and 7 with lane 1, and data not shown). BAPTA treatment prevented such an increase (Fig. 3A, lane 6) because of inhibition of transcription of the Stat1 gene (data not shown) and probably also inhibition at the level of protein translation (35). As a control, the steady-state level of Stat3 was unchanged (Fig. 3A). Northern blot analyses of total RNA showed that BAPTA inhibited the IFN-γ-induced production of a Stat1-dependent gene, IFN regulatory factor-1 (IRF-1; refs. 36 and 37; Fig. 3B, compare lanes 6 and 12 with lanes 2 and 8). Treatment with the solvent DMSO had no effect (Fig. 3A, lanes 4 and 7, and B, lanes 3, 4, 9, and 10). These results indicate that a Ca2+ flux is involved and required for INF-γ-induced serine phosphorylation of Stat1 and gene activation. To determine further which serine kinase is involved in IFN-γ-induced S727 phosphorylation, NIH 3T3 cells were treated with the following specific serine kinase inhibitors: KN93 for CaMKII, bisindolylmaleimide 1 for PKC, and SB203580 for p38 MAP kinase. Only the CaMKII inhibitor, KN93, inhibited IFN-γ-induced Stat1 S727 phosphorylation (Fig. 3C, lane 4). KN-93 also inhibited IFN-γ-induced IRF-1 transcription (data not shown). Altogether, these results indicate that Ca2+-activated CaMKII but not PKC or p38 MAP kinase is involved in the IFN-γ-induced phosphorylation of Stat1 S727.
Figure 3.
Requirement of Ca2+ and CaMKII for IFN-γ-induced phosphorylation of Stat1 S727 and gene activation. (A) Whole-cell extracts were prepared from NIH 3T3 cells with the indicated treatment and analyzed by Western blotting. Pretreatments with DMSO (D) or 50 μM BAPTA-AM (B) were for 30 min followed by 200 units/ml mouse IFN-γ for the length of time indicated. PS, phospho-S727; PY, phospho-Y701. (B) Total RNAs were prepared from NIH 3T3 cells with the indicated treatment as described for A and analyzed by Northern blotting. (C) NIH 3T3 cells were pretreated with the indicated reagents for 30 min followed by 200 units/ml mouse IFN-γ for 30 min, and whole-cell extracts were analyzed by Western blotting. 93 , KN93 (5 μM); BIS, bisindolylmaleimide 1 (20 μM); SB, SB203580 (10 μM); P-PKC, activated phospho-PKC.
To demonstrate that Stat1 could interact with a functional CaMKII, in vitro kinase assays were performed with three substrates: a known CaMKII substrate, autocamtide-3 (Fig. 4A), Stat1 TAD (Fig. 4B), and histone H3 (Fig. 4C). Two types of CaMKIIs were used: the purified CaMKIIα/β from rat brain as a positive control and the CaMKIIγ/δ that was partially purified by using a GST-Stat1 TAD affinity column (8). The concentration of CaMKIIγ/δ in the eluate was estimated to be at 20 ng/μl by Western blotting (Fig. 4A Lower, compare lanes 2 and 3) and silver staining (data not shown). The partially purified CaMKIIγ/δ showed a Ca2+/calmodulin-dependent activity comparable to that of the purified CaMKII-α/β from rat brain (Fig. 4A Upper). Stat1 TAD could be phosphorylated by both CaMKIIα/β and CaMKIIγ/δ (Fig. 4B, lanes 4 and 8). Mutation of S727 in the Stat1 TAD resulted in a significant decrease in 32P incorporation (Fig. 4B, lanes 8 and 9). These results suggest that of the seven serine residues in the Stat1 TAD, S727 is the major site for CaMKII in these in vitro kinase assays. As a positive control, histone H3 also could be phosphorylated by CaMKII (Fig. 4C, lanes 4 and 7) as suggested previously (38). Altogether, these results demonstrate that there is a functional enzyme–substrate interaction between Stat1 and CaMKII.
To demonstrate further the specific role of CaMKII in Stat1 S727 phosphorylation and transcription activation, DN mutants of CaMKII were generated: CaMKIIγ-A (see Fig. 1D), and the ATP-binding-deficient kinase with a K43M mutation (DN). Their effects on Stat1 S727 phosphorylation and transcription activity were tested by transient transfections in both NIH 3T3 cells and U3A cells. NIH 3T3 cells transfected with the indicated plasmids were grown first in G418-containing medium for 6 days to eliminate nontransfected cells, and G418-resistant cells then were pooled and used for the experiment shown in Fig. 5A. Overexpression of these CaMKII mutants in NIH 3T3 cells inhibited IFN-γ-induced Stat1 S727 phosphorylation but not Y701 phosphorylation (Fig. 5A, lanes 4 and 6). Furthermore, to demonstrate that the CaMKII mutants can inhibit IFN-γ-induced transcription of an endogenous gene, U3A cells were transiently transfected with Stat1 and the WT or the CaMKII DN mutant followed by reverse transcription–PCR analyses of total RNA. CaMKII DN inhibited IRF-1 transcription by 70%, whereas the WT CaMKII had no effect (Fig. 5B, compare lanes 4, 6, and 8). The CaMKII WT and DN were overexpressed similarly as described for Fig. 5A (data not shown). The gels in Fig. 5B also were quantitated by a PhosphorImager, and the relative intensities of the IRF-1 bands normalized by those of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) bands are shown in Fig. 5C. Because of the nature of the CaMKII as a multienzyme complex in vivo (21), the DN effect of the CaMKII mutants would depend on the stoichiometry of the WT and mutant enzyme subunits in the complex, and therefore complete inhibition was not seen. However, consistent inhibition by ≈50% or more was observed with multiple experiments using different approaches (data not shown). Together, these results indicate that CaMKII is necessary for Stat1 S727 phosphorylation and Stat1-mediated transcription activation in response to IFN-γ.
Discussion
Our findings suggest that a Ca2+ flux induced by the IFN-γ signaling pathway, a separate event from the tyrosine phosphorylation by Janus kinases, is required for phosphorylation of S727 in Stat1. We also found that reagents that cause [Ca2+]i increase can induce phosphorylation of S727 without concurrent Y701 phosphorylation in lymphocytes (J.S.N. and J.J.Z., unpublished data). UV, lipopolysaccharide, and tumor necrosis factor have been reported to induce Stat1 S727 phosphorylation without Y701 phosphorylation (12). Together with earlier observations that Ca2+/calmodulin is required for IFN-γ-induced MHCII expression (27) and lipopolysaccharide-stimulated inducible NO synthase (iNOS) production (39), these results suggest that signaling events that elicit Ca2+ flux can result in Stat1 S727 phosphorylation, which is critical for Stat1-mediated gene activation. Ca2+ is a ubiquitous messenger involved in a multitude of cellular processes (16). The frequency and magnitude of Ca2+ flux elicited by various signaling ligands confers specificity to the many different signaling pathways, leading to activation of different transcription activators such as NFAT, Oct-1/OAP, or NF-κB (18, 19, 40). Our results suggest that in NIH 3T3 cells, the Ca2+ flux caused by IFN-γ activates the multifunctional CaMKII, which in turn phosphorylates Stat1 on S727 for maximum transcription activation.
The specific interaction between Stat1 and CaMKII also suggests the possibility that CaMKII may phosphorylate sites in chromatin to participate in chromatin remodeling near the Stat1 binding sites, which may occur in addition to the acetylation of histones by CBP/p300, also recruited by Stat1 (6, 41). Phosphorylation on serine residue 10 of histone H3 is correlated directly with the induction of immediate-early genes such as c-jun, c-fos, and c-myc (42). Our results indicate that CaMKII can phosphorylate histone H3 directly, although it remains to be determined which serine residue in histone H3 is phosphorylated.
The search for the serine kinase responsible for phosphorylating the serine residue in STAT TADs has revealed the involvement of several kinases. The ERK MAP kinase was shown to phosphorylate Stat3 S727 (43). The p38 MAP kinase is required for UV-induced phosphorylation of Stat1 S727 (13), whereas the involvement of p38 MAP kinase in IFN-γ-induced Stat1 S727 is controversial (13, 14). Our results suggest that CaMKII but not p38 MAP kinase or PKC is involved in IFN-γ-induced Stat1 S727 phosphorylation. Two types of CaMKII substrates have been identified: one group with an RXXS/T motif and the other group with no definitive motif except a preference of negative-charged residue at +2 position (25). The Stat1 sequence LPMSPEEF would be consistent with the second type of motif. It was reported recently that the phosphatidylinositol 3-kinase (PI3K)/Akt pathway could be involved also in the IFN-γ-induced phosphorylation of Stat1 S727 in human tumor cell lines or mouse embryonic fibroblasts (15). PI3K has been shown to facilitate antigen-stimulated Ca2+ flux in Jurkat T cells and mast cells (44, 45), which could result in activation of Ca2+-dependent serine kinases. It seems likely that in different cellular contexts, more than one serine kinase pathway can converge on Stat1 during or after its activation through tyrosine phosphorylation on the cell surface.
Acknowledgments
We thank G. Stark and I. Kerr for the U3A cell line, H. Schulman for the CaMKIIγ and δ cDNAs, and A. Nairn for the purified CaMKIIα/β. We thank J. Ding for technical assistance and S. Cvejic for help with the Ca2+ flux assay. This work is supported by National Institutes of Health (NIH) Grant GM61652 and American Cancer Society Grant RPG-00-264-GMC (to J.J.Z.), NIH Grants AI34420 and AI31489 (to J.E.D.), and NIH Grant RR00862 (to B.T.C.). J.J.Z. is a recipient of a Scientist Development Award from the American Heart Association.
Abbreviations
- Stat1
signal transducers and activators of transcription 1
- MAP
mitogen-activated protein
- TAD
transcription activation domain
- CaMK
Ca2+/calmodulin-dependent kinase
- PKC
protein kinase C
- HA
hemagglutinin
- BAPTA
1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate
- AM
acetoxymethyl ester
- GST
glutathione S-transferase
- CMV
cytomegalovirus
- WT
wild type
- DN
dominant-negative
- IRF-1
IFN regulatory factor
References
- 1.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 2.Leonard W J, O'Shea J J. Annu Rev Immunol. 1998;16:293–322. doi: 10.1146/annurev.immunol.16.1.293. [DOI] [PubMed] [Google Scholar]
- 3.Becker S, Groner B, Muller C W. Nature (London) 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Jr, Kuriyan J. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston D M. Nature (London) 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath C M, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moriggl R, Berchtold S, Friedrich K, Standke G J, Kammer W, Heim M, Wissler M, Stocklin E, Gouilleux F, Groner B. Mol Cell Biol. 1997;17:3663–3678. doi: 10.1128/mcb.17.7.3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J J, Zhao Y, Chait B T, Lathem W W, Ritzi M, Knippers R, Darnell J E., Jr EMBO J. 1998;17:6963–6971. doi: 10.1093/emboj/17.23.6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu B, Reichel M, Fisher D A, Smith J F, Rothman P. J Immunol. 1997;159:1255–1264. [PubMed] [Google Scholar]
- 10.Paulson M, Pisharody S, Pan L, Guadagno S, Mui A L, Levy D E. J Biol Chem. 1999;274:25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 11.Wen Z, Zhong Z, Darnell J E., Jr Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 12.Kovarik P, Stoiber D, Novy M, Decker T. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovarik P, Stoiber D, Eyers P A, Menghini R, Neininger A, Gaestel M, Cohen P, Decker T. Proc Natl Acad Sci USA. 1999;96:13956–13961. doi: 10.1073/pnas.96.24.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh K C, Haque S J, Williams B R. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nguyen H, Ramana C V, Bayes J, Stark G R. J Biol Chem. 2001;276:33361–33368. doi: 10.1074/jbc.M105070200. [DOI] [PubMed] [Google Scholar]
- 16.Bootman M D, Collins T J, Peppiatt C M, Prothero L S, MacKenzie L, De Smet P, Travers M, Tovey S C, Seo J T, Berridge M J, Ciccolini F, Lipp P. Semin Cell Dev Biol. 2001;12:3–10. doi: 10.1006/scdb.2000.0211. [DOI] [PubMed] [Google Scholar]
- 17.Santella L, Carafoli E. FASEB J. 1997;11:1091–1109. [PubMed] [Google Scholar]
- 18.De Koninck P, Schulman H. Science. 1998;279:227–230. doi: 10.1126/science.279.5348.227. [DOI] [PubMed] [Google Scholar]
- 19.Dolmetsch R E, Xu K, Lewis R S. Nature (London) 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 20.Soderling T R. Trends Biochem Sci. 1999;24:232–236. doi: 10.1016/s0968-0004(99)01383-3. [DOI] [PubMed] [Google Scholar]
- 21.Braun A P, Schulman H. Annu Rev Physiol. 1995;57:417–445. doi: 10.1146/annurev.ph.57.030195.002221. [DOI] [PubMed] [Google Scholar]
- 22.Mellor H, Parker P J. Biochem J. 1998;332:281–292. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tombes R M, Krystal G W. Biochim Biophys Acta. 1997;1355:281–292. doi: 10.1016/s0167-4889(96)00141-3. [DOI] [PubMed] [Google Scholar]
- 24.Srinivasan M, Edman C F, Schulman H. J Cell Biol. 1994;126:839–852. doi: 10.1083/jcb.126.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.White R R, Kwon Y G, Taing M, Lawrence D S, Edelman A M. J Biol Chem. 1998;273:3166–3172. doi: 10.1074/jbc.273.6.3166. [DOI] [PubMed] [Google Scholar]
- 26.Cruzalegui F H, Bading H. Cell Mol Life Sci. 2000;57:402–410. doi: 10.1007/PL00000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koide Y, Ina Y, Nezu N, Yoshida T O. Proc Natl Acad Sci USA. 1988;85:3120–3124. doi: 10.1073/pnas.85.9.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kung A W, Lau K S, Wong N S. Endocrinology. 1995;136:5028–5033. doi: 10.1210/endo.136.11.7588238. [DOI] [PubMed] [Google Scholar]
- 29.Aas V, Larsen K, Iversen J G. Cell Signal. 1999;11:101–110. doi: 10.1016/s0898-6568(98)00040-0. [DOI] [PubMed] [Google Scholar]
- 30.Yang E, Wen Z, Haspel R L, Zhang J J, Darnell J E., Jr Mol Cell Biol. 1999;19:5106–5112. doi: 10.1128/mcb.19.7.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali B R, Tjernberg A, Chait B T, Field M C. J Biol Chem. 2000;275:33222–33230. doi: 10.1074/jbc.M003845200. [DOI] [PubMed] [Google Scholar]
- 32.Singla S I, Hudmon A, Goldberg J M, Smith J L, Schulman H. J Biol Chem. 2001;276:29353–29360. doi: 10.1074/jbc.M101744200. [DOI] [PubMed] [Google Scholar]
- 33.Hanson P I, Schulman H. J Biol Chem. 1992;267:17216–17224. [PubMed] [Google Scholar]
- 34.Pine R, Canova A, Schindler C. EMBO J. 1994;13:158–167. doi: 10.1002/j.1460-2075.1994.tb06245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palfrey H C, Nairn A C. Adv Second Messenger Phosphoprotein Res. 1995;30:191–223. doi: 10.1016/s1040-7952(05)80008-4. [DOI] [PubMed] [Google Scholar]
- 36.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 37.Meraz M A, White J M, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, et al. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 38.Whitlock J P, Jr, Galeazzi D, Schulman H. J Biol Chem. 1983;258:1299–1304. [PubMed] [Google Scholar]
- 39.Park Y C, Jun C D, Kang H S, Kim H D, Kim H M, Chung H T. Immunology. 1996;87:296–302. doi: 10.1046/j.1365-2567.1996.456544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li W, Llopis J, Whitney M, Zlokarnik G, Tsien R Y. Nature (London) 1998;392:936–941. doi: 10.1038/31965. [DOI] [PubMed] [Google Scholar]
- 41.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T M, Rose D W, Rosenfeld M G, Glass C K. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strahl B D, Allis C D. Nature (London) 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 43.Chung J, Uchida E, Grammer T C, Blenis J. Mol Cell Biol. 1997;17:6508–6516. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu A L, Ching T T, Sen G, Wang D S, Bondada S, Authi K S, Chen C S. J Biol Chem. 2000;275:16242–16250. doi: 10.1074/jbc.M002077200. [DOI] [PubMed] [Google Scholar]
- 45.Ching T T, Hsu A L, Johnson A J, Chen C S. J Biol Chem. 2001;276:14814–14820. doi: 10.1074/jbc.M009851200. [DOI] [PubMed] [Google Scholar]