Abstract
Molecular combing is a powerful procedure for aligning a large array of DNA molecules onto a surface. This technique usually leads to an overstretching of about 150% of the molecules' contour length. By changing the magnitude of capillary forces during the combing process, we were able to reduce the relative extension of the DNA molecules. Thus we achieved combing of T7 DNA with an extension close to its molecule contour length. We checked the ability of combed DNA to interact with DNA binding proteins. Using the T7 bacteriophage transcription system, we investigated the transcription activity of RNA polymerase on combed DNA by direct visualization of newly synthesized fluorescent RNAs. Our experiments show that no transcription activity occurs on overstretched DNA molecules, whereas we observe a transcription activity for nonoverstretched molecules. This activity is observed both in multiple initiation experiments and for one immobilized T7 RNA polymerase per promoter. These results open possibilities for the study of single enzyme actions on combed DNA by optical methods.
Transcription is a fundamental process in gene expression that allows the regulation of cellular adaptation and differentiation. This function is carried out by RNA polymerases (RNAPs) that produce an RNA copy of a given DNA strand (1). The interaction between the DNA and the RNAP is complex because RNAP must recognize a promoter, a sequence-specific region of double-stranded DNA before polymerization. After isomerization of the nucleoprotein complex, resulting in local melting of the double helix, the enzyme transcribes DNA into RNA following the double-stranded DNA until it reaches a terminator (2). Motion is therefore part of the intrinsic activity of RNAP (3). Having the possibility to follow and visualize the movement of an RNAP on the DNA template will open a new area of investigation for understanding the mechanisms of transcription and its regulation.
Our goal is to detect the activity of RNAP along a DNA molecule during the transcription process. Several groups have reported single molecule investigation of the transcription process (4–9). Because we use fluorescence microscopy techniques, we cannot use DNA in its normal aqueous solution state, which is a Brownian fluctuating coil. Therefore the first steps we have to perform are to stretch the DNA molecule to avoid conformational fluctuations and to hinder its Brownian motion. Several techniques permit researchers to obtain both immobilized and stretched DNA molecules. The techniques developed so far are micromanipulation with optical or magnetic tweezers (10–14), elongation in a flow (15) or in an electric field (16), and molecular combing (17). Although micromanipulation has provided much insight it has an inherent disadvantage because the observation is limited to one molecule at a time and large statistics are difficult to obtain. By contrast, molecular combing is of particular interest because it allows direct observation of a large array of immobilized and aligned DNA by fluorescence microscopy. This latter technique was successfully applied to the identification of genomic DNA regions using fluorescent in situ hybridization (18, 19), to optical mapping (20, 21), or to the identification of replication origins (22, 23). Nevertheless, this method presents several drawbacks. Molecular combing usually leads to an overstretching of DNA molecules of about 150% of their molecule contour length (24). Furthermore, combed DNA molecules are in close proximity to the surface.
Is transcription activity possible on immobilized and stretched DNA? To answer this question, we decided to use the combing technique while overcoming its drawbacks. We found a way to avoid DNA overstretching during the combing process. Thus we obtained combed DNA with an extension close to its contour length.
We focused our research on a basic transcription system. We worked on the T7 bacteriophage transcription system, using T7 RNAP and the whole T7 DNA. Abundant literature already has been dedicated to the well-described T7 RNAP transcription system (25). T7 RNAP is a 110-kDa single unit enzyme that recognizes a 17-bp promoter. No transcription factor is needed to initiate, catalyze, or terminate the transcription. The average speed for T7 RNAP is 250–300 nt/s (corresponding to 100 nm/s along DNA). T7 DNA is a linear double-strand molecule containing 17 T7 promoters and one terminator and having an expected crystallographic length of 13 μm (26).
We detected the enzymatic activity of T7 RNAP by labeling newly synthesized RNA with fluorescent nucleotides. The first observation was that no transcription activity occurs on overstretched combed DNA. On the other hand, we were able to observe transcription activity of T7 RNAP on nonoverstretched combed T7 DNA. During these experiments T7 RNAP is in large excess compared with the promoters, and multiple initiations are possible. To prevent multiple initiations, we stabilized one RNAP per promoter and removed the free RNAP before transcription.
Materials and Methods
Surface Treatments.
Glass surfaces were rendered hydrophobic by two different methods. The first method involved coating the surfaces with polymethylmetacrylate (PMMA), which is a hydrophobic polymer. A droplet of PMMA in chlorobenzene [20% (wt/wt), molecular weight = 8,000 g⋅mol−1] is put down onto a clean glass cover slide and spread by spin-coating at 2,500 rpm for 1 min. Surfaces are then baked at 165°C for 20 min and stored at room temperature in a dust-free environment. The second method results in a silanated surface (according to the protocol in ref. 17). In a reactor, a clean glass cover slide is exposed to 7-octenyltrichlorosilane in the gas phase. This process allows the reaction of the hydroxyl groups of the glass surface with the chlorosilane, leading to the formation of a silane monolayer on the surface. This latest process leads to more hydrophobic surfaces as probed by measuring the contact angle between a drop of water and the treated surface (90° for silane and 65° for PMMA).
DNA Preparations.
T7 DNAs (38 kbps, 7.8 nM, Sigma) were prepared to a final concentration of 6.5 pM in appropriate buffer. For PMMA surfaces, we used bis-Tris buffer (50 mM, Sigma) pH = 6.6, and with silanated surfaces, we used Mes buffer (50 mM, Roche Molecular Biochemicals) pH = 5.5. When needed, DNAs were stained with a fluorescent intercalating agent, the dimmer yellow oxozalone (YOYO-1, Molecular Probes) in a 1 dye for 30 bases ratio.
Molecular Combing.
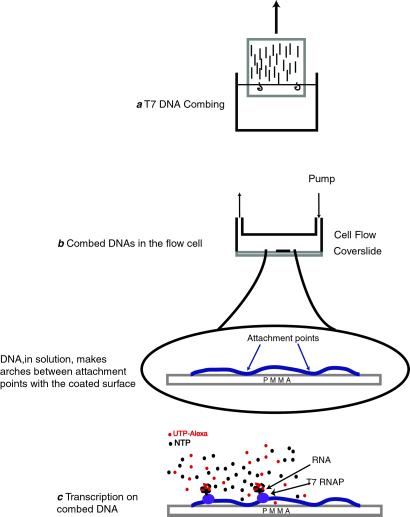
The combing process consists of the stretching of the DNA by the passage of an air/water meniscus (17, 24, 27). A hydrophobic surface is dipped into a DNA solution (DNA concentration 6.5 pM) at pH 5.5 for silanated surfaces and pH 6.6 for PMMA surfaces. DNA adsorbs onto this hydrophobic surface by one extremity in a “mushroom” state. This means that the adsorbed DNA has only one attachment point with the surface and retains its fluctuating coil conformation. The reason the DNA binds to the hydrophobic surface is still a matter of discussion. One explanation relies on pH-induced denaturation of the DNA ends, which then expose the hydrophobic part of bases and thus strongly interact with the surface (27). Whatever its origin, it should be mentioned that this interaction is very strong, so the DNA can be considered as grafted onto the surface.
When the slide is pulled out of the solution the anchored DNA molecules pass through the air/water interface. There, capillary forces pull down the DNA perpendicularly to the meniscus. Because once in contact with air DNA sticks onto the surface, there is no retraction of the molecules, which remain stretched onto the surface once out of the solution (Fig. 1).
Figure 1.
Sketch of the successive experimental steps. (a) T7 DNA combing. (b) Schematic of the flow cell used to study RNAP activity in solution. (c) Transcription on combed DNA. In the presence of NTPs and labeled-UTPs, RNAP synthesizes a fluorescent RNA. This fluorescent signal is used as a sensitive probe to report the efficiency of transcription.
To vary the magnitude of the capillary forces, the combing process has been performed under two different conditions: first, with the normal air/water surface tension, and second, with a lower surface tension obtained by spreading a monolayer of fatty alcohol at the air/water interface. For combing in the presence of fatty alcohol, we added a droplet of 1-dodecanol at the air/water interface just before pulling out the cover slide (28). Because of the low solubility of 1-dodecanol in water (1 ppm), this addition of alcohol did not interfere with the DNA molecules or glass surface in solution. The presence of a reservoir at the surface maintained the dodecanol monolayer in a dense phase. After the combing process the fatty alcohol that was adsorbed on the cover slide during the retraction spontaneously evaporated in the air. We designed our experimental set-up with a motorized translation stage so that the slides were pulled out of the solution at a constant speed (200 μm⋅s−1).
Rehydration in the Flow Cell.
After molecular combing DNA is on a dry surface. Thus, before biochemical investigations, we needed to rehydrate it. By rehydration we mean that the combed DNAs are put back in contact with an aqueous solution (Fig. 1). For such a purpose, we built a special cell with a small internal volume (a few tens of microliters) that permits circulation of fluids with a peristaltic pump. The rehydration of the combed DNA was performed with buffered solutions, the pH of which varied from 5.5 to 8.0. The bottom of the flow cell is the combed cover slide that can be placed above a microscope objective, allowing a direct visualization of the inner surface.
T7 RNAP Preparation.
T7 RNAP was overexpressed from cultures of Escherichia coli BL21 carrying plasmid pBH161 (29). These plasmids encode RNAPs with a hexa-His tag at the N terminus, allowing a single-step purification by absorption on Ni2+ column (30). The polymerase was stored at −20°C in 50% glycerol at a concentration of 33 μM.
Plasmid Preparation.
The plasmid pT361 (12 kb) carrying a T7 promoter was a generous gift of T. Lamonerie (Ecole Normale Supérieure Lyon). The plasmid was linearized by digestion at a unique NotI restriction site with the corresponding nuclease. The linear plasmid gives 9,171 bases transcript with T7 RNAP.
Transcription on Combed DNA.
Combed DNAs on the cover slide were placed at the bottom of a flow cell and rehydrated with a coating solution containing 1 mg/ml BSA, 250 μM nucleoside triphosphates (NTPs), 1 mM DTT, 50 mM Tris (pH 7.8), and 5 mM MgCl2. This coating solution was allowed to incubate for 30 min to prevent nonspecific binding of RNAPs or fluorescent nucleotides on the surface. The cell's volume was rinsed with a transcription buffer containing 20 mM Tris (pH 7.8), 5 mM MgCl2, 10 mM NaCl, 1 mM DTT, 0.25 mM EDTA, and 0.05% Tween 20. Combed T7 DNAs were then incubated with 200 nM T7 RNAP for 2, 5, and 25 min at room temperature in the presence of 200 μM NTPs and 100–500 units/ml RNase inhibitor (Roche, Molecular Biochemicals). Fluorescent UTP-Alexa (10 μM, Molecular Probes) in a 1/25 ratio versus UTP was added in the transcription solution for RNA labeling. The presence of UTP-Alexa in solution results in a high fluorescent background noise, which prevents the real-time visualization of the transcription complex. To overcome this problem, the cell volume was flushed for 1 min at 0.3 μl/s until the fluorescent background signal reached the initial level, permitting observation. Simultaneously, the enzyme activity was stopped because of starvation of NTPs. The minimum time necessary to visualize the transcription pattern is around 2 min, corresponding to the dead time caused by experimental manipulation and the flush time of the cell.
RNase Digestion.
Transcription on combed nonoverstretched DNA in the experimental cell was performed as described above. Then, the combed DNAs were incubated in transcription buffer for 2 min with, respectively, RNase T1 (50 units/μl, Ambion), which cleaves the phosphodiester backbone of RNA at 3′ G residues, or RNase H (0.02 units/μl, Promega), which specifically digest RNA in DNA–RNA hybrids (note that the definition of unit is not similar for the two enzymes; the enzymes were used in the company-recommended concentration).
Nuclease Digestion.
DNase I.
Combed DNA was incubated with DNase I (0.2 mg/ml) for 2 min in transcription buffer containing 20 mM Tris (pH 7.8), 5 mM MgCl2, 10 mM NaCl, 1 mM DTT, 0.25 mM EDTA, and 0.05% Tween 20 at room temperature.
DraI.
T7 DNA has nine restriction sites for DraI endonucleases at positions 274, 439, 5971, 6499, 10721, 16869, 20295, 31368, and 39745. DraI recognizes a specific AAA/TTT site and releases blunt DNA ends. Combed DNA was incubated with DraI (0.4 units/μl, Roche Molecular Biochemicals) for 2 min in buffer M (Roche Molecular Biochemicals).
HindIII.
HindIII cleaves DNA specifically at A/AGCTT sites. This sequence is not present on the T7 DNA. Combed DNA was incubated with HindIII (0.4 units/μl, NEB) for 2 min in buffer NEB2.
Computer Image Analysis and Video Microscopy.
Samples were observed by using an inverted microscope (Leica DM IRBE, Deerfield, IL) by epifluorescence. Both ×63 and ×100 infinity-corrected 1.4 numerical aperture oil objectives (Leica) were used, and a cooled charge-coupled device (CCD) camera (C4880 Hamamatsu, Ichinocho, Japan) was mounted on the microscope. For fluorescence observations, a Hg lamp was used in combination with a filter set for fluorescein (Leica I3). The images were acquired by using hipic software (Hamamatsu), with an exposition time of 500 ms. Acquisitions by the cooled CCD camera allowed a quantitative analysis of the fluorescent signal intensity. The integrated intensity of each dot was measured by analyzing the region of interest. We analyzed only dots that belonged to a string of at least three aligned dots. For visualization of retractions of stained combed DNA in solution, an intensified camera replaced the CCD camera. Video signal was recorded with a S-VHS video recorder (Panasonic) and digitized with a frame grabbing system (National Institutes of Health image program).
Calibration of the 9-kb Fluorescent RNAs.
We measured the intensity of a labeled RNA of known length, synthesized in bulk solution, to calibrate the fluorescence intensity of the transcribed RNA. A linearized plasmid of 12 kbps (pT361) with a T7 promoter was used to guarantee a mono-disperse-transcribed RNA population of 9 kbs in length. In addition, we used the same transcription procedure as that used for experiments on combed DNA. After stopping the bulk enzymatic reaction, the solution was injected in a flow cell on a hydrophobic surface. The synthesized fluorescent RNA adsorbs onto the surface. Thus we were able to measure the intensity of fluorescent RNAs in conditions similar to those of experiments on combed DNA. This process allows us to take into account quenching and bleaching of the fluorescent dyes.
Results
Molecular Combing of T7 DNA.
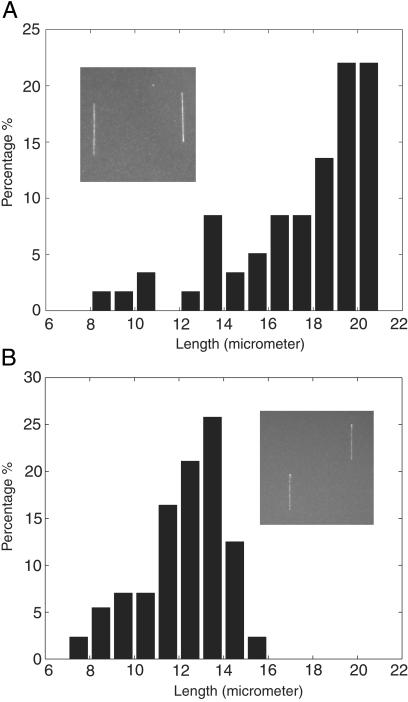
Length extension distributions of combed T7 DNA on glass cover slides coated with PMMA are shown in Fig. 2. Fig. 2a shows the distribution of length extension for T7 DNA combed in the absence of 1-dodecanol at the air/water interface. Fig. 2b shows the distribution of length extension for T7 DNA combed in the presence of 1-dodecanol at the air/water interface. In the absence of 1-dodecanol, the length of most combed DNA is between 17 μm and 21 μm, and the mean extension is close to 19.5 μm. Smaller combed DNAs are caused by double-strand breaks during the combing process. These data confirm the previous studies, i.e., the relative extension corresponds to 150% of the molecule's contour length (24) therefore the DNA molecules are overstretched. By contrast (Fig. 2b), in the presence of 1-dodecanol, most of the combed DNAs have a length between 12 and 14 μm. The mean length is 13 μm, which is close to the predicted crystallographic length of T7 DNA. The presence of the 1-dodecanol monolayer at the air/water interface leads to a diminution of the molecule extension. The same effect is observed on silanated surfaces (data not shown). It should be noted that this decrease in extension also has been observed with hydrophilic surfaces (24).
Figure 2.
Distribution of the DNA lengths in the absence (a, 120 molecules) or presence (b, 150 molecules) of 1-dodecanol during the combing process. (Insets) Combed T7 DNA on PMMA surface. DNA was stained with YOYO-1 and visualized with an epifluorescence microscope with a ×63 oil objective.
Behavior of the Combed Molecules in Solution.
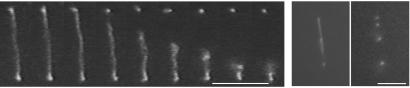
The state of rehydrated combed DNA has been determined with fluorescence microscopy. After prolonged observation (several seconds) under 450–490 nm irradiation, the double strand of DNA stained with YOYO-1 breaks. These breaks are caused by the release of free radicals during the photo-bleaching process of the intercalated dye molecules (31). The DNA then retracts spontaneously toward its closest attachment points onto the surface, as shown in Fig. 3. This retraction allowed an estimation of the density of attachment points along combed DNA. We analyzed DNA behaviors depending on the nature of the surface used for the combing process. On PMMA surfaces, we observed the DNA retraction after the double-strand break (as shown in Fig. 3). In this case, we can conclude that between two attachment points combed DNA molecules are not in contact with the surface, as sketched in Fig. 1. Only a few attachment points onto the surface per combed DNA were observed. The distance between two attachment points was always observed as being larger than 1 μm. In this case the density of attachment points is low. For combed DNA on silanated surfaces no DNA retraction was ever observed. Nevertheless, it is likely that double-strand breaks occur in this case as well. However, our optical observations failed to detect them because of the very high density of attachment points. Therefore we concluded that combed DNA binds strongly onto silanated surfaces and so remains stuck to the surface all along its length after rehydration. It should be mentioned that similar results on silanated surfaces already had been shown in previous studies (24). It should be noted that varying the pH of the solution needed for the rehydration procedure does not change these results in the pH range of 6 to 8. We found that the presence or absence of 1-dodecanol during the combing had no effect on the pattern of attachment points.
Figure 3.
Double-strand break and retraction of a combed T2 DNA molecule on PMMA in solution (Left). This retraction occurs in a few hundred ms. Time-lapse recordings of DNA retraction are seen in Movie 1, which is available as supporting information on the PNAS web site, www.pnas.org. After retraction is complete, the number of attachment points between the DNA and the surface can be visualized (Right). The DNA was stained with YOYO-1 and observed with an epifluorescence microscope with a ×100 objective. (Bars = 5 μm.)
Transcription on Combed DNA.
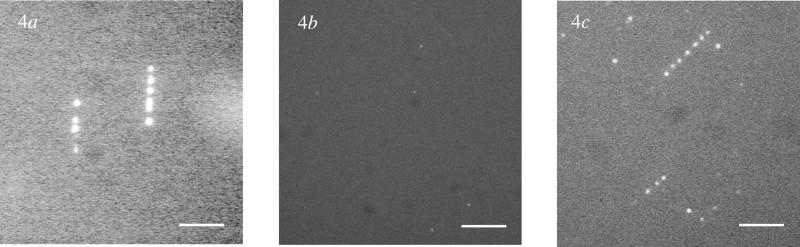
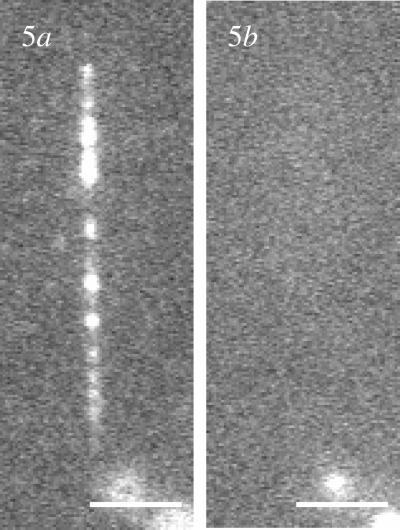
T7 RNAP has been used to perform transcription on unlabeled T7 DNA. We used DNA combed in the presence of a monolayer of 1-dodecanol on a PMMA surface. In the presence of both T7 RNAP and labeled nucleotides in transcription buffer aligned bright dots in the direction of the combing process appear (Fig. 4a). The bright dots form strings over a distance of 7–8 microns. Subsequent addition of nonfluorescent NTPs, a chase experiment, does not change the pattern.
Figure 4.
Transcription patterns representative of the transcription efficiency of T7 RNAPs on combed nonoverstretched DNA (a), combed overstretched DNA (b), and combed nonoverstretched DNA after the immobilization of one RNAP per promoter (c). The bright dots are the labeled RNAs aligned along DNA (a and c) or dispersed on the surface (b). Observations are performed by fluorescence microscopy [objective ×100, CCD acquisition time = 500 ms; bars = 5 μm (a and c) and 10 μm (b)].
To identify the nature of these dots we performed further control experiments. No fluorescent dots were observed when we carried out the transcription experiment in the absence of the enzyme. Thus, we concluded that the observed dots result from the RNA polymerization by the enzyme transcribing combed DNA. To probe the state of the RNA produced, whether it was as a DNA–RNA hybrid or coiled RNA, we incubated the RNA with RNase H and RNase T1. After transcription RNase H had no effect on the pattern of bright dots (data not shown) whereas RNase T1 completely eliminated the bright dots (Fig. 5). Thus, the RNA synthesized is accessible in solution. We also investigated the influence of the DNA template: using lambda DNA as a substrate, instead of T7 DNA, drastically changes the transcription pattern. Only a few dispersed points appear with a very low intensity (data not shown). Because lambda DNA template does not have T7 promoters, from these observations we conclude that no efficient transcription occurs on nonspecific DNA. The same procedure was applied to overstretched DNA, and again only a few points appeared with a very low intensity (Fig. 4b). Therefore transcription is efficient only for nonoverstretched DNAs bearing T7 promoters. To probe the accessibility of the overstretched DNA to enzymes, we studied the efficiency of their digestion by nonspecific endonucleases. DNase I is an enzyme that cuts nonspecifically double-stranded DNA in solution (32). Labeled combed DNAs exposed to DNase I disappear completely and small pieces of DNA are released in solution (data not shown). DNase I is thus able to recognize and digest combed overstretched DNA. The same experiments performed with DraI, a specific endonuclease that cleaves T7 DNA at nine sites, did not lead to the same results. That time, the DNA was unaffected by the action of the enzyme (data not shown). As for T7 RNAP, a specific site was not recognized on overstretched DNA. On the other hand, DraI was fully active on nonoverstretched DNA. We observed only a few dots remaining on the surface after digestion with DraI (data not shown). A control experiment with the restriction endonuclease HindIII, which is not supposed to cleave T7 DNA (there is no HindIII restriction site present on the T7 DNA), gave the expected result: the combed nonoverstretched DNA was unaffected by the presence of the enzyme. Thus, overstretched DNA is accessible for nonspecific enzymes and not recognized by specific endonucleases. Whereas nonoverstretched DNA is digested by nonspecific endonucleases, and restriction sites are recognized specifically by specific endonucleases.
Figure 5.
(a) Aligned bright dots forming the transcription pattern. (b) The same area after digestion by RNase T1. The dust at the lower right allows us to identify the area. Observations are performed by fluorescence microscopy (objective ×100, CCD acquisition time = 500 ms, bars = 2 μm).
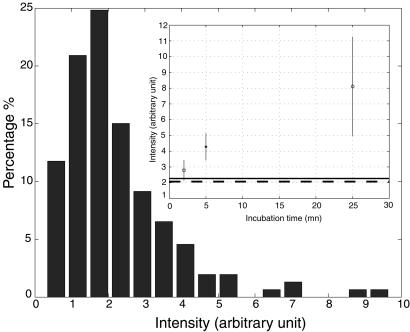
In these experiments a large excess of T7 RNAP vs. DNA was used. T7 RNAP is able to restart from a free promoter to form a new nucleo-protein complex; thus several initiations can take place at the same promoter. To prevent this reinitiation, we needed to stabilize one T7 RNAP per promoter before the addition of NTPs. Unfortunately, T7 RNAP on linear DNA is not stable on the promoter (30, 33). To bypass this property of T7 RNAP, we stabilized the enzyme in an abortive initiation complex by synthesizing a poly(G) RNA chain (25, 34). We added GTPs together with the RNAP and then rinsed the cell volume extensively. That time the washing solution contained GTPs as substrate for the enzyme. Thus only proteins bound to a promoter stayed in the cell. We then injected the classical transcription buffer, used in the previous experiments, to initiate transcription and label the RNA. The results of the transcription are given in Fig. 4c. The transcription pattern is similar to that observed in Fig. 4a even though the intensity of bright dots is less intense. We analyzed the total intensity of each dot for both single initiation and multiple initiation experiments (with three different incubation times, 2, 5, and 25 min). The mean intensity and the SD for these experiments are plotted in Fig. 6. Measurements of the fluorescence intensity of 9-kb labeled RNAs (see calibration in Materials and Methods) are also reported. This intensity scales with the intensity of the single initiation experiment, suggesting that the mean size of the RNAs is close to 9,000 bases. For multiple initiation experiments, we observed an increase of the intensity of the bright dots with the time of incubation. This increase seems to be nonlinear, which suggests a loss of activity of transcription with time. But a clear interpretation of the intensity is rendered difficult because of the quenching or bleaching of the dyes.
Figure 6.
Histogram of intensity of the bright dots observed on combed DNA for the single initiation experiment. (Inset) Mean value and SD of the intensity of the bright dots observed on combed DNA for the multiple initiations experiments. The dashed line and plain line represent the average intensity of, respectively, the single initiation experiment and a 9-kb labeled RNA. Acquisition time: 500 ms, numbers of analyzed dots 100–150 for each value.
Discussion
Physical mapping of the human genome by in situ hybridization was an important application of molecular combing (18, 19). Here we show that we can extend the combing process to image protein–DNA interactions. Molecular combing is well characterized even if the exact physical processes involved are unknown. The capillary forces exerted by the air/water receding meniscus lead to an overstretching of DNA molecules. Previous studies of elastic measurements on a single DNA molecule yield a well-defined force/extension curve (11, 12, 14). For a molecule having a relative extension of 150% this curve gives an estimation of the applied force close to 65 pN. The situation of such overstretched DNA is in the “plateau” transition between the B form and the S form (11, 35). This finding implies that domains of both B form and S form coexist along overstretched DNA. At the beginning of these studies we inferred that the presence of S-form domains along DNA could suppress enzymatic activity. Therefore, control of DNA elongation appears to be important. Previous studies (24) demonstrated that the reduction of the DNA molecule extension could be achieved by decreasing the surface hydrophobicity. In our hands this procedure led to a drastic loss of density of combed DNA (data not shown). Thus we changed the air/water interface rather than the water/solid interface to keep a high density of DNA fixation. Our experiments demonstrate that the decrease of the air/water surface tension, by the addition of an alcohol monolayer, leads to a decrease in molecular extension.
As DNA is dry behind the meniscus, it has to be rehydrated to interact with the protein. After addition of the solution, the combed molecules remain strongly bound to the silanated surface. We have shown that by using PMMA surfaces rather than silanated surfaces there were much fewer attachment points with the surface. The distance between two attachment points was observed as being larger than 1 μm. Combed DNA between two attachment points is not in contact with the surface and fully hydrated. This characteristic was demonstrated by the observation of cleaved DNA retraction to its remaining attachment points (Fig. 3). This retraction is caused by the entropic elasticity of the DNA. Several factors can explain these different behaviors between silanated and PMMA surfaces. The first factor could be the difference in surface hydrophobicity. PMMA surfaces are less hydrophobic than silanated surfaces, which can induce a decreased affinity with denatured parts of DNA. Thus, the number of contact points is closely related to the nature of the surface. Moreover, the combing process on PMMA was optimized at pH 6.6, higher than for silanated surfaces (pH 5.5). We know that there are more denaturation bubbles along the DNA molecules at low pH. Because these bubbles are likely to participate to the anchoring of the DNA onto the surface, this could explain why the number of attachment points is much higher for silanated surfaces than for PMMA surfaces.
We observed transcription on combed DNA at an extension close to the full contour length, in contrast to what is observed on overstretched DNA. The dots strings are 7–8 μm long, scaling with the transcription region located before the terminator on the T7 DNA. Why doesn't transcription occur on overstretched DNA? Basically three successive steps are needed for a specific transcription with T7 RNAP: the recognition of the promoter by the enzyme, the initiation of the transcription, and the escape from the promoter to an elongation complex (25). The use of combed DNA can imply a non-negligible influence of the surface on this process. Activity of DNase I on overstretched DNA permits us to conclude that the DNA is still accessible to nonspecific interactions. The prevention of activity of DRAI endonuclease on overstretched DNA strongly supports the idea that the state of the DNA is crucial for specific interactions to take place. A reported case on λ DNA and EcoRI gives more complicated results (because the authors changed several parameters before digestion) but leads to the conclusion that extension and conformation of straight DNA influence the efficiency of specific endonucleases cutting (21). Numerical simulations predict major structural modifications of the DNA under high tension (65 pN), both for bases stacking and the deformation of the grooves (36). Another model of overstretched DNA was proposed by Bloomfield and coworkers (37). They proposed that overstretched DNA consists of the coexistence of the single-stranded DNA and double-stranded DNA region. T7 RNAP recognizes T7 promoters by means of specific interactions in the major groove of B-DNA (38, 39), so strong deformation or a single-strand state of this region could prevent specific DNA sequence recognition. That is probably the main reason transcription is hindered on overstretched DNA and RNA synthesis is seldom observed. But we cannot rule out the hypothesis that the overstretching prevents the processivity of the enzyme rather than the initiation. Further experiments with fluorescent-labeled enzymes will shed light on this point.
We have shown that transcription can occur on combed DNA in nonoverstretching conditions. However, a question remains on the state of the ternary complex RNAP–DNA–RNA at the end of the experiment. In classical bulk experiments a chase leads to the restart of the paused transcription elongation complex (25). The chase experiment on combed DNA, performed by addition of unlabeled NTPs, leads to an unchanged transcription pattern. Therefore the ternary complex is no longer active because labeled RNA is not released in solution as expected. We propose that complexes stop and accumulate on the DNA attachment points as pictured in Fig. 7 as suggested by the increased intensity of the fluorescent dots of RNA observed when multiple transcription initiations are allowed. Alternatively, the observed RNA could be bound to the surface after dissociation with the RNAP.
Figure 7.
Sketch of the accumulation of elongation complexes at attachment points on combed DNA on PMMA cover slide.
Finally, we overcame the difficulty of having several RNAPs initiating per promoter during the experiments. In the presence of GTPs as a sole substrate the T7 RNAP can be stabilized on the promoter while transcribing poly(G) RNA. This is caused by the slippage of the enzyme at the promoters that initiate with a sequence + 1 CCC (25, 34). It is therefore possible to have one enzyme per promoter before the addition of the NTPs. The subsequent addition of NTPs drives the initiation complex into an elongation complex. Thus a characteristic specificity of the T7 transcription system is preserved on combed DNA. In addition, being able to observe one RNAP per promoter opens the way for future investigations with a single protein on a DNA bearing one promoter. This configuration could rule out confusion about the numbers of RNAP per dots.
We definitively think that combed DNA will be a very powerful assay to image RNAP–DNA interactions. Moreover, the use of evanescent wave excitation should give access to the processivity of the enzymes in real time. Likewise, enzymes cross-linked with dyes or beads could be followed with a high spatial and temporal precision. In that case, we can have access to data such as rate of transcription, formation of paused complexes, efficiency of initiation, and termination using wild-type enzyme or mutants. This method also can be extended to other proteins interacting with DNA.
Conclusions
We demonstrate that transcription is possible on combed DNA when DNA stretching is reduced. Furthermore, the T7 RNAP keeps its biochemical specificity similarly to bulk experiments: recognition of the promoter and stabilization on the promoter during poly(G) synthesis. One major advantage of molecular combing for visualization of protein–DNA interactions is that several events can be observed in the microscope field at the same time. Combed DNA is therefore a valuable substrate for the study of enzyme–DNA interactions, especially when the interactions imply a movement of the enzyme along the DNA as it is for RNAP. The main disadvantage of this method is probably the lack of controlled number of DNA attachment points on the surface, which can hinder the enzymes' processivity. We have shown, however, that we were able to determine this parameter.
Supplementary Material
Acknowledgments
We thank G. Brun and E. Goillot for biological lab facilities; W. T. McAllister, H. Buc, and M. Buckle for sharing their knowledge on RNAP; F. Vittoz, T. Lamonerie, J. Bechhoefer, and J. C. Geminard for helpful discussions; B. Sclavi for critical reading of the manuscript; and P. Oswald for his encouragement. C.P. and Z.G. were supported, respectively, by a grant from the Fondation pour La Recherche Médicale and a grant from the Ministère de l'Education Nationale, de la Recherche et de la Technologie. We also thank Centre National de la Recherche Scientifique-Nano-objet Individuel and Action Thématique et Incitative sur Programme et Equipe for financial support.
Abbreviations
- RNAP
RNA polymerase
- NTP
nucleoside triphosphate
- PMMA
polymethylmetacrylate
- CCD
cooled charge-coupled
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Kornberg A, Baker T. DNA Replication. New York: Freeman; 1992. pp. 231–259. [Google Scholar]
- 2.Upstain S M, Kane C M, Chamberlin M J. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 3.Gelles J, Landick R. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 4.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson S A, Glass R E, Shimamoto N. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 5.Yin H, Wang M D, Svoboda K, Landick R, Block S M, Gelles J. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 6.Wang M D, Schnitzer M J, Yin H, Landick R, Gelles J, Block S M. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 7.Davenport R J, Wuite G J, Landick R, Bustamante C. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 8.Guthold M, Zhu X, Rivetti C, Yang G, Thomson N H, Kasas S, Hansma H G, Smith B, Hansma P K, Bustamante C. Biophys J. 1999;77:2284–2294. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada Y, Funatsu T, Murakami K, Nonoyama Y, Ishihama A, Yanagida T. Biophys J. 1999;76:709–715. doi: 10.1016/S0006-3495(99)77237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang M D, Yin H, Landick R, Gelles J, Block S M. Biophys J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cluzel P, Lebrun A, Heller C, Lavery R, Viovy J L, Chatenay D, Caron F. Science. 1996;271:792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- 12.Bustamante C, Smith S B, Cui Y. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 13.Smith S B, Finzi L, Bustamante C. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 14.Strick T R, Allemand J F, Bensimon D, Bensimon A, Croquette V. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 15.Perkins T T, Quake S R, Smith D E, Chu S. Science. 1994;264:822–826. doi: 10.1126/science.8171336. [DOI] [PubMed] [Google Scholar]
- 16.Washizu M, Kurosawa O, Arai I, Suzuki S, Shimamoto N. IEEE Trans Ind Appl. 1994;31:447–456. [Google Scholar]
- 17.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Science. 1994;265:2096–2098. doi: 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 18.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann J S, Bensimon A. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 19.Herrick J, Michalet X, Conti C, Schurra C, Bensimon A. Proc Natl Acad Sci USA. 2000;97:222–227. doi: 10.1073/pnas.97.1.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng X, Benson K, Chada K, Huff E J, Schwartz D C. Nat Genet. 1995;9:432–438. doi: 10.1038/ng0495-432. [DOI] [PubMed] [Google Scholar]
- 21.Yokota H, Johnson F, Lu H, Robinson R M, Belu A M, Garrison M D, Ratner B D, Trask B J, Miller D L. Nucleic Acids Res. 1997;25:1064–1070. doi: 10.1093/nar/25.5.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrick J, Bensimon A. Biochimie. 1999;81:859–871. doi: 10.1016/s0300-9084(99)00210-2. [DOI] [PubMed] [Google Scholar]
- 23.Herrick J, Stanislawski P, Hyrien O, Bensimon A. J Mol Biol. 2000;300:1133–1142. doi: 10.1006/jmbi.2000.3930. [DOI] [PubMed] [Google Scholar]
- 24.Bensimon D, Simon A J, Croquette V, Bensimon A. Phys Rev Lett. 1995;74:4754–4757. doi: 10.1103/PhysRevLett.74.4754. [DOI] [PubMed] [Google Scholar]
- 25.McAllister W T. In: Mechanisms of Transcription. Eckstein F, Lilley D M J, editors. Berlin: Springer; 1997. pp. 15–26. [Google Scholar]
- 26.Dunn J J, Studier F W. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 27.Allemand J F, Bensimon D, Jullien L, Bensimon A, Croquette V. Biophys J. 1997;73:2064–2070. doi: 10.1016/S0006-3495(97)78236-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berge B, Renault A. Europhys Lett. 1993;21:773–777. [Google Scholar]
- 29.He B, Rong M, Lyakhov D, Garteistein H, Diaz G, Castagna R, McAllister W T, Durbin R K. Protein Expression Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 30.Place C, Oddos J, Buc H, McAllister W T, Buckle M. Biochemistry. 1999;38:4948–4957. doi: 10.1021/bi982689e. [DOI] [PubMed] [Google Scholar]
- 31.Akerman B, Tuite E. Nucleic Acids Res. 1996;24:1080–1090. doi: 10.1093/nar/24.6.1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz S C, Lahm A, Oefner C. Nature (London) 1998;322:464–468. [Google Scholar]
- 33.Villemain J, Guajardo R, Sousa R. J Mol Biol. 1997;273:958–977. doi: 10.1006/jmbi.1997.1358. [DOI] [PubMed] [Google Scholar]
- 34.Martin C T, Muller D K, Coleman J E. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- 35.Marko J F. Phys Rev E. 1998;57:2134–2149. [Google Scholar]
- 36.Lebrun A, Lavery R. Nucleic Acids Res. 1996;24:2260–2267. doi: 10.1093/nar/24.12.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams M C, Wenner J R, Rouzina I, Bloomfield V A. Biophys J. 2001;80:874–881. doi: 10.1016/S0006-3495(01)76066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cheetham G M, Jeruzalmi D, Steitz T A. Nature (London) 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 39.Li T, Ho H H, Maslak M, Schick C, Martin C T. Biochemistry. 1996;35:3722–3727. doi: 10.1021/bi9524373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.