Abstract
Precision medicine has become a cornerstone in modern therapeutic strategies, with nucleic acid aptamers emerging as pivotal tools due to their unique properties. These oligonucleotide fragments, selected through the Systematic Evolution of Ligands by Exponential Enrichment process, exhibit high affinity and specificity toward their targets, such as DNA, RNA, proteins, and other biomolecules. Nucleic acid aptamers offer significant advantages over traditional therapeutic agents, including superior biological stability, minimal immunogenicity, and the capacity for universal chemical modifications that enhance their in vivo performance and targeting precision. In the realm of osseous tissue repair and regeneration, a complex physiological process essential for maintaining skeletal integrity, aptamers have shown remarkable potential in influencing molecular pathways crucial for bone regeneration, promoting osteogenic differentiation and supporting osteoblast survival. By engineering aptamers to regulate inflammatory responses and facilitate the proliferation and differentiation of fibroblasts, these oligonucleotides can be integrated into advanced drug delivery systems, significantly improving bone repair efficacy while minimizing adverse effects. Aptamer-mediated strategies, including the use of siRNA and miRNA mimics or inhibitors, have shown efficacy in enhancing bone mass and microstructure. These approaches hold transformative potential for treating a range of orthopedic conditions like osteoporosis, osteosarcoma, and osteoarthritis. This review synthesizes the molecular mechanisms and biological roles of aptamers in orthopedic diseases, emphasizing their potential to drive innovative and effective therapeutic interventions.
Subject terms: Bone, Bone cancer
Introduction
In the pursuit of novel and efficacious therapeutic strategies for disease treatment, scientific researchers are continuously striving for innovation. In recent years, precision medicine has emerged as a pivotal approach in the management of numerous diseases. Notably, nucleic acid aptamer, a closely associated technology, has attracted widespread attention. These aptamers are oligonucleotide fragments derived from the systematic evolution of ligands by exponential enrichment (SELEX) technology, characterized by their high affinity and specificity towards target molecules.1,2 They can be either DNA or RNA sequences, with some studies also exploring nucleic acid analogs and peptides. Aptamers possess notable advantages, including considerable biological stability, potential for universal chemical modification, low immunogenicity, and rapid tissue penetration.3 In recent years, significant advancements have been achieved in targeted gene therapy research. Current research focuses on delivering therapeutic genes to target cells via specific antibodies or ligands, along with the use of disease-specific gene promoters to tightly regulate gene expression within the disease microenvironment.3,4 Although viral vectors demonstrate high efficacy in gene delivery, concerns regarding their safety, immunogenicity, and manufacturing complexities restrict their advancement in clinical applications. In contrast, nonviral gene delivery systems exhibit considerable potential owing to their reduced immunogenicity and simpler manufacturing processes.5 Nonviral systems possess a longer shelf-life, lower production costs, and reduced batch-to-batch variability. Moreover, their physicochemical properties and target specificity can be tailored by modifying the nucleic acid composition. They are readily amenable to chemical synthesis and modification to enhance enzyme resistance and in vivo pharmacokinetics.6 The incorporation of aptamers into nonviral vectors or their combination with other nonviral vectors can improve the targeting, cellular uptake efficiency, and endosomal escape capability of nucleic acid therapeutics. For example, aptamers can be modified or conjugated with other materials to facilitate improved loading and protection of nucleic acid drugs, thereby enabling more precise delivery to specific cells or tissue sites and achieving enhanced therapeutic efficacy.6–8 This review aims to summarize and elucidate the molecular mechanisms and biological roles of nucleic acid aptamers in orthopedic diseases, proposing innovative and effective strategies for future medical interventions.
The research history and advantages of nucleic acid aptamers
The development of SELEX technology in 1990 marked the emergence of nucleic acid aptamers.2,9 Subsequent discoveries included RNA aptamers with catalytic activity in vitro and microRNAs (miRNAs).10–13 From the mid-1990s to the early 21st century, research primarily focused on developing aptamers for small molecule sensing applications.14–16 A significant advancement occurred in 2001 with the development of aptamer-based protein labeling and detection techniques.17,18 The introduction of Cell-SELEX in 2003 was followed by the successful development of the first aptamer-based drug.19–21 Further innovations emerged in 2006 with non-covalently coupled aptamer-DOX conjugates and the establishment of aptamer-based optical and electrochemical sensing platforms during the mid-2000s.15,16,22 The concept of liquid biopsy gained prominence around 2010, leading to preliminary research on circulating tumor cell release and separation methods using nucleic acid aptamers.23–25 Investigations into miRNA detection methods utilizing aptamers commenced in 2015.26 During the SARS-CoV-2 pandemic, researchers successfully isolated DNA aptamers targeting the viral spike protein.27–31 Recent advancements have focused on developing aptamer-drug conjugates for cancer therapy and diagnostic methods for periprosthetic joint infection (PJI).32–35
Nucleic acid aptamers demonstrate enhanced stability through their secondary structures, including stem-loops, hairpins, and G-quadruplexes, with base modifications and stable phosphate backbones contributing significantly to their structural integrity.36–38 Protein binding further protects aptamers from external influences.39 Environmental factors, particularly ion concentration and pH levels, significantly influence aptamer stability.9 Technological advancements have introduced nucleotide analogs to enhance binding affinity and nanotechnology applications for co-delivery with therapeutic molecules.7,40–43 Chemical modifications prevent enzymatic degradation and alter binding properties. Optimization of connection modes, length, and density is essential for developing functional aptamer conjugates.38,42–48 Multivalent aptamers improve detection sensitivity, while pre-structured DNA libraries expand screening diversity.7,49–51 The formation of aptamer-drug complexes through therapeutic agent conjugation improves drug targeting and efficacy.52–56 Advanced technologies, including microfluidic technology and high-throughput sequencing, have optimized separation efficiency and the SELEX process.57–59
The three-dimensional (3D) conformation of an aptamer, derived from its secondary structure, is pivotal in determining its binding affinity and specificity to homologous targets.60 This structural diversity endows aptamers with the remarkable ability to recognize a wide array of target molecules, including proteins, RNA, DNA, and small molecules, with exceptional specificity and affinity.60–62 The functionality of aptamer-target complexes is underpinned by a network of intramolecular interactions, such as hydrogen bonds, hydrophobic interactions, van der Waals forces, and complementary base pairing.62 These interactions synergistically facilitate the precise recognition and binding of aptamers to their targets.63,64 SELEX, a widely utilized method for aptamer screening, has found extensive applications, particularly in the biomedical domain, where it is instrumental in selecting aptamers against proteins, tumor markers, and other biomolecules (Fig. 1a). The aptamers are invaluable in diagnostics, therapeutics, and research due to their high affinity and specificity. Furthermore, SELEX can be refined through various iterations, such as Cell-SELEX (Fig. 1b) and in vivo SELEX, which are tailored for the selection of aptamers in a cell-specific context and within living organisms, respectively.65,66 The counter-SELEX strategy introduces a negative selection step to exclude aptamers that bind to non-target molecules, thereby enhancing specificity by distinguishing between highly similar structures.66,67 Beyond SELEX, other methodologies can also be employed to augment the specificity and affinity of aptamers. Competitive binding assays, for instance, allow for the assessment of the binding affinity between an aptamer and its target, enabling the optimization of experimental conditions to improve aptamer performance.68,69 This approach is particularly useful in selecting aptamers that can effectively compete for target binding sites, thereby enhancing their efficacy in practical applications. In recent years, advancements in separation techniques, such as fluorescence-activated cell sorting (FACS), capillary electrophoresis, solid-phase, and chip-based microfluidic methods, have emerged to refine the affinity and specificity of candidate aptamers.70,71 The field of aptamer screening has witnessed the advent of innovative techniques aimed at overcoming the limitations of traditional methods. Particle display technology transforms solution-phase aptamers into aptamer particles, enabling quantitative screening based on affinity through FACS. It effectively separates high-affinity aptamers in fewer selection rounds, minimizing selection bias and human error.72,73 Multiparameter particle display further refines this process by employing dual-color FACS to simultaneously evaluate affinity and specificity, facilitating the identification of high-performance aptamers even in complex environments like serum, thus mitigating the risk of discarding potentially valuable aptamers.73,74 Click chemistry particle display integrates click chemistry with particle display, simplifying the generation and high-throughput screening of “non-natural” aptamers with extensive base modifications. This innovation addresses the chemical diversity limitations of natural nucleic acid aptamers, opening new avenues for aptamer selection against challenging targets.73,75 Collectively, these advancements have significantly propelled the development and application of aptamer technology.
Fig. 1.
The SELEX process diagram. a Schematic representation of SELEX process. b Schematic representation of cell-SELEX process
The applications of nucleic acid aptamers in bone-related diseases
Bone repair and healing constitute a complex and highly coordinated physiological process aimed at restoring or maintaining osseous function.76 The biological importance of this process encompasses several aspects. Primarily, bones provide structural support and protection for internal organs, making bone repair essential for maintaining anatomical integrity. Investigation of the cellular and molecular mechanisms involved in this process enhances understanding of general tissue repair and regeneration processes.76 With global population aging, the escalating risk of osteoporosis-related fractures underscores the significance of optimizing bone healing to improve geriatric quality of life. Current treatments frequently struggle to completely restore pre-fracture biomechanical properties, particularly in complex open fractures or elderly patients.77 Factors such as chronic inflammation, diabetes, vitamin deficiency, aging, and polytrauma can impede healing progression.78 Although various technologies and pharmacological agents have been developed to facilitate bone repair and healing, clinical validation remains insufficient for many applications.79 Moreover, bone grafting and substitute materials face challenges such as immunogenicity, infection risks, and donor source limitations.80
Aptamers exert crucial regulatory effects on bone repair through modulation of key molecular pathways, including parathyroid hormone (PTH), bone morphogenetic proteins (BMPs), and the Wnt/β-catenin signaling pathway.81–83 PTH promotes osteogenic differentiation by reducing osteoblast apoptosis and increasing the number of osteoblasts.84 Under specific conditions, PTH activates bone lining cell transformation into active osteoblasts.85–89 Besides, PTH expands osteoprogenitor cell reservoirs in bone marrow and direct early osteoblast lineage cells away from the adipocyte lineage, thereby promoting their differentiation into osteoblasts.90,91 On the other hand, PTH stimulates osteoclastogenesis through non-cell-autonomous mechanisms, enhancing bone resorption to liberate growth factors from the bone matrix that subsequently promote osteoblast migration, differentiation, and functional activation.92,93 BMPs synergize with angiogenic factors (eg, VEGF) and metallic cofactors to induce mesenchymal stem cells (MSCs) osteoblastic differentiation while coordinating osteogenesis and angiogenesis during repair.94–102
The Wnt signaling pathway can determine the differentiation of mesenchymal precursor cells into osteoblasts or chondrocytes, with β-catenin serving as a vital molecular switch.103–105 Concurrently, the expression of key components within the Wnt signaling pathway is regulated during osteoblast differentiation, influencing cellular proliferation, apoptosis, and functional maturation.81 Furthermore, the Wnt signaling pathway promotes the differentiation of osteoblasts by inhibiting adipogenic transcription factors CCAAT/enhancer binding protein alpha and peroxisome proliferator-activated receptor-γ (PPARγ).106,107 Osteoprotegerin (OPG), a soluble decoy receptor, competitively inhibits receptor activator of nuclear factor-κB ligand (RANKL)–RANK interaction by binding RANKL trimers, thereby blocking osteoclast differentiation and inducing apoptosis.108 RANKL, a tumor necrosis factor (TNF) superfamily member upregulated in postmenopausal women, can be primarily modulated by estrogen supplementation.109 It stimulates osteoclastogenesis while suppressing osteoclast apoptosis. OPG competes with RANK for binding to RANKL, thus preventing the generation and maturation of osteoclasts induced by RANKL.110 Therefore, the OPG/RANKL ratio is intimately associated with osteoclastogenesis, ultimately influencing bone mineral density and mechanical strength. In β-catenin-deficient osteoblasts, RANKL expression is upregulated and OPG expression is downregulated, whereas adenomatous polyposis coli (Apc)-deficient osteoblasts demonstrate inverse regulation.111,112 This regulatory network enables Wnt signaling to indirectly govern osteoclast differentiation through osteoblast-mediated mechanisms, thus coordinating bone resorption during repair processes.113
Fracture
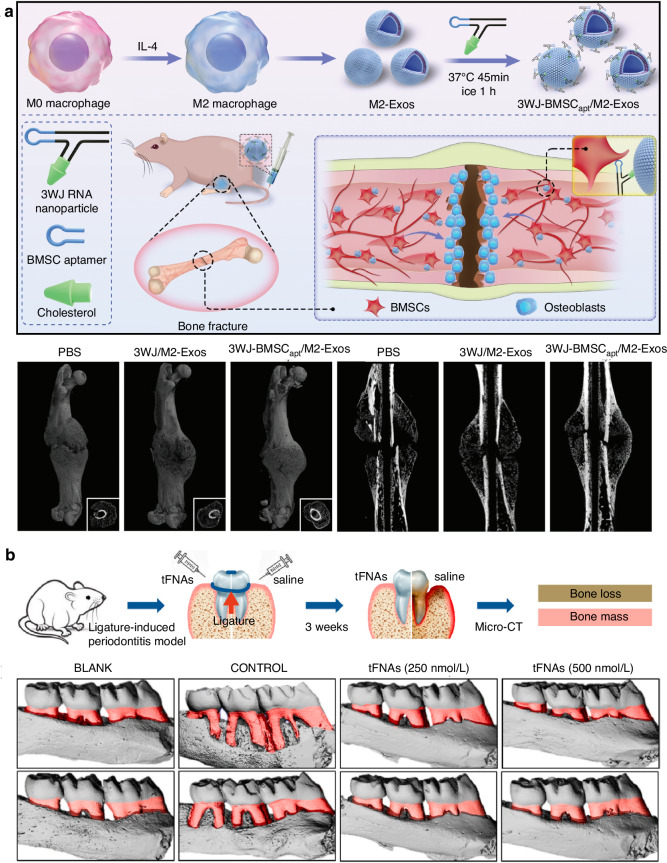
Fractures refer to pathological conditions characterized by the disruption of bone continuity or integrity due to external forces. These injuries can occur in any bone. The etiology of fractures is multifactorial, including direct or indirect external forces, chronic repetitive stress, and osteoporosis secondary to diseases or medications. Clinically, the healing process of fractures is typically divided into three stages: hematoma formation, callus formation, and callus remodeling. The injury results in the bone breaking into two or more fragments, accompanied by damage to the surrounding periosteum, blood vessels, and other tissues. Extravasation from bone marrow and vasculature leads to the formation of a localized hematoma containing bone-derived and immune cells.114 Immediately following a fracture, the inflammatory response is activated, serving as a critical initiator of the bone healing process. The inflammatory period is a crucial stage characterized by hypoxia, impaired perfusion, and the migration of various growth factors.115 The fracture hematoma is essential for effective bone regeneration due to its high osteogenic potential.116,117 This characteristic was initially observed in non-specialized bone progenitor cells, now identified as MSCs.118,119 MSCs are multipotent stem cells that can be isolated from many sources in both animals and humans, such as bone marrow, adipose tissue, amniotic fluid, and periosteum. Bone marrow contains the highest abundance of these cells, which are collectively termed bone marrow mesenchymal stromal cells (BMSCs).120,121 These cells possess the potential for osteogenic, chondrogenic, and adipogenic differentiation.122 Aptamer-loaded delivery systems can stimulate the osteogenic differentiation of these stem cells, thereby promoting the repair and regeneration of bone tissue.123 The fracture hematoma also contains platelets, macrophages, and other cell types.124 Cytokines triggering coagulation within the hematoma simultaneously activate local phagocytic cells, facilitating debris clearance at the fracture site.125 Successful fracture healing demands coordinated interactions between macrophages and BMSCs. During the process of bone repair, the polarization of macrophages plays a key role in regulating the differentiation of BMSCs. M1 macrophages predominantly mediate pro-inflammatory responses, while M2 macrophages promote tissue repair by enhancing BMSC osteogenesis.126 Meanwhile, M2 macrophages are increasingly recognized as positive regulatory factors for fracture healing.127 Macrophages facilitate fracture healing primarily through paracrine mechanisms mediated by exosomes.128,129 Exosomes derived from M2 macrophages (M2-Exos) serve as key paracrine effectors, driving BMSC osteogenic differentiation.128,130,131 Nevertheless, current therapeutic applications of exosomes are limited by nonspecific delivery and rapid clearance by the reticuloendothelial system.132–134 Shou et al.132 utilized interleukin (IL)-4 to induce the differentiation of the murine macrophage cell line RAW264.7 into M2 macrophages, isolated exosomes (M2-Exos) from osteoblasts, and characterized these exosomes using transmission electron microscopy, nanoparticle tracking analysis, and Western blot. After identifying a BMSC-specific aptamer sequence validated by flow cytometry and confocal microscopy, they engineered 3WJ RNA nanoparticles with three functional components: a3wj-Cholesterol, b3wj-BMSC aptamer, and c3wj-Alexa 647 (Table 1). Chain a was conjugated with cholesterol for anchoring to the exosome membrane, chain b was coupled with the BMSC aptamer serving as a targeting ligand, and chain c was labeled with Alexa 647 for imaging purposes. Chains were mixed at a 1:1:1 molar ratio and self-assembled via gradual cooling from 95 °C to 4 °C at 0.1 °C/s. The resulting 3WJ-BMSCapt/M2-Exos system combines BMSC-targeting aptamers with M2-Exos through 3WJ RNA nanoparticles. This functionalized exosome platform demonstrated precise BMSC targeting in vitro and significant fracture site accumulation in vivo, accelerating bone regeneration in a murine femoral fracture model (Fig. 2a). This cell-free strategy offers a novel therapeutic approach, with potential applications in clinically targeted drug delivery. The acute inflammatory response peaks within 24 h and resolves by day 7.135 Concurrently, the hematoma transitions to granulation tissue by about day 7.114 As inflammation subsides, the repair phase initiates with the formation of granulation tissue.124 With the emergence of MSCs and their differentiation into fibroblasts, the hematoma tissue gradually organizes and forms fibrous tissue,136,137 known as fibrous callus, which initially bridges the fracture ends.
Table 1.
Summary of the application of relevant aptamers in orthopedic diseases
| Aptamer | Material/Drug | Molecular target | Function | Diseases/Therapeutic effect | Research progress | Ref. |
|---|---|---|---|---|---|---|
|
3WJ-BMSCapt/M2-Exos a3wj-Cholesterol b3wj-BMSCaptamer c3wj-Alexa 647 |
Drug | BMSC | Modification of M2-Exos enhances their targeting to BMSCs | In vitro, targeting BMSCs more effectively enhances their proliferation, migration, and osteogenic differentiationIn vivo, it stimulates callus formation, increases bone volume/total volume (BV/TV), bone mineral density (BMD), and trabecular number (Tb. N), reduces trabecular separation (Tb. Sp), promotes new bone formation, and elevates the expression of osteogenic markers ALP and OCN | Under development | 132 |
|
Apt Apt01 Apt02 |
Material | VEGF receptor | They exhibit high affinity for VEGFR-1 and VEGFR-2 | Simulating the function of VEGF-A, it has the potential to activate VEGR receptors and promote angiogenesis | Under development | 159 |
| Aptamer-agomiR-195 | Drug | Endothelial cell | It specifically targets endothelial cells and increases miR-195 levels in cells | Increasing the number of CD31hiEmcnhi vessels in aged murines stimulates bone formation and improves bone strength | Under development | 297 |
| aptamer-antagomiR-188 | Drug | BMSC | It regulates the expression of miR-188 in BMSCs | Reduce miR-188 levels in BMSCs to increase trabecular volume, quantity, and cortical bone thickness; decrease trabecular separation and intimal circumference; enhance bone strength; elevate the bone formation rate; reduce the number and area of adipocytes in the bone marrow; and increase the number and surface area of osteoblasts on the trabecular and intimal bone surfaces, thereby preventing bone loss and fat accumulation in the bone marrow of elderly murines | Under development | 298 |
| OS-7.9 | Material | MG-63 | It shows high affinity and specific binding to MG-63 cells, as well as recognition of lung and colon colorectal adenocarcinoma cell lines | It enables early diagnosis and targeted treatment of osteosarcoma and metastatic disease | Under development | 317 |
| SL1067 | Material | IL-1α | It demonstrates high affinity and specific recognition of IL-1α | Inhibit the IL-1α signaling pathway to treat related inflammatory reactions and diseases | Under development | 272 |
| VR11 | Material | TNFα | It exhibits high affinity binding and specific recognition of TNFα, inhibiting its receptor binding and blocking TNFα-induced cytotoxicity and NO production | Treat inflammatory diseases and avoid immune reactions | Under development | 273 |
| 8A-35 | Drug | IL-8 | It shows high affinity binding to IL-8, inhibiting IL-8 receptor binding, regulating the IL-8-induced intracellular signaling pathway, and inhibiting neutrophil migration | Potential anti-cancer effects in treating inflammatory diseases | Under development | 274 |
|
Apc001PE aptscl56 |
Drug | The loop3 region of sclerostin | It inhibits the antagonistic effect of sclerostin on Wnt signaling pathway and osteogenic potential | It promotes bone formation without increasing cardiovascular risk | Under development | 507 |
Fig. 2.
Evaluation of bone repair based on nucleic acid aptamer intervention. a Overview of 3WJ-BMSCapt/M2-Exos in fracture model. Representative 3D reconstruction images, 2D cross-sectional and sagittal images from micro-CT scanning of the murine femur fracture model. PBS: phosphate buffered saline. Reproduced form ref. 132 with permission from Oxford University Press, copyright 2023. b tFNAs significantly protected the alveolar bone from periodontitis in vivo. Schematic representation of the rat periodontitis experiment. The micro-CT 3D reconstruction images of the left maxillary alveolar bone. The red area indicates the exposure of the root. Reproduced form ref. 190 with permission from KeAi Communications Co, copyright 2020
The primary hematoma undergoes reorganization into granulation tissue, thereby initiating endochondral ossification. This process generates a cartilaginous callus in the endosteum, intramedullary region, and periosteum. Subsequently, the cartilaginous callus is replaced by a hard callus.138 Endochondral ossification occurs both between fracture ends and externally in the periosteal region. Proliferating osteoblasts produce ossified tissue, forming new bones. These zones remain mechanically unstable until cartilaginous tissue forms a soft callus that stabilizes the fracture structure.139 Intramembranous ossification beneath the periosteum generates new bone adjacent to the fracture ends, contributing to hard callus formation. The bridging of this central hard callus ultimately provides a semi-rigid structure capable of weight-bearing.140 The osteoblast-specific aptamer CH6, identified via Cell-SELEX screening, enables the preparation of functionalized lipid nanoparticles encapsulating osteoblast-targeted Plekho1 siRNA (CH6-LNPs-siRNA). Targeted delivery of this siRNA to bone formation surfaces enhances osteogenesis.67,141–143 CH6 exhibits high-affinity binding to osteoblasts with minimal interaction with rat osteoclasts, human osteoclasts, or hepatocytes. CH6-LNPs-siRNA possesses excellent in vitro osteoblast selectivity, promoting siRNA uptake while significantly reducing liver/kidney accumulation compared to conventional vectors.67 This system achieves osteoblast-specific siRNA delivery, minimizing the uptake of siRNA by liver cells, Kupffer cells, and peripheral blood mononuclear cells.67,144–148 As markers of osteoblast activity, alkaline phosphatase (Alp) and osteocalcin (OCN) are closely associated with bone formation.146,147 CH6-LNPs-siRNA shows enhanced gene silencing efficiency in Alp+/OCN+ cells, with Plekho1 mRNA reduction exhibiting dose-dependent and sustained effects that facilitate bone formation.67 During callus maturation, hypertrophic chondrocytes undergo extracellular matrix calcification, synergized by macrophage colony-stimulating factor (M-CSF), RANKL, OPG, and TNF-α. These factors mediate mineralized cartilage resorption, osteocyte/osteoclast recruitment, and woven bone formation.149,150 TNF-α additionally promotes MSC recruitment and initiates chondrocyte apoptosis.150 Aptamer-based delivery systems can regulate osteoclast activity by targeting specific molecules, achieving balanced bone metabolism. This system can simultaneously promote BMSC-mediated bone formation and inhibit osteoclast-mediated resorption. This approach effectively counteracts post-traumatic bone loss such as osteoporosis while improving trabecular microstructure and mechanical properties.151
Although hard callus provides biomechanical stability, complete restoration requires remodeling into lamellar bone with a central medullary cavity.150 This remodeling phase involves marrow space reconstruction, hematopoietic tissue regeneration, vascular bed normalization, and blood flow restoration to pre-injury levels.152,153 The hard callus is progressively remodeled; periosteal and endosteal calluses are absorbed, and the medullary cavity is restored.150 As discussed, aptamers targeting osteoblast differentiation factors could accelerate callus formation, while those suppressing osteoclast activity may protect bone integrity. Successful remodeling depends critically on adequate vascular supply and progressive mechanical loading.154–156 These principles are extensively validated in fracture studies. VEGF, a hypoxia-inducible angiogenic regulator, plays pivotal roles in vascular development, angiogenesis and fracture healing.157,158 Among these, VEGF-A is an essential molecule that activates and binds to VEGF receptor-1 (VEGFR-1) and VEGFR-2. Due to the limitations of recombinant VEGF-A, including low stability, significant batch-to-batch variations, and high production costs, Apt01 and Apt02 (Table 1) have been successfully isolated via SELEX. These aptamers exhibit a high affinity for VEGFR-1 and VEGFR-2, with dissociation rate constants comparable to those of VEGF-A. Notably, their binding affinity hierarchy (VEGFR-1>VEGFR-2) mirrors that of VEGF-A, suggesting DNA aptamers may functionally substitute for VEGF-A.159 In summary, aptamer-mediated precision regulation of bone repair processes represents a novel therapeutic paradigm. By orchestrating tissue regeneration pathways, this approach could advance targeted treatments for orthopedic pathologies.
Alveolar bone
Alveolar bone is the protrusion located on the lower border of the maxilla and the upper border of the mandible, surrounding the root of the tooth. Through the periodontal ligament, it is closely connected to the tooth root and plays a critical role in tooth development, eruption, and mastication processes.160 Under pathological conditions including oral inflammatory diseases, trauma, tumors, hereditary diseases, and systemic diseases, the balance between alveolar bone resorption and formation is disrupted, resulting in alveolar bone defects. These defects can cause tooth loosening and loss, trigger masticatory dysfunction, and endanger the physical and mental health of patients.161 In dental implant therapy, alveolar bone resorption following tooth extraction or defect leads to insufficient bone volume and altered alveolar ridge dimensions, thereby affecting dental implant placement and complicating treatment.162,163 Furthermore, alveolar bone defects can also impact facial appearance, masticatory function, nutrient intake, and cause counterclockwise rotation of the mandible.164–166
In the process of alveolar bone repair and reconstruction, signaling pathways including Notch, Wnt, Toll-like receptor, and NF-κB regulate vital activities such as proliferation, differentiation, apoptosis, and autophagy of osteoclasts, osteoblasts, osteocytes, periodontal ligament cells, macrophages, and adaptive immune cells. These pathways also modulate the expression of inflammatory mediators and influence the balance of the RANKL/RANK/OPG system, thereby participating in the repair and reconstruction of alveolar bone.167–176 The Notch signaling pathway functions through a complex network of pro-inflammatory cytokines and bone resorption regulators. Notably, Notch1 signaling promotes alveolar bone repair, whereas Notch2 signaling inhibits it.177,178 Notch2 upregulation coincides with increased IL-1β and IL-6 levels, creating an osteoclast-favorable environment.168 In periapical periodontitis lesions where RANKL expression exceeds OPG, Notch2, Jagged1, Hey1, and TNF-α are overexpressed and strongly correlated, collectively driving extensive alveolar bone resorption.169
In a murine periapical periodontitis model, systemic administration of the Wnt inhibitor IWR-1 via the caudal vein significantly exacerbates alveolar bone lesions. Conversely, lithium chloride (a Wnt activator) applied for root canal sealing enhances collagen type I α1 and Runx2 expression, increases CD45R-positive cells, and accelerates alveolar bone healing via bone formation and immune responses.170 Furthermore, estrogen activates the Wnt signaling pathway through estrogen receptors, stimulating osteoblast differentiation, inhibiting RANKL-induced osteoclast activity, promoting osteocyte autophagy, and preventing apoptosis, directly contributing to alveolar bone reconstruction. Additionally, estrogen indirectly regulates the metabolic homeostasis of alveolar bone by suppressing oxidative stress and inflammatory responses.171,172 TNF-α stimulates gingival MSCs to secrete exosomes rich in miRNA-1260b, which inhibits non-canonical Wnt5a and JNK signaling, downregulates RANKL, upregulates OPG, reduces osteoclastogenesis, polarizes M2 macrophages, and enhances alveolar bone repair in murine periodontitis.172 IL-6 activates canonical Wnt signaling via Wnt2b/Wnt10b and non-canonical signaling via Wnt5a, collectively inducing osteogenic differentiation of human periodontal ligament stem cells to promote alveolar bone regeneration.179
The NF-κB pathway directly influences inflammatory alveolar bone repair by regulating cytokines and mediators expression. In a ligature and Escherichia coli lipopolysaccharide induced murine periodontitis model, dimethyloxaloylglycine enhances hypoxia-inducible factor-1α expression, suppresses NF-κB phosphorylation, downregulates pro-inflammatory cytokines, upregulates anti-inflammatory cytokines, reduces the M1/M2 macrophages ratio, inhibits osteoclast differentiation, attenuates alveolar bone resorption, and increases alveolar bone volume and density.174 Intraperitoneal injection of pirfenidone inhibits the NF-κB signaling pathway in bone marrow-derived macrophages, suppressing the expression of pro-inflammatory cytokines including IL-1β, IL-6, and TNF-α induced by lipopolysaccharide, as well as inhibiting RANKL-induced osteoclastogenesis, which significantly mitigates alveolar bone loss.175 NF-κB also indirectly affects bone repair by regulating inflammasome assembly, pro-inflammatory factor maturation and pyroptosis. IL-37 inhibits NF-κB to suppress nucleotide-binding oligomerization domain-like receptor protein 3 inflammasome, reducing osteoclast numbers, calcitonin receptor/RANKL/IL-10 expression, while increasing OPG/IL-1β/IL-6/TNF-α levels, ultimately alleviating alveolar bone resorption.180,181
Periodontal ligament stem cells (PDLSCs), a subset of dental pulp stem cells, exhibit unique multi-lineage differentiation into cementum, osteoid, and periodontal ligament-like tissues. This multi-lineage differentiation potential has not been observed in other MSCs, making it a unique characteristic of PDLSCs.182 PDLSCs are ideal seed cells for true periodontal regeneration, but inflammatory microenvironments threaten this process by exacerbating tissue destruction and impairing the osteogenic differentiation and migration of PDLSCs.183,184 Tetrahedral framework nucleic acids (tFNAs), with unique biological properties, enhance proliferation and osteogenic/odontogenic differentiation of dental pulp stem cells (DPSC)/PDLSC and exert anti-inflammatory/antioxidant effects by inhibiting MAPK phosphorylation in macrophages.185–189 Thereupon, Yunfeng Lin et al.190 discovered in their research that tFNAs can suppress the release of pro-inflammatory factors in cells, along with the production of cellular reactive oxygen species (ROS), facilitating the migration and osteogenic differentiation of PDLSCs in vitro. It is indicated that tFNAs may exert a protective effect on the osteogenic differentiation of PDLSCs in inflammatory conditions. Furthermore, in the murine periodontitis model, tFNAs reduce inflammatory cell infiltration, downregulate IL-6/IL-1β, inhibit osteoclastogenesis, and protect periodontal tissues (Fig. 2b). Moreover, tFNA activates Wnt/β-catenin signaling to drive proliferation and osteogenic differentiation of PDLSCs. Since its development, tFNA functionalization strategies have been extensively explored. Professor Yunfeng Lin’s team has applied tFNA to orthopedic, metabolic diseases, sarcopenia, bladder obstruction, and cancer,191–196 establishing new frameworks for nucleic acid aptamer research. This material offers novel tools for fundamental studies on nucleic acid functions and structure-activity relationships, while holding diagnostic and therapeutic potential for previously intractable diseases.
Articular cartilage and osteochondral tissue
Articular cartilage is a type of hyaline cartilage primarily composed of proteoglycans and type II collagen within the matrix. It is categorized into deep, middle, and superficial zones.197,198 The meniscus is one of the most significant articular cartilages in the human body, playing an extremely crucial position in protecting the health and integrity of the knee joint during long-term weight-bearing activities. Cartilages like the meniscus lack the capacity to generate adequate healing responses independently.199 Previously, research on the reconstruction and regeneration of cartilages and osteochondral tissues posed considerable challenges. Yet currently, with the emergence and maturity of new technologies, many of these difficulties are gradually being conquered.
The meniscus can be divided into three different zones: the highly vascularized (red) peripheral region, the avascular (white) inner region, and the red-white transitional region, which possesses characteristics of both.200 Meniscus tears are generally recognized to heal poorly due to inadequate vascular supply and reduced cell proliferation.200–202 Vascular supply is critical for the healing of the meniscus. The superior, lateral, and medial genicular arteries, branching from the popliteal artery, provide the primary blood supply to the meniscus, forming peripheral capillary plexuses within the synovial and capsular tissues of the knee joint.202–206 In adults, vascularization is limited to the peripheral 10%–25% of the meniscus, and the extent of this vascularized area determines the healing potential of tears.202 Because of the supply of oxygen, essential factors, and nutrients from adjacent blood vessels, tears in the vascularized area can facilitate tissue healing, while injuries in the avascular region remain irreparable.207 Insufficient blood supply leads to deficiencies in nutrients, oxygen, and other critical elements, restricting the repair capacity of the meniscus’s avascular region.208 In terms of biomechanics, clinical studies have shown that cartilage damage always extends deeply into the subchondral bone, resulting in osteochondral defects in the knee joint. These defects alter joint biomechanics and impair the long-term performance of cartilage tissue,199 underscoring the vital importance of simultaneous articular cartilage and subchondral bone restoration for successful knee joint repair.
Currently, treatment strategies mainly consist of arthroscopic suture, partial or total meniscectomy, and meniscal allograft transplantation (MAT). Nevertheless, arthroscopic meniscectomy inevitably results in progressive cartilage degeneration and osteoarthritis.209 While MAT has been clinically implemented, it faces challenges such as donor scarcity, implant shrinkage, and disease transmission risks.210 Aptamers are widely used in various biomedical applications, including disease diagnosis, drug delivery, and biosensors, because of their high affinity, specificity, and stability. In meniscus research, aptamers may be utilized to recognize and target specific biomarkers of meniscus injury or degeneration, thereby facilitating early diagnosis and treatment of the condition. Moreover, nucleic acid technologies like PCR and metagenomic next-generation sequencing (mNGS) have demonstrated significant potential in pathogen identification for joint prosthesis infections, with mNGS showing 95% sensitivity and 94.7% specificity in diagnosing PJI.211–213 These findings highlight nucleic acid technologies’ diagnostic precision, further exemplified by aptamers’ ability to target and diagnose specific biomarkers.
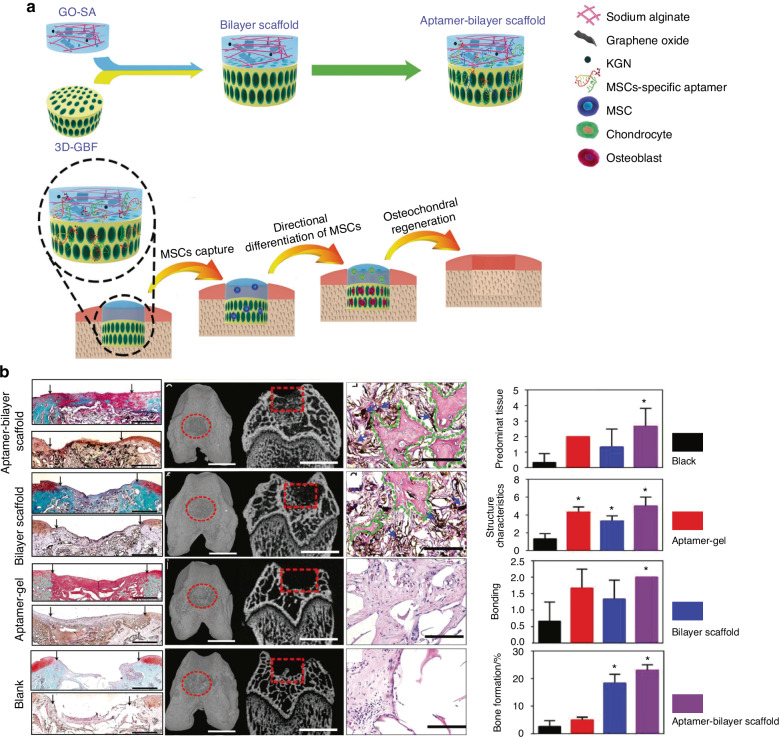
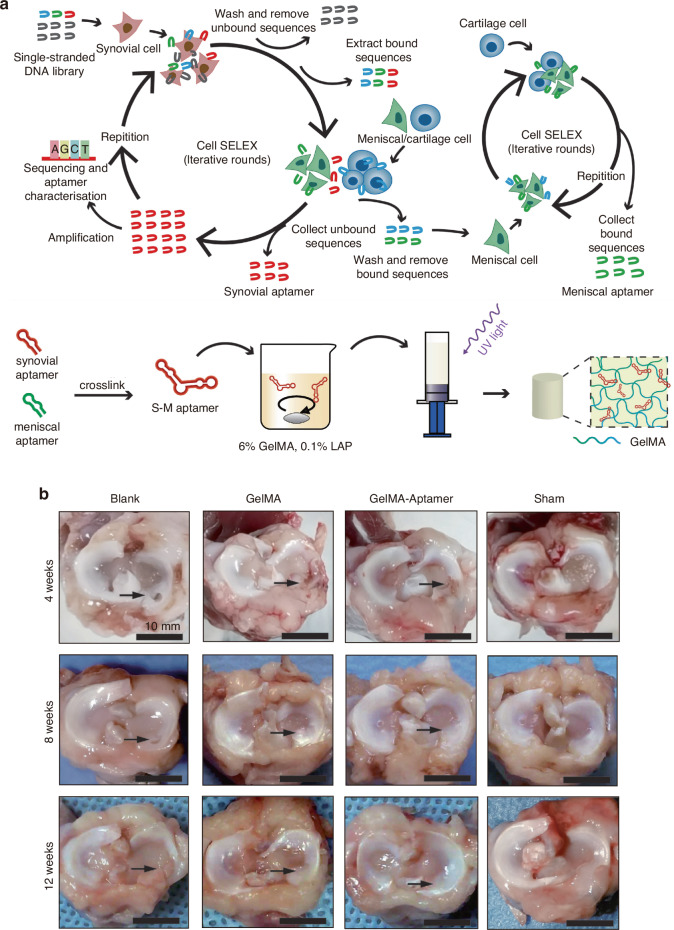
In recent years, MSC transplantation and their directed differentiation into chondrocytes have become a preferred approach for cartilage repair.214 Nevertheless, exogenous stem cell transplantation frequently faces challenges in cartilage tissue engineering, such as the absence of a targeted delivery system for MSCs, the low survival rate of exogenous cells, and the risk of exogenous infection during in vitro culture and cell proliferation.67,215,216 Currently, an aptamer-bilayer scaffold for knee joint repair has been developed,217 comprising an aptamer-gel for cartilage regeneration and an aptamer-functionalized 3D graphene oxide-based biomineral framework for bone regeneration. MSC-specific aptamers anchored on this scaffold enable targeted MSC recognition, binding, and recruitment to osteochondral defect sites (Fig. 3a). The aptamer-gel incorporates kartogenin, a chondrogenic factor, which is released sustainably to accelerate MSC differentiation into chondrocytes for cartilage regeneration. As type II collagen abundance reflects regenerated cartilage quality and maturity,218,219 comparisons with the aptamer-bilayer scaffold reveal inferior type II collagen staining, reduced cell density, and limited subchondral bone formation in standalone aptamer-gel applications (Fig. 3b).217 This demonstrates the bilayer scaffold’s superior repair efficacy. Based on Apt19S, a DNA aptamer targeting pluripotent stem cells,220 Hao et al.221 proposed a meniscus regeneration strategy combining endogenous stem/progenitor cell (ESPC) recruitment with fibrocartilage formation. They developed a 3D scaffold featuring a biomimetic microstructure and the MSC-specific aptamer Apt19S. This scaffold can sequentially activate the homing of ESPCs and the differentiation of fibrocartilage, achieving superior meniscus regeneration and protection of articular cartilage. It presents a potential alternative for meniscus tissue regeneration and a ready-made cell-guided meniscus product for clinical applications. In addition, gelatin methacryloyl (GelMA) hydrogel loaded with circular bispecific synovial-meniscal aptamers has been proven to be effective for avascular meniscus repair by recruiting endogenous synovial and meniscus cells and facilitating fibrocartilage regeneration (Fig. 4a). In a rabbit meniscus defect model, this approach demonstrated fibrocartilage-like tissue formation, reduced cartilage degeneration, and improved mechanical strength 12 weeks post-operation (Fig. 4b).222 Aptamers’ role in articular cartilage and osteochondral therapy lies in their potential as novel biomaterials for repair and regeneration. Engineered aptamers can modulate post-injury inflammation by suppressing pro-inflammatory cytokines or accelerate repair by promoting fibroblast proliferation and differentiation. They may also serve as drug delivery components to enhance therapeutic precision and minimize side effects. However, translating these concepts into clinical practice requires further validation of aptamer safety, efficacy, and optimal application methods, including thorough investigation of in vivo biocompatibility, long-term stability, and tissue interactions. In summary, aptamers hold diverse therapeutic potential for cartilage repair, spanning inflammation regulation, tissue regeneration, and targeted drug delivery.
Fig. 3.
Preparation process of a nucleic acid aptamer-bilayer scaffold and its therapeutic evaluation for osteochondral defects. a Overview of the aptamer-bilayer scaffold in osteochondral defect repair. b The scaffold was implanted into the osteochondral defect in rats. After an eight-week healing period, histomorphological analyses were performed, including safranin-O staining for glycosaminoglycans (GAG) and immunohistochemical staining for collagen type II to evaluate cartilage regeneration. Concurrently, microcomputed tomography (μCT) reconstruction and hematoxylin and eosin (H&E) staining of articular joint samples were conducted to assess subchondral bone regeneration. The aptamer-bilayer scaffold demonstrated significantly higher scores across all evaluated parameters compared to other groups. Reproduced form ref. 217 with permission from Wiley, copyright 2017
Fig. 4.
Preparation process of a bispecific GelMA-aptamer system and its therapeutic evaluation for meniscus defects. a Technological procedure for the filtration of aptamers and construction of the bispecific GelMA-aptamer system. b Observations of menisci at 4, 8, and 12 weeks post-surgery. Black arrows indicate the defect sites. Blank group, no repair; GelMA group, repair with GelMA hydrogel; GelMA-aptamer group, repair with GelMA hydrogel and bispecific aptamer; sham group, uninjured meniscus. Reproduced form ref. 222 with permission from SAGE Publications Inc., copyright 2023
Osteonosus
Osteonosus encompasses a broad spectrum of bone-related disorders involving diverse pathological mechanisms, including genetic, metabolic, inflammatory, or tumorigenic processes. These diseases may be congenital such as achondroplasia and osteogenesis imperfecta.223,224 Alternatively, these disorders may be acquired, such as diabetic bone disease, cancer-associated bone disorders, or osteoarthritis resulting from sports injuries.225–227 Similar to osteoporosis, osteonosus often involves genetic predisposition interacting with non-genetic factors, including endocrine imbalances, chronic inflammation, or systemic diseases. Under physiological conditions, bone tissue develops via intramembranous and endochondral ossification, initially forming weaker woven bone that is subsequently replaced by stronger lamellar bone.228 However, pathological states disrupt the balance between bone resorption and formation, leading to net bone loss and reduced bone mass.229 In summary, osteonosus represents a complex group of disorders frequently linked to genetic mutations, multifactorial pathogenesis, and impaired bone function.
Osteoporosis
In 1984, osteoporosis was defined as an age-related disease characterized by reduced bone mass and increased susceptibility to fractures.230 In 2001, the definition was revised to characterize osteoporosis as a skeletal disorder marked by compromised bone strength and an increased susceptibility to fractures, where bone strength is primarily determined by bone mineral density and bone quality.231,232 As the extent of population aging continues to deepen, osteoporosis emerges as an extremely crucial public health concern, imposing escalating socioeconomic burdens.233 In the USA, ~1.5 million fragility fractures occur annually,234 while UK epidemiology predicts lifetime osteoporotic fracture risks of 50% for women and 20% for men aged ≥50.235 Fracture risk escalates with age, particularly among the elderly, and is associated with substantial healthcare costs, functional impairment. The risk of hip fractures is particularly significant and is correlated with higher rates of disability and mortality.231
Osteoporosis pathogenesis centers on disrupted bone remodeling-resorption equilibrium. Estrogen potently inhibits osteoclast recruitment/activity in early menopause, suppressing bone resorption. Estrogen deficiency increases osteoclast numbers/lifespan while promoting osteoblast apoptosis, leading to trabecular thinning, cortical porosity, and accelerated remodeling, thereby causing bone mass loss.236–238 Remodeling initiates via osteoblast/osteocyte signals activating osteoclast-mediated resorption followed by bone formation.231,239,240 Excessive remodeling reduces bone mass and strength, promotes microdamage accumulation, and predicts fracture risk; lowering remodeling rates mitigates this risk.231,241,242 In osteoporosis, excessive remodeling degrades bone material properties, including bending resistance, elasticity, toughness, and strength.231,243,244 Moreover, osteoporosis is related to a decrease in the quantity and size of trabecular bones, their thinning, and transformation into a rod-like shape, replacing the stronger plate-like structures found in non-osteoporotic bone. In most osteoporosis patients, excessive remodeling may be the primary factor contributing to alterations in bone microarchitecture, as it leads to the loss of trabecular bone connectivity and further undermines the structural stability of the bone.231,237 In terms of mechanical loading, a deficiency in load will cause bone loss and an increase in remodeling activity.245 On the other hand, excessive loading can lead to an increase in microdamage.246,247 Microdamage is a reaction to repeated submaximal loads. It can sever lamellae and canaliculi, disrupt osteocytes communication, and induce osteocyte apoptosis. Subsequently, microdamage targeted for removal by osteoclasts is removed and repaired by osteoblasts.231,248,249 The optimal range of mechanical loading should fall between the two extremes, ensuring effective repair of microdamage without becoming excessive. Microdamage accumulates with advancing age, and it increases at a more rapid pace in women.250 In healthy postmenopausal women, bone remodeling doubles in the early stage and triples after 10–15 years.251 With increasing age, the body’s capacity to defend against oxidative stress diminishes. Some scholars have identified ROS as a contributing factor to the development of osteoporosis.252,253 Furthermore, the bone microstructure is disrupted to varying degrees, which impacts both bone quality and fracture resistance. Meanwhile, cortical bone undergoes structural changes because of aging and the menopausal transition.254,255 The occurrence of osteoporosis is also linked to genetic factors. Research has revealed that a single gene or a group of genes may determine bone mass at different skeletal locations. This suggests that genetic influences may affect an individual’s ability to accumulate and maintain bone mass throughout their lifetime, thus affecting the risk of developing osteoporosis.
A deficiency in estrogen increases the rate of bone remodeling. Even before menopause, a negative remodeling balance between bone resorption and formation can result in bone loss and compromise bone structure and strength256. Anti-osteoporosis medications reduce the rate of bone remodeling by either diminishing bone resorption or enhancing bone formation. For example, estrogen, selective estrogen receptor modulators, and bisphosphonates exert their effects primarily by inhibiting bone resorption, whereas intact parathyroid hormone functions as a powerful anabolic agent that increases bone formation.257,258 Currently, bisphosphonates are effective in reducing the risk of fractures and are relatively cost-effective. However, they are associated with gastrointestinal side effects. More critically, long-term application of bisphosphonates may simultaneously suppress the functions of both osteoblasts and osteoclasts. Even worse, prolonged administration may lead to osteonecrosis of the jaw and increase the risk of certain malignancies.259,260 In recent years, the exploration of various biological mechanisms in scientific research has deepened significantly. In the realm of osteoporosis research, an increasing number of studies indicate that the gut microbiota may serve as a potential target for the prevention and treatment of osteoporosis. Against this backdrop, the research concept of the “gut-bone axis” has gradually emerged and gained widespread attention. The nutrients ingested by the human body, along with the dietary patterns adopted, can influence the gut microbiota. In turn, changes in the gut microbiota may affect the host’s metabolic state, thereby regulating the process of bone metabolism. According to the gut-bone axis theory, dietary intake can participate in the regulatory mechanisms of bone metabolism by altering the abundance, diversity, and composition of the gut microbiota.261–264 An imbalance in the gut microbiota can result in intestinal inflammation, which releases various inflammatory factors such as TNF-α, IL-6, and IL-1β. These factors can further exacerbate the inflammatory response, causing adverse reactions such as damage to the intestinal mucosal barrier and immune dysfunction in the host, leading to the occurrence and progression of a series of diseases.265–267 These factors activate the RANKL signaling pathway, enhancing osteoclast function, which results in osteoporosis and bone loss.268–270 Nucleic acid aptamers can effectively modulate these inflammatory factors, alleviate inflammation, and subsequently influence bone metabolism.271–275 Multiple studies have shown that probiotics can enhance the integrity of the intestinal epithelium, prevent barrier degradation, and reduce pro-inflammatory responses.276,277 After effective probiotic intake in murines, there is an increase in the populations of bifidobacteria and lactobacilli in the gut, along with elevated levels of short-chain fatty acids (SCFAs), significant enhancements in bone density, and improvements in bone microstructure.278–281 SCFAs are key regulatory factors in osteoclast metabolism and bone mass.282 Propionate and butyrate can induce metabolic reprogramming in osteoclasts, leading to a downregulating of bone resorption and the protection of bone mass.267 Probiotic supplementation can significantly improve the gut environment and reduce the expression of inflammation-related factors.283–286 Research also indicates that a high-calcium diet combined with probiotic intervention can further amplify protective effects on murine bones, suggesting a potential synergistic relationship between diet and the microbiota. Probiotics can improve the efficiency of calcium absorption in the gut and enhance calcium utilization through the regulation of the gut microbiota, thereby increasing bone density and bone mineral content.287 For nucleic acid aptamers, their capacity to efficiently promote calcium absorption and foster the generation and survival of probiotics is a critical factor in determining their potential to improve the intestinal microenvironment. If nucleic acid aptamers demonstrate strong efficacy in these two areas, they could provide a novel and potent approach to optimizing gut microecology and boosting intestinal health.
Advancements in molecular biology and genetic engineering have promoted the development of aptamers, which serve as novel biomolecular tools that demonstrate significant potential in the study of osteoporosis. Furthermore, aptamers can regulate gene expression by specifically targeting nucleic acid sequences, thus affecting cellular functions related to bone formation and resorption. miRNAs represent a category of non-coding small RNAs that bind to target messenger RNAs (mRNAs). This binding results in either the degradation of the target mRNAs or the inhibition of their translation, ultimately regulating the expression of specific genes.288–290 Previous research has indicated a significant role of miRNAs in the process of bone remodeling, particularly in regulating the differentiation and function of both osteoblasts and osteoclasts.291,292 Furthermore, long non-coding RNAs are implicated in various signaling pathways and in the regulation of miRNAs during the process of bone formation.293 Three polymorphic loci within the FGF2 gene have been identified as being significantly correlated with femoral neck bone mineral density. Acting as potential binding sites for specific miRNAs, these loci may influence the expression of the FGF2 gene by altering the binding affinity between miRNAs and their target mRNAs.294 This suggests that modulating the activity of specific miRNAs could potentially serve as a novel therapeutic approach for osteoporosis. Circular RNAs (circRNAs), as a novel type of RNA molecule, have demonstrated significant importance in osteoporosis research. circRNAs influence bone metabolism by participating in the regulation of the differentiation, proliferation, and apoptosis of BMSCs.295,296 These research findings offer a new direction for the development of novel diagnostic markers and therapeutic targets. The previously mentioned CH6-LNPs-siRNA directly injects osteogenic Plekho1 siRNA into osteoblasts, leading to better bone microstructure and increased bone mass in tissues of osteopenic and healthy rats. Injecting an endothelial-specific aptamer-agomiR-195 system into aged murines can enhance the formation of CD31hiEmcnhi blood vessels and reverse age-related osteoporosis (Fig. S1). Utilizing SELEX technology, endothelial-cell-specific aptamers are screened and aptamer candidate 2 (Table 1) with higher binding ability and a satisfactory secondary structure is selected. A small RNA molecule, agomiR-195, which mimics the function of miR-195, is synthesized. Subsequently, 1 volume of polyethyleneimine solution (100 μg/mL, pH 6.0) is mixed with six volumes of 4.2 μmol/L sodium citrate to form a polyethyleneimine-citric acid core structure (nanocore). Then, three volumes of the synthesized aptamers (50 nmol/L) and agomiR-195 (1 μmol/L) are added into the nanocore, and the mixture is reacted for 5 min to assemble into a nano-complex, namely aptamer-agomiR-195.297 Furthermore, an aptamer-antagomiR-188 targeting system has successfully and selectively silenced miR-188 in BMSCs, which in turn increases bone formation and reduces the accumulation of bone marrow fat in aged murines (Fig. S2).298 This system assembles into a nano-complex (Table 1) after obtaining the aptamer sequence with the highest binding affinity to murine BMSCs through the same technology previously described. Despite the significant potential of aptamers in osteoporosis research, several challenges persist regarding their clinical applications. Rapid renal filtration and metabolic instability result in the rapid clearance, degradation or structural alteration of aptamers, ultimately influencing their therapeutic efficacy. This occurs due to the relatively low molecular weight of aptamers, which enables them to easily traverse the glomerular filtration membrane, leading to a reduced half-life in the body and an inability to maintain adequate concentrations at the site of action, significantly impacting therapeutic efficacy.299,300 The glomerular filtration membrane has a specific pore size, typically around 4–5 nm, allowing small molecules to pass freely.301 The molecular weight of nucleic acid aptamers generally ranges from 5 to 15 kDa, which is considerably lower than the retention threshold of the glomerular filtration membrane (30–50 kDa), facilitating their smooth passage through the membrane and rapid clearance.60,299 Previous studies have chemically modified aptamers by conjugating them with macromolecules like polyethylene glycol (PEG) or cholesterol to enhance their molecular weight, which slows their passage through the glomerular filtration membrane and prolonging their half-life in the body.302–304 Furthermore, new drug delivery systems, such as nanoparticle carriers, can be developed to encapsulate aptamers, shielding them from rapid renal clearance and achieving targeted delivery, thus enhancing their stability and therapeutic effectiveness in the body. Additional issues such as polyanion effects, unexpected tissue accumulation, and non-specific immune activation further elevate the risk of potential side effects. Therefore, future studies must focus on strategies to enhance the biological properties and efficacy of aptamers while exploring effective methods to translate these findings into clinical therapeutic approaches.
Bone tumor
From a historical perspective, the classification of bone tumors has experienced numerous changes and amendments. As early as the 1950s, researchers had already initiated revisions to the classification of bone tumors; however, this classification still possessed certain limitations. Subsequently, additional research and findings have prompted further refinement of the classification. In 2013, the fourth edition of the World Health Organization (WHO) Classification of Soft Tissue and Bone Tumors updated the classification of bone tumors. The classification categorizes bone tumors into three categories based on their biological behavior: benign, intermediate (including subgroups of locally invasive and rarely metastatic), and malignant.305 Benign tumors only recur sporadically and are non-destructive. These tumors can be removed or curetted locally. The intermediate type is subdivided into locally invasive and rarely metastatic subtypes. Locally invasive tumors have a high recurrence propensity and show invasive and destructive growth, yet there is no risk of metastasis. Generally, extensive resection or the implementation of adjuvant measures is necessary for effective treatment. The rarely metastatic subgroup typically demonstrates locally invasive growth with a relatively low metastatic propensity of less than 2%. Malignant tumors are characterized by destructive growth, a high recurrence propensity, and a significant risk of metastasis, typically exceeding 20%. These tumors necessitate surgical intervention in conjunction with radiotherapy and/or chemotherapy.306 The most recent classification standard for bone tumors is the fifth edition released by the WHO in 2020. This edition does not introduce significant changes to the classification of bone tumors; however, it includes new tumor entities and provides descriptions of subtypes for existing tumor types. Meanwhile, it integrates new molecular and genetic data.307
Osteosarcoma
Osteosarcoma (OS) is a primary malignant skeletal tumor characterized by malignant mesenchymal cells that directly form immature bone or osteoid tissue.308,309 Typically, 80%–90% of osteosarcomas occur in long tubular bones. Conventional osteosarcoma accounts for ~15% of all primary bone tumors subjected to biopsy analysis. Among primary malignant bone tumors, its incidence is surpassed only by that of multiple myeloma.309 Patients are generally younger, with a higher prevalence in males than females, while those under 6 years old or over 60 years old rarely develop this disease.309–311 Its characteristics include a high propensity for metastasis, mainly to the lungs, and pathological fractures resulting from bone destruction.312,313 The etiology of osteosarcoma remains unclear, although evidence suggests that human osteosarcoma can be induced by viruses or cell-free extracts from human osteosarcoma, and ionizing radiation is a recognized environmental factor contributing to its development.314 Besides, osteosarcoma also shows genetic susceptibility, as several families have been reported with multiple members affected.315 Screening of a significant number of children with osteosarcoma reveals that around 3% to 4% carry germline mutations in the p53 gene, indicating a family history consistent with Li-Fraumeni syndrome.316
Through Cell-SELEX, the specific DNA aptamer OS-7.9 (Table 1) targeting MG-63 osteosarcoma cells has been successfully identified. It has been verified that the OS-7.9 aptamer has a greater affinity for osteosarcoma cancer cells, shows no affinity for human bone marrow neuroblastoma, and can bind to lung cancer and colorectal cancer cell lines.317 This suggests that there may be common markers shared between osteosarcoma cells and these cells, with the OS-7.9 aptamer potentially serving as a valuable probe in cancer research. It could be applicable in developing early diagnostic approaches and treatment methods for metastatic diseases, as well as in identifying common membrane proteins present in different cancer types. To improve the diagnostic capabilities for osteosarcoma, the selected single-stranded DNA aptamer LP-16 can specifically bind to osteosarcoma cells and is the first aptamer to identify metastatic osteosarcoma cells.318 Research has shown that combining aptamers with salinomycin or clustered regularly interspaced short palindromic repeat (CRISPR)-associated Cas9 nuclease (CRISPR/Cas9) can decrease tumor volume and malignancy in osteosarcoma.319,320
Metastatic bone tumor: Bone is a primary site for many tumor metastasis.321 The occurrence and metastasis of high-grade and high-stage cancers are associated with the heterogeneity deletion of specific gene loci.322,323 For instance, miR-15a and miR-16-1 are tumor suppressor genes located on chromosome 13q14 and are correlated with the expression of genes like BCL2, CCND1, and WNT3A, which play direct roles in cell survival, proliferation, and invasion processes.324–328 The loss of these genes may facilitate the survival, proliferation, and invasion of cancer cells. Aberrant expression of these miRNAs in tumor cells can lead to a more aggressive metastatic capacity. On the other hand, bone metastasis may also be influenced by interactions within the bone microenvironment. During the invasion of tumor cells, a variety of adhesion receptors are implicated. These adhesion events are of great significance for the invasion, metastasis, and ultimate establishment of new tumors by tumor cells. These adhesion receptors connect the extracellular environment with intracellular signaling by binding to extracellular ligands in the tumor microenvironment, thus enhancing tumor cell migration, invasion, proliferation, and survival.329,330 Tumor cells can colonize and proliferate within bone tissue through adhering to specific components. They are capable of stimulating the activity of osteoclasts, resulting in enhanced bone resorption. Conversely, they may also impact the function of osteoblasts and facilitate bone formation.331 This imbalance in bone metabolism creates a conducive microenvironment for tumor cells growth. Moreover, tumor cells secrete large amounts of pro-angiogenic factors, giving rise to an aberrant, disorganized, immature, and poorly permeable vascular network.332 The blood vessels supply oxygen and nutrients to tumor cells, and tumor angiogenesis remarkably accelerates growth and increases the metastatic potential of the tumor.333
Numerous aptamers have been shown to suppress tumor metastasis. For angiogenesis, Rana et al.334 demonstrated an aptamer-based dynamic platform for the spatiotemporal control of the bioavailability of an angiogenic growth factor by utilizing affinity interactions within a biofabrication-friendly polymer matrix, aiming to realize a mature and stable vascular network. The RNA aptamer A10-3.2 is a new ligand for bone-metastatic prostate tumors that expresses the prostate-specific membrane antigen and is conjugated with atelocollagen (ATE) to deliver miR-15a and miR-16-1. It synergistically induces selective death of prostate cancer cells and augments the anticancer efficacy of the system.324 As a specific aptamer targeting complement C5a, AON-D21 can block the C5a/C5aR1 signal axis, effectively reducing the bone metastasis and tumor burden in lung cancer (Fig. S3).335 Research indicates that BMSCs are capable of migrating from the primary tumor site to the bone marrow, with the migration dependent on osteopontin (OPN). When OPN is blocked by the R3 aptamer, BMSCs are unable to migrate to the bone marrow.336
Multiple myeloma
Multiple myeloma (MM) is a malignant plasma cell neoplasm characterized by clonal proliferation within the bone marrow microenvironment. Although it is classified as a hematologic malignancy, it exhibits a distinct propensity to develop almost exclusively within the bone marrow, leading to severe bone destruction. This occurs by augmenting bone resorption through osteoclasts while simultaneously suppressing bone formation.337 Consequently, MM is specifically addressed here for discussion and analysis.
The occurrence of bone disease in MM is analyzed at the cellular level. The predominant pathogenesis of MM-induced bone disease is attributed to the up-regulation of osteoclast differentiation and activity, resulting in unbalanced bone resorption and the initiation of characteristic lytic bone lesions.338 Several factors mediate the activation of osteoclasts in MM patients. MM cells directly secrete cytokines capable of generating osteoclasts, including IL-1, IL-3, IL-6, TNF-α, MIP-1α, MIP-1β, and decoy receptor 3.339–344 It has been previously mentioned that RANKL and OPG play a core role in osteoclast differentiation. The adhesion between MM cells and BMSCs triggers the expression of RANKL in osteoblasts and advances osteoclast differentiation by stimulating the NF-κB and Jun N-terminal kinase pathways. Therapeutic methods that normalize the RANKL/OPG ratio by increasing OPG and/or decreasing RANKL can effectively impede bone destruction and MM growth in vivo.345 Both MM cells and osteoclasts secrete elevated levels of the pro-inflammatory cytokine MIP-1α.346 MIP-1α facilitates the survival and migration of MM cells, stimulates the generation of osteoclasts by activating the ERK and AKT signaling pathways, and downregulates bone formation-related transcription factors (eg, osterix) to inhibit osteoblast differentiation, thereby reducing bone formation.347–349 Moreover, a recent study indicated that osteoclasts protect MM cells from T cell-mediated cytotoxicity by directly inhibiting proliferating T cells.350 This defective T cell-mediated reaction is a crucial mechanism for tumors to evade immune surveillance.351 The immunosuppressive function of osteoclasts and their key role in lytic bone diseases could be considered important targets for the treatment of MM.
The differentiation of MSCs into bone cells is governed by the activation of two principal transcription factors: runt-related transcription factor 2 (RUNX2) and osterix.352 These transcription factors are vital for the maturation and ossification of osteoblasts and depend on the typical Wnt signaling pathway.353 Whereas, myeloma cells secrete various Wnt antagonists like dickkopf-1 (DKK1) and sclerostin (SOST). These proteins compete for the same Wnt signaling receptors, suppressing the differentiation and function of osteoblasts.354–358 Studies conducted both in vitro and in vivo have demonstrated that blocking DKK1 and augmenting Wnt signaling can restore the quantity of osteoblasts and trabecular bone, along with a reduction in tumor burden.359–362 After treating myeloma murines in vivo with a SOST-neutralizing antibody, results indicated a decrease in bone loss and lytic lesions, although there was no impact on tumor burden.363
In terms of bone marrow adipocytes, while there is no direct connection with MM bone disease, emerging evidence shows that adipocytes support the growth and survival of MM, which is inversely related to bone mass.353 Adipocytes and osteoblast lineage commitment originate from common progenitor cells. Wnt signaling acts as the primary initiator for the osteoblast lineage while simultaneously suppressing adipocyte lineage commitment regulated by the PPARγ signaling pathway.364,365 As people age, bone marrow adipocytes increase, and the rate of bone formation declines.353,366 Obesity is positively correlated with the risk of MM occurrence and is associated with an increase in bone marrow lipid as well.367–369 Furthermore, bone marrow adipocytes are capable of secreting free fatty acids, which serve as an energy source for the proliferation of tumor cells.370 Many lipids function as ligands for nuclear receptors involved in the PPARγ signaling pathway.371 The levels of free fatty acids are generally elevated in MM patients.372 This may result in the up-regulation of the PPARγ signaling pathway, thereby enhancing bone marrow adipogenesis while inhibiting the differentiation of osteoblasts and supporting the survival of MM. A recent in vitro research has indicated that saturated fatty acids, specifically stearate and palmitate, exert a direct lipotoxic effect on human osteoblasts.371,373 Furthermore, the data illustrate that increased adipogenesis during obesity and aging undermines bone regeneration.374 In conclusion, bone marrow fat is emerging as a crucial regulatory factor in the development of MM and bone disorders, potentially representing a novel and promising therapeutic target.
In the treatment of cancer, aptamers can serve as free molecules targeting specific cancer biomarkers as either agonists or antagonists (Fig. 5).375 Both AS1411 and NOX-A12, which have reached the clinical trial stage, are antagonistic aptamers that specifically target nucleolin and CXCL12, respectively.376–379 Nucleolin is highly expressed in multiple tumor cells. Through binding to the nucleolin receptor, AS1411 can play an anti-tumor effect.380 CXCL12 (also known as SDF-1) and CXC receptor 4 (CXCR4) are a chemokine and its receptor.381 CXCR4 is overexpressed in more than 75% of cancer cells, including those of myeloma.382 The CXCL12/CXCR4 pathway is a known crucial regulatory factor for tumor proliferation, cell spread, and migration both within and outside the bone marrow.383,384 NOX-A12 has been evaluated in research studies when used in combination with bortezomib and dexamethasone for treating relapsed MM.377 It produces a therapeutic effect on MM by preventing the interaction between CXCL12 and its receptor, thereby interfering with the CXCL12/CXCR4 signaling pathway and inhibiting the growth and proliferation of MM cells. What’s more, the specific aptamer ola-PEG for stromal cell-derived factor-1 (SDF-1) can neutralize SDF-1 and block SDF-1-dependent signal transduction, effectively suppressing the progression of MM.385
Fig. 5.
The potential application of nucleic acid aptamers in MM. a Aptamer against AXII. b BCMA targeted aptamer. c Aptamer against C-MET. d Conjugated CD38-doxorubicin aptamer and e NOX-A12 RNA aptamer for CXCL12. All the aptamers with the exception of the NOX-A12 spiegelmer are targeted at the receptors that are expressed on the plasma cell membrane of MM. BM bone marrow, Doxo doxorubicin, OBL osteoblast, sAXII soluble annexin A2. Reproduced form ref. 387 with permission from MDPI, copyright 2022
Annexin A2 (AXII) is overexpressed in the plasma cell membrane of MM, and its expression correlates negatively with patient survival rates.386 AXII, a member of the annexin family, is calcium-dependent and binds to phospholipids. The interaction between AXII and its receptor AXIIR augments the adhesion and growth of MM cells within the bone marrow microenvironment,387 potentially supporting the homing and growth of MM cells in this niche. AXII can be secreted by diverse cell types within the bone marrow, facilitating the growth of MM cells by establishing a pro-tumorigenic niche. Therefore, targeting the AXII/AXIIR axis represents a promising strategy for developing treatments against the MM niche.388 Zhou et al.389 have identified an ssDNA aptamer (wh6) that can specifically bind to MM cells expressing AXII both in vitro and vivo, and it can also suppress the adhesion and progression of MM cell lines induced by AXII. This shows that the wh6 aptamer is suitable for targeted MM therapy.
B cell maturation antigen (BCMA), a member of the TNF receptor superfamily, is preferentially expressed by mature B lymphocytes but is rarely expressed in hematopoietic stem cells.390 The ligands for BCMA, such as B-cell activating factor and a proliferation-inducing ligand (APRIL), can activate the NF-κB pathway.387,391,392 Nevertheless, the overexpression of BCMA and increased activation correlate with the progression of MM. These key genes exhibit a state of overexpression, facilitating the upregulation of the NF-κB pathway as well as the growth and survival of MM.390,393 In this context, Catuogno et al.393 selected a BCMA-targeted internalizing RNA aptamer (apt69.T) via Cell-SELEX, which demonstrated the inhibitory capacity on the APRIL-induced NF-κB pathway.
Hepatocyte growth factor (HGF) constitutes a specific ligand for the tyrosine kinase receptor C-MET.394 In MM, the expression of C-MET progressively augments during disease progression, and its elevated expression is correlated with a poorer prognosis for MM patients. Upon binding of HGF, C-MET dimerizes and initiates the processes that promote cell growth, migration and angiogenesis while inhibiting apoptosis, ultimately enhancing the development of MM.395 SL1 is the truncated form of the original CLN0003 ssDNA aptamer. This aptamer was obtained via the filtered SELEX method against purified C-MET to identify tumors with overexpressed C-MET.396,397 Previous research has proven that targeting C-MET with the SL1 aptamer can inhibit HGF-dependent C-MET signaling and restrain the growth of MM cells.
CD38 is a kind of cell surface glycoprotein that shows especially extensive and high expression levels in MM, contributing to the disease’s progression.387,398 Consequently, it has become one of the primary targets for developing anti-MM targeted therapy in recent years. Wen et al.399 identified a CD38-specific ssDNA aptamer using a hybridization protein and Cell-SELEX method. Subsequently, doxorubicin was non-covalently coupled to create a CD38-specific aptamer drug conjugate. It can be readily internalized by MM cells. Following pH-dependent release in lysosomes, doxorubicin is capable of exerting its specific anti-tumor activity by inhibiting tumor growth in both in vitro and vivo models.387
Osteoarthritis
Osteoarthritis (OA) is a degenerative joint disease characterized by joint pain, tenderness and restricted mobility. As the most prevalent form of arthritis globally, it exhibits the highest incidence among all arthritis types.400 Evidence suggests that ~240 million individuals worldwide suffer from symptomatic OA, making it a leading contributor to physical disability and diminished quality of life.401,402 The primary therapeutic goals are pain management and avoidance of treatment-related toxicity.403 The knee joint is the most frequently affected site, with higher prevalence among the elderly, particularly females.400,404–406 Originally, OA was attributed to mechanical wear of articular surfaces;407 however, it is now recognized to involve genetic predisposition, aging, hormonal influences, lifestyle factors, and other contributors. Its typical pathological characteristics include degeneration of articular cartilage, sclerosis of subchondral bone, synovial lesions, and joint inflammation.408,409 In the synovial joint of the knee, critical structures such as the meniscus, articular cartilage, subchondral bone, and synovium collectively mediate these pathological changes while maintaining joint functionality. Ji et al.410 revealed significant dysregulation of miR-141/200c in OA cartilage tissue. Regulating miR-141/200c levels profoundly affects chondrocyte proliferation, apoptosis, and metabolic markers. Utilizing a Cell-SELEX system, researchers identified chondrocyte-specific aptamers (tgg2, tgg5, tgg8) (Fig. S4). The tgg2 aptamer possesses a more satisfactory secondary structure and shorter nucleotide sequence for nanoparticle conjugation. Based on this, an aptamer (tgg2)-PEG2000-PAMAM6.0-cy5.5 nanoplatform for delivering miR-141/200c was developed (Fig. 6a). In the destabilized medial meniscus model of OA, intra-articular injections of miR-141/200c inhibitors significantly alleviate the OA pathology via SIRT1/IL-6/STAT3 pathway modulation, reduces chondrocyte apoptosis, and increases pain thresholds (Fig. 6b, c).
Fig. 6.
The mechanism of nucleic acid aptamers on chondrocytes in the pathological process of OA. a Schematic representation of the synthesis of tgg2-PEG2000-PAMAM6.0-Cy5.5. b Differentiated diffusion models of cationic nanocarriers binding to the negatively charged extracellular matrix (ECM) of chondrocytes via electrostatic interactions, followed by delivery into chondrocytes in normal and OA cartilage. c Modulation of the SIRT1/IL-6/STAT3 signaling pathway by miR-141/200c. Reproduced form ref. 410 with permission from Elsevier, copyright 2021
The synovium is composed of fibroblasts and macrophages. It produces synovial fluid containing lubricants and hyaluronic acid, which lubricates the joints and nourishes the articular cartilage.411–415 Synovitis is characterized by the proliferation of fibroblast-like synoviocytes (FLS) and the recruitment of macrophages, resulting in the hyperplasia of the synovial lining.416 Among the numerous factors that activate FLS in OA, follistatin-like protein 1 (FSTL1) is overexpressed in the synovium of OA patients, and its level correlates with OA severity.417 Activated FLS in OA secrete pro-inflammatory cytokines, chemokines, and proteolytic enzymes, contributing to inflammation propagation and cartilage matrix degradation.418 These responses and elevated production of inflammatory cytokines give rise to leukocyte recruitment, macrophage accumulation, and subsequent osteoclast activity enhancement.419 Primary FLS isolated from OA patients for in vitro experiments confirmed that most OA-FLS are senescent and capable of inducing chondrocyte apoptosis and an OA-like phenotype.420 As people age, the accumulation of senescent cells in the body has been identified as a contributing factor to OA.421 Chen et al.420 utilized Cell-SELEX technology to screen aptamers (CX1, CX2, and CX3) targeting FLS, with CX3 showing superior binding specificity. Consequently, CX3-conjugated liposomes (LS) were developed to eliminate senescent FLS, reducing non-specific toxicity and side effects while providing a novel therapeutic strategy for OA (Fig. 7).
Fig. 7.
Schematic diagram illustrating the pathogenesis of OA and the effects of targeted eliminating of senescent FLS on the progression of OA. a FLS were predominantly senescent during the progression of OA, promoting chondrocyte dysfunction. b FLS-targeted aptamers were screened using Cell-SELEX system, and aptamer CX3 was identified as a specific targeting ligand for FLS, not chondrocytes. c Intra-articular injection of CX3-modified liposomes (CX3-LS) loaded with senolytics could effectively induce the apoptosis of senescent FLS without affecting the cartilage, thereby suppressing cartilage degradation. Reproduced form ref. 420 with permission from Wiley-VCH Verlag GmbH, copyright 2022
Furthermore, inflammatory factors play a pivotal role in the pathological process of arthritis. In a specific study, the function of inflammatory factors in post-traumatic OA was examined using an “idealized“ anterior cruciate ligament reconstruction (IACL-R) model. The research discovered that although the IACL-R surgery restored normal gait characteristics, the concentrations of all tested inflammatory factors, including IL-1α, IL-1β, IL-2, IL-6, IL-8, IL-18, TNF-α, and granulocyte-macrophage colony-stimulating factor, in synovial fluid significantly increased on days 3, 7, and 15 following the surgery.422 These factors can activate multiple cell types within the joint, including fibroblasts, macrophages, and T cells, initiating a cascade of inflammatory reactions.423 Inflammatory cytokines are crucial in the pathogenesis of OA, significantly contributing to the disruption of metabolic homeostasis within joint tissues by promoting catabolism and destruction. In the pathogenesis of OA, these cytokines influence the production of other cytokines, inflammatory compounds and enzymes through combinatorial intracellular signaling pathways.423
IL-1 is a crucial cytokine involved in the pathogenesis of OA, capable of inducing inflammatory responses and catabolic effects.423 IL-1 inhibits chondrocytes from synthesizing ECM components and disrupts the synthesis of key structural proteins, such as type II collagen and aggrecan.424–426 Furthermore, it facilitates the production of two ECM-degrading enzymes: a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTS) and matrix metalloproteinases (MMPs). It induces chondrocyte apoptosis and stimulates the production of other cytokines in an autocrine manner, thereby promoting hydrolysis.427–430 IL-1α, a member of the IL-1 family, is a cytokine that exerts a crucial function in the modulation of immune responses and participates in various diseases. Ren et al.272 obtained the aptamer SL1067 (Table 1), which exhibits high affinity and remarkable specificity for human IL-1α through cell-SELEX screening. Comprised of only 22 nucleotides, it achieves high-affinity binding, representing the smallest modified aptamer to be crystallized, and acts as an effective inhibitor of the IL-1α/IL-1 receptor pathway signaling. IL-18 is another representative of the IL-1 family.431 Research has revealed that the levels of Caspase 1 in the articular cartilage and synovium of patients with OA are elevated, significantly promoting the formation of IL-1β and IL-18.432 The production of IL-18 in joints is determined by chondrocytes, osteoblasts, FLS and macrophages.433–435 The level of IL-18 is elevated in the articular cartilage and synovium of OA patients and is positively correlated with disease severity.436–438 IL-18 can induce the upregulation of IL-18 receptors on chondrocyte surfaces and stimulate the synthesis of MMP-1, MMP-3, and MMP-13.439 In addition to increasing cartilage-degrading enzyme concentrations, it suppresses the production of proteoglycans, aggrecan, and type II collagen. Furthermore, chondrocytes exhibit typical apoptotic morphological changes.440–442 It facilitates the generation of multiple compounds and enzymes by chondrocytes and synoviocytes in an autocrine fashion.433,443–446
TNF-α and IL-1 are collectively recognized as crucial inflammatory cytokines involved in the pathophysiological process of OA. They hinder the synthesis of proteoglycan components, protein-bound proteoglycans, and type II collagen in chondrocytes.447,448 TNF-α exerts effects similar to those of IL-1β. During the progression of OA, there is a distinct synergistic interaction between these two cytokines.449 They promote MMP and ADAMTS-4 production, reduce the efficiency of the mitochondrial respiratory chain, and induce chondrocyte death and aberrant progenitor cell migration.430,450–453 Furthermore, chondrocytes influenced by IL-1β and TNF-α exhibit accelerated senescence and apoptosis.451,452,454 Additionally, TNF-α enhances the synthesis of IL-6, IL-8, regulated upon activation normal T cell expressed and secreted factor, and VEGF.455–458 Using SELEX technology, Orava et al.273 screened a DNA aptamer with a specific variable region (VR) sequence that binds to TNF-α. Among these, VR11 (Table 1) is the sole aptamer capable of inhibiting human TNF-α function without inducing the innate immune response associated with the production of inflammatory cytokines.273
IL-6 is a cytokine with multifaceted interactions, capable of activating the immune system and intensifying the inflammatory response.423 Its primary role involves promoting the formation of osteoclasts, thereby facilitating bone resorption, and it exhibits synergistic effects with IL-1β and TNF.459,460 Polymorphisms in its gene may predict the development of OA.461,462 IL-6 acts in concert with other factors to decrease type II collagen production and increase MMP synthesis.463–465 Furthermore, experiments on IL-6-deficient murines have demonstrated that, compared with wild-type murines, they tend to present more severe degenerative alterations.466 Gupta et al.271 isolated IL-6 SOMAmers through the SELEX process, which possess high affinity, slow complex dissociation, nuclease resistance, and effectively inhibit IL-6 signaling.
IL-8 belongs to the C-X-C chemokine family and predominantly exhibits chemotactic activity towards neutrophils. Although these cells typically do not produce high levels of IL-8 under quiescent conditions, they generate elevated concentrations upon agonist stimulation. Nevertheless, IL-8 is highly expressed in some tumor cells.467 IL-1 and TNF are the most potent inducers of IL-8, while other cytokines such as IL-6 do not effectively signal its production. The expression of IL-8 is cell-specific and stimulus-specific.468,469 Sung et al.274 have successfully developed and characterized the 20-fluoro-pyrimidine-modified RNA aptamer 8A-35 (Table 1), which can recognize human IL-8. This aptamer demonstrates highly specificity/affinity and effectively neutralizes human IL-8. The 8A-35 aptamer modulates various biological activities of IL-8 in human neutrophils, including intracellular signal transduction and chemotaxis.
IL-17 represents a family of cytokines with inflammatory functions. Its primary sources include stimulated CD4 + T cells and mast cells, which infiltrate the synovial membrane and the entire joint via blood vessels.470–472 Several studies have demonstrated that these T cells mediate direct immune response against chondrocyte and fibroblast membrane antigens during OA.473 IL-17 has been demonstrated to suppress proteoglycan synthesis in chondrocytes and promote the production of MMPs.474–476 It also secretes other cytokines and compounds that negatively affect cartilage.477–479 Besides, IL-17 facilitates VEGF secretion, contributing to synovial hypertrophy.458,480 IL-17 gene polymorphisms may correlate with OA susceptibility.481 As a member of the IL-17 family, IL-17A is associated with a variety of autoimmune diseases. Ishiguro et al.275 screened aptamers Apt3 and Apt21 and obtained optimized short aptamer derivatives (Apt3-4 and Apt21-2) through chemical and functional manipulation. In normal human dermal fibroblasts, Apt21-2 significantly blocks IL-17A-induced phosphorylation of signaling factors and IL-6 expression, suppressing inflammatory lesions and neurological symptoms, thus demonstrating therapeutic potential for autoimmune diseases. Adachi et al.482 prepared the RNA aptamer AptAF42dope1, which exhibits a higher affinity for IL-17A/F, offering novel biotechnological and medical applications.
IL-15 is one of the crucial cytokines in rheumatoid arthritis pathogenesis.483,484 Its function primarily involves stimulating the differentiation and proliferation of T cells and natural killer cells.485 During the pathogenesis of OA, elevated IL-15 concentrations in synovial fluid at early disease stages stimulate secretion of specific MMP-group metalloproteinases.486
Osteogenesis imperfecta
Osteogenesis imperfecta (OI) is a hereditary connective tissue disorder primarily characterized by fragile bones.487 The etiology of OI is mainly linked to genetic mutations, particularly those involving the COL1A1 or COL1A2 genes, which encode the alpha-1 and alpha-2 chains of type I collagen, a key component of the skeletal structure. Abnormal synthesis of these proteins impacts bone strength and architecture.488,489 The clinical manifestations of OI are highly heterogeneous. Depending on the severity and type, patients may present symptoms such as dentinogenesis imperfecta, blue sclera, short stature, hearing loss and cardiac anomalies.487,490 OI is typically categorized into four main types: Type I (the most common and mild form), Type II (associated with neonatal mortality), Type III (severe), and Type IV (moderate to severe).491 The incidence of OI is ~1 in 15 000 to 20 000 live births, with no gender, racial, or ethnic predilection.488,492,493 The primary treatment goals for OI are to prevent fractures, manage symptoms, and increase bone mass; however, currently available therapeutic options have limited efficacy.494,495 Commonly used medications, such as bisphosphonates, denosumab, synthetic parathyroid hormone, and growth hormone for children, can improve bone density and reduce fracture incidence to some extent. Nonetheless, their effectiveness remains suboptimal, and they may be ineffective in certain patients or induce cytotoxic side effects.494
Sclerostin, a glycoprotein secreted by mature osteocytes, antagonizes the Wnt/β-catenin signaling pathway by binding to low-density lipoprotein receptor-related proteins 5 and 6 (Lrp 5/6), thereby inhibiting bone formation.496–499 In clinical trials targeting low bone mass disorders such as osteoporosis and OI, sclerostin antibody (Scl-Ab) therapy has been shown to interfere with sclerostin, consequently increasing bone formation and mass.500,501 Clinically, the humanized therapeutic Scl-Ab (romosozumab) has demonstrated bone anabolic potential in postmenopausal osteoporosis, though serious cardiovascular adverse events have been reported.502–504 Given that cardiovascular abnormalities are a secondary feature in OI patients, Scl-Ab treatment may elevate cardiovascular risks, particularly in those with preexisting cardiovascular conditions.487,505,506 Ge Zhang et al.507 developed next-generation sclerostin inhibitors to promote bone formation without exacerbating cardiovascular risks in OI. The study designed multiple animal models. The Col1a2+/G610C murine model was established for OI investigation. Another model, hSOSTki:Col1a2+/G610C, was generated by knocking in human sclerostin (hSOST) into Col1a2+/G610C. The Δloop3-hSOSTki:Col1a2+/G610C model was further modified from hSOSTki:Col1a2+/G610C, where Δloop3 denotes the deletion of the loop3 domain of sclerostin. This model was utilized to examine the role of the loop3 domain of sclerostin in bone formation and the cardiovascular system. Additionally, an aortic aneurysm (AA) and atherosclerosis murine model, ApoE−/−, was constructed based on the previous three models.
In vitro primary osteoblast experiments demonstrated that overexpression of full-length sclerostin (FL hSOST) significantly inhibited the Wnt signaling pathway and downregulated osteogenic markers, confirming the sclerostin’s anti-anabolic role. In contrast, cells overexpressing loop2 and/or loop3 deficient sclerostin exhibited increased Wnt signaling and osteogenic marker expression, underscoring loop3’s critical role in sclerostin-mediated bone inhibition (Fig. S5a). Micro-CT and bone histomorphometric analysis revealed that sclerostin’s loop3 domain is pivotal in bone regulation. Compared to Col1a2+/G610C murines, hSOSTki:Col1a2+/G610C murines (expressing FL hSOST) displayed significantly poorer trabecular bone volume and microstructure. In contrast, Δloop3-hSOSTki:Col1a2+/G610C murines demonstrated notable improvements in these bone parameters relative to the hSOSTki:Col1a2+/G610C murine model, indicating that loop3 deletion mitigates sclerostin’s inhibitory effects (Fig. S5b, c).
The construction of AA and atherosclerosis models and measurement of relevant indicators revealed that, compared to Col1a2+/G610C:ApoE−/−, both hSOSTki:Col1a2+/G610C:ApoE−/− and Δloop3-hSOSTki:Col1a2+/G610C: ApoE−/− exhibited significantly reduced AA incidence, smaller maximum diameters of the aortic arch and renal artery, lower proportions of atherosclerotic lesions at the aortic root, and decreased serum levels of IL-6, TNFα and monocyte chemoattractant protein-1 (MCP-1) (Fig. S6). No significant differences in these cardiovascular-related parameters were observed between the hSOSTki:Col1a2+/G610C:ApoE−/− and Δloop3-hSOSTki:Col1a2+/G610C:ApoE-/ groups. These findings indicate that sclerostin lacking the loop3 domain confers a cardiovascular protective effect similar to FL hSOST, suggesting that sclerostin’s cardiovascular protection is independent of the loop3 domain. Apc001PE, a PEG40k-conjugated modified aptamer targeting the loop3 domain of sclerostin (Table 1), was developed based on its precursor aptscl56. Aptscl56 binds specifically to the loop3 domain of sclerostin in the serum of OI patients, inhibiting sclerostin’s antagonistic effects on the Wnt signaling pathway and osteogenic potential. Notably, this inhibition does not interfere with sclerostin’s suppressive effects on inflammatory cytokines/chemokines or elevate cardiovascular risk. Furthermore, Apc001PE demonstrated no toxicity in healthy murine and rat models, highlighting its potential as a novel therapeutic agent with significant clinical implications for OI treatment.
In subsequent research, Ge Zhang’s team further addressed the issue of lower affinity in the aptamer Aptscl56.508 They introduced seven distinct quinoline derivatives (spanning 2-quinoline to 8-quinoline) at key sites of Aptscl56 to synthesize a modified aptamer library. The binding ability of these derivatives to sclerostin was evaluated against the unmodified Aptscl56, revealing that the 5-quinoline-modified aptamers enhanced binding ability by 6 to 16 times at multiple critical sites, outperformed other modifications, and exhibited a lower dissociation constant. Following enzyme-resistant modification, the 5-quinoline-modified aptamer demonstrated further improved binding efficacy. In vitro cell experiments showed that the 5-quinoline-modified aptamer more robustly restored Wnt signaling pathway activity and bone formation marker expression. In vivo animal experiments confirmed its pronounced bone-forming effects in murine models compared to the unmodified aptamer. Molecular dynamics simulations and experimental analyses revealed that the 5-quinoline modification altered the aptamer-sclerostin binding conformation, diverging significantly from the natural Aptscl56 interaction.
The other applications of nucleic acid aptamers
Nucleic acid aptamers not only demonstrate significant potential in treating orthopedic diseases but also exhibit broad applications across biomedical fields. These applications highlight the multifunctionality of aptamer technology and provide novel approaches for disease research and therapy. For instance, advancements in aptamer-based biosensing enable precise diagnostic methods, offering powerful tools for early disease detection and progression monitoring. Biological detection utilizes biological principles and techniques to detect, quantify, or monitor various substances, biological processes, or environmental conditions, through analysis of cells, proteins, nucleic acids, and metabolites. This technology is critical for early health threat detection, drug safety/efficacy evaluation, bioequivalence assessment, and dosing regimen optimization.509,510 With Next-Generation Sequencing (NGS) technology, it plays an increasingly important role in diagnosing genetic diseases like cancer.511 However, population-wide implementation remains limited, necessitating new biomarkers to bridge gaps between exposure and disease onset. Success in biological detection hinges on data quality and actionable insights from analyses. As personalized medicine advances, rapid genomic testing becomes increasingly vital. Overall, biological detection is a multidisciplinary field essential to public health, environmental protection, drug development, and personalized medicine. Detection technologies such as fluorescence, electrochemical, and nucleic-acid-aptamer-based biosensors have gradually matured.512–516 Nucleic-acid-aptamer-based detection technology relies on the high affinity and specific binding between aptamers and specific target molecules. SELEX-derived aptamers recognize specific molecules and, when integrated with other technologies, generate quantifiable signals.512,513,515,516 Their unique 3D structures ensure specificity and sensitivity, while in vitro synthesis enables scalable production and functional modifications.512,515 Compared to antibodies, aptamers exhibit superior stability and environmental adaptability.515 Aptamer biosensors offer the advantages of rapid response, simple operation, and low cost.512,513,516 Moreover, they have been successfully applied in the detection of various substances, demonstrating promising application prospects.513,515 Furthermore, in further research, efficient signal transduction can be readily achieved, enabling sensitive and selective detection as well as high-throughput drug screening.515 Aptamers possess extensive target-binding capabilities and can be utilized to construct sensors for multi-substance detection.514,516 They demonstrate significant potential in disease diagnosis, food safety detection and environmental monitoring.517–526
During the SARS-CoV-2 pandemic, early nucleic acid and antigen tests faced limitations including reagent shortages, complex procedures, and high costs.527,528 Polymerase chain reaction (PCR) could not confirm viral infectivity, and antigen detection lacked sensitivity.529 To address this, a macromolecular DNA nanoscaffold (DNA Net) was designed based on the trimeric spike protein structure of SARS-CoV-2 (Fig. 8a). A tri-aptamer targeting the spike’s receptor-binding domain (RBD) enabled rapid, sensitive virus detection and inhibition (Fig. 8b).27,527 Moreover, an aptamer switch was designed to transform DNA Net-aptamers into a highly sensitive biosensor for virus detection via fluorescence readings (Fig. 8c). Subsequently, molecular docking and dynamics simulations validated the design, while SPR confirmed that DNA Net aptamers (especially larger constructs) enhanced binding affinity.527 Chauhan et al.527 identified the optimal aptamer-lock candidate pair (Apta-lock-1) and utilized a 4 × 4 DNA Net for their research, demonstrating that multivalent aptamer-spike interactions improve SARS-CoV-2 detection sensitivity. This system detects viruses without nucleic acid extraction and amplification, fulfills early-infection sensitivity requirements, and differentiates infectious from inactivated viruses, reducing false positives. Increased DNA scaffold flexibility can enhance virus detection sensitivity. More importantly, it exhibits selectivity, with no cross-reaction against H1N1, OC43 and Zika virus, and it can detect SARS-CoV-2 variants, albeit with relatively lower signal intensity and sensitivity for these variants. DNA Net aptamers also show therapeutic potential, inhibiting SARS-CoV-2 in vitro with nearly 1 000-fold greater efficacy than monomeric aptamers.
Fig. 8.
The potential of nucleic acid aptamers in biological detection. a The technological procedure and construction of the DNA Net-aptamers. b The complex of DNA Net-aptamers are attached to a virion can block the interactions between the spike protein and ACE2 receptors on the surface of a host cell, inhibiting viral infection due to its high neutralization potency. c The principle of signal output for DNA Net aptamers. Reproduced form ref. 527 with permission from American Chemical Society, copyright 2023
Conclusion and perspective
Nucleic acid aptamers, as short oligonucleotide sequences, form specific secondary/tertiary structures enabling high-affinity target binding. Ranging from traditional SELEX technologies to novel techniques such as Pro-SELEX and microfluidic-chip-SELEX, screening techniques continue to evolve, improving efficiency and specificity. Furthermore, aptamer-target interactions involve conformational changes and multimodal binding, with tunable affinity. Regarding applications, nucleic acid aptamers have demonstrated substantial potential across various fields, including biosensors, repair, oncology, and therapy.
Despite significant advancements in the field of nucleic acid aptamers, several areas warrant further investigation and exploration. Research on DNA, siRNA, and miRNA within the scope of nucleic acid aptamers has been relatively comprehensive and well-documented. However, research concerning circRNA remains somewhat limited. Given that RNA aptamers and RNA-aptamer-based devices are constrained by low expression levels and rapid degradation in mammalian cells.530 circRNAs are characterized by a covalently closed loop structure resulting from a specialized alternative splicing process known as back-splicing, where the originally linear mRNA molecules form loops.531–534 This unique structure endows circRNAs with diverse roles within organisms, such as acting as miRNA sponges, serving as protein scaffolds, and functioning as translation templates. While it is known that they play roles in various diseases, many mysteries remain concerning the specific functions of most circRNAs and their mechanisms of gene expression regulation.535 The complexity of circRNA research is also reflected in their expression specificity; circRNAs exhibit both cell-type and tissue-specific expression, which can be largely independent of their corresponding linear host genes' expression levels.536 Even within the same species, across different tissues or cell types, certain circRNAs may exhibit divergent expression patterns, further complicating the research. Furthermore, although some circRNAs contain start codons, experimental results indicate that circRNAs with AUG sequences cannot bind to ribosomes and thus are not translated.537 However, whether all circRNAs are incapable of participating in protein translation remains currently under investigation, suggesting they may not function through the conventional translational pathway. The biosynthetic pathways of circRNAs also present a research challenge. Their synthesis is complex and exhibits low efficiency. While chemical cyclization methods can facilitate the synthesis of circRNA, they involve strict reaction conditions, low yields, and a propensity to generate by-products, such as 2'-5' phosphodiester bonds, which restrict their application in nucleic acid aptamers.538,539 Enzymatic strategies can enhance cyclization efficiency but necessitate specific enzymes and complex reaction conditions, while also imposing length restrictions on precursor RNA, which are typically suitable only for sequences shorter than 500 nucleotides.538,539 Currently, researchers have proposed several solutions to these challenges. For example, optimizing synthesis methods and employing novel enzymatic strategies can improve the efficiency and quality of circRNA synthesis.540–542 Lee et al.543 have designed RNA constructs derived from Tetrahymena Group I introns that enable end to end self-targeting and splicing reactions to facilitate efficient circRNA generation. They have also optimized target sequence selection to enhance circularization efficiency. Additionally, enhancing the protein expression efficiency of circRNA remains a significant challenge in circRNA-related nanomedicine. Existing methods include RNA modifications, optimization of homologous arms, incorporation of internal ribosome entry sites (IRES), and the use of aptamers.544–547 Recently, a novel circRNA platform has been developed, incorporating an optimized vector topology, 5' and 3' untranslated regions (UTRs), IRES, and synthetic aptamers. This advancement has achieved a highly efficient and stable protein production process, with yields increased by several hundred times.544,547
Regarding screening techniques, existing methods must be continuously optimized to improve screening efficiency and accuracy while reducing costs. For instance, the development of more intelligent screening platforms capable of automatically adjusting screening parameters according to the characteristics of target molecules is necessary. While chemical modification strategies have improved the stability of nucleic acid aptamers, further exploration of more effective approaches remains imperative to address the requirements of diverse application scenarios. In terms of application fields, it is essential to further broaden the scope of applications for nucleic acid aptamers. Furthermore, in-depth investigations into the pharmacokinetics and biosafety of nucleic acid aptamers in vivo are required to establish a stronger foundation for their clinical applications. If these challenges can be addressed, exploring more valuable nucleic acid aptamers could signify a significant qualitative leap forward in this field’s research, potentially offering novel directions for future disease treatment strategies.
Supplementary information
Acknowledgements
Key research and development projects of Sichuan Science and Technology Plan Project (2024YFFK0135), Fujian Provincial Natural Science Foundation of China (2024J011450).
Author contributions
Zhenhong He: Investigation, Writing-original draft, Table and figure editing, Writing—review & editing. Qingping Peng: Investigation, Writing—review & editing. Wenying Bin: Investigation, Writing—review & editing. Luyao Zhao: Investigation, Writing—review & editing. Yihuang Chen: Investigation. Yuanqun Zhang: Investigation. Weihu Yang: Conceptualization, Supervision, Project administration, Writing—review & editing. Xingchen Yan: Supervision, Project administration, Writing—review & editing. Huan Liu: Conceptualization, Supervision, Project administration, Funding acquisition, Writing—review & editing. All authors approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Zhenhong He, Qingping Peng
Contributor Information
Weihu Yang, Email: yangweihu@cqu.edu.cn.
Xingchen Yan, Email: yanxingchen@gdinm.com.
Huan Liu, Email: 20016040@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41413-025-00447-8.
References
- 1.Ma, H. et al. Nucleic acid aptamers in cancer research, diagnosis and therapy. Chem. Soc. Rev.44, 1240–1256 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Tuerk, C. & Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science249, 505–510 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Gao, F. et al. Recent advances in aptamer-based targeted drug delivery systems for cancer therapy. Front. Bioeng. Biotechnol.10, 972933 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu, X. et al. Structure-guided designing pre-organization in bivalent aptamers. J. Am. Chem. Soc.144, 4507–4514 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Di Matteo, M., Belay, E., Chuah, M. K. & VandenDriessche, T. Recent developments in transposon-mediated gene therapy. Expert Opin. Biol. Ther.12, 841–858 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Sivakumar, P., Kim, S., Kang, H. C. & Shim, M. S. Targeted siRNA delivery using aptamer‐siRNA chimeras and aptamer‐conjugated nanoparticles. WIREs Nanomed. Nanobiotechnology11, e1543 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Yan, Y. et al. Non-viral vectors for RNA delivery. J. Control. Release342, 241–279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weng, Y., Xiao, H., Zhang, J., Liang, X.-J. & Huang, Y. RNAi therapeutic and its innovative biotechnological evolution. Biotechnol. Adv.37, 801–825 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature346, 818–822 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Robertson, D. L. & Joyce, G. F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature344, 467–468 (1990). [DOI] [PubMed] [Google Scholar]
- 11.Bartel, D. P. MicroRNAs. Cell116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Kuersten, S. & Goodwin, E. B. The power of the 3′ UTR: translational control and development. Nat. Rev. Genet.4, 626–637 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell75, 843–854 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Jenison, R. D., Gill, S. C., Pardi, A. & Polisky, B. High-resolution molecular discrimination by RNA. Science263, 1425–1429 (1994). [DOI] [PubMed] [Google Scholar]
- 15.Stojanovic, M. N., de Prada, P. & Landry, D. W. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc.123, 4928–4931 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Baker, B. R. et al. An electronic, aptamer-based small-molecule sensor for the rapid, label-free detection of cocaine in adulterated samples and biological fluids. J. Am. Chem. Soc.128, 3138–3139 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Strehlitz, B., Nikolaus, N. & Stoltenburg, R. Protein detection with aptamer biosensors. Sensors8, 4296–4307 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamaguchi, N., Ellington, A. & Stanton, M. Aptamer beacons for the direct detection of proteins. Anal. Biochem.294, 126–131 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Daniels, D. A., Chen, H., Hicke, B. J., Swiderek, K. M. & Gold, L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl. Acad. Sci.100, 15416–15421 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov.9, 537–550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayyed, S. G. et al. Podocytes produce homeostatic chemokine stromal cell-derived factor-1/CXCL12, which contributes to glomerulosclerosis, podocyte loss and albuminuria in a mouse model of type 2 diabetes. Diabetologia52, 2445–2454 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Bagalkot, V., Farokhzad, O. C., Langer, R. & Jon, S. An aptamer–doxorubicin physical conjugate as a novel targeted drug‐delivery platform. Angew. Chem. Int. Ed.45, 8149–8152 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Poudineh, M., Sargent, E. H., Pantel, K. & Kelley, S. O. Profiling circulating tumour cells and other biomarkers of invasive cancers. Nat. Biomed. Eng.2, 72–84 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Phillips, J. A., Xu, Y., Xia, Z., Fan, Z. H. & Tan, W. Enrichment of cancer cells using aptamers immobilized on a microfluidic channel. Anal. Chem.81, 1033–1039 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu, J., Nguyen, T., Pei, R., Stojanovic, M. & Lin, Q. Specific capture and temperature-mediated release of cells in an aptamer-based microfluidic device. Lab Chip12, 3504–3513 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J., Lee, E., Kang, Y. Y. & Mok, H. Multivalent aptamer–RNA based fluorescent probes for carrier-free detection of cellular microRNA-34a in mucin1-expressing cancer cells. Chem. Commun.51, 9038–9041 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Song, Y. et al. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal. Chem.92, 9895–9900 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Liu, X. et al. Neutralizing aptamers block S/RBD‐ACE2 interactions and prevent host cell infection. Angew. Chem.133, 10361–10366 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmitz, A. et al. A SARS‐CoV‐2 spike binding DNA aptamer that inhibits pseudovirus infection by an RBD‐independent mechanism.Angew. Chem.133, 10367–10373 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun, M. et al. Aptamer blocking strategy inhibits SARS‐CoV‐2 virus infection. Angew. Chem. Int. Ed.60, 10266–10272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, R., He, L., Hu, Y., Luo, Z. & Zhang, J. A serological aptamer-assisted proximity ligation assay for COVID-19 diagnosis and seeking neutralizing aptamers. Chem. Sci.11, 12157–12164 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He, J. et al. Molecularly engineering triptolide with aptamers for high specificity and cytotoxicity for triple-negative breast cancer. J. Am. Chem. Soc.142, 2699–2703 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Li, Y. et al. A new paradigm for artesunate anticancer function: considerably enhancing the cytotoxicity via conjugating artesunate with aptamer. Signal Transduct. Target. Ther.6, 327 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandotra, R. et al. Aptamer selection against alpha-defensin human neutrophil peptide 1 on an integrated microfluidic system for diagnosis of periprosthetic joint infections. Lab Chip22, 250–261 (2022). [DOI] [PubMed] [Google Scholar]
- 35.Chen, T.-W. et al. Automatic detection of two synovial fluid periprosthetic joint infection biomarkers on an integrated microfluidic system. Anal. Chem.95, 7693–7701 (2023). [DOI] [PubMed] [Google Scholar]
- 36.Lipps, H. J. & Rhodes, D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol.19, 414–422 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Wang, R. E., Wu, H., Niu, Y. & Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem.18, 4126–4138 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Shukla, S., Sumaria, C. S. & Pradeepkumar, P. I. Exploring chemical modifications for siRNA therapeutics: a structural and functional outlook. Chem. Med. Chem.5, 328–349 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Uppal, G. K., Poolsup, S., Zaripov, E., Gu, Y. & Berezovski, M. V. Comparative analysis of aptamers binding to SARS-CoV-2 N protein using capillary electrophoresis and bio-layer interferometry. Anal. Bioanal. Chem.416, 1697–1705 (2024). [DOI] [PubMed] [Google Scholar]
- 40.Gelain, F. et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS ONE5, e15004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davies, D. R. et al. Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc. Natl. Acad. Sci. USA109, 19971–19976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kimoto, M., Yamashige, R., Matsunaga, K.-I., Yokoyama, S. & Hirao, I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol.31, 453–457 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Vaught, J. D. et al. Expanding the chemistry of DNA for in vitro selection. J. Am. Chem. Soc.132, 4141–4151 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Moore, M. D. et al. Protection of HIV neutralizing aptamers against rectal and vaginal nucleases. J. Biol. Chem.286, 2526–2535 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu, L. et al. Aptamer-based detection of circulating targets for precision medicine. Chem. Rev.121, 12035–12105 (2021). [DOI] [PubMed] [Google Scholar]
- 46.Gramlich, P. M. E., Wirges, C. T., Manetto, A. & Carell, T. Postsynthetic DNA modification through the copper‐catalyzed azide–alkyne cycloaddition reaction. Angew. Chem. Int. Ed.47, 8350–8358 (2008). [DOI] [PubMed] [Google Scholar]
- 47.Lao, Y.-H., Peck, K. & Chen, L.-C. Enhancement of aptamer microarray sensitivity through spacer optimization and avidity effect. Anal. Chem.81, 1747–1754 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Gu, F. et al. Precise engineering of targeted nanoparticles by using self-assembled biointegrated block copolymers. Proc. Natl. Acad. Sci. USA105, 2586–2591 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vorobyeva, M., Vorobjev, P. & Venyaminova, A. Multivalent aptamers: versatile tools for diagnostic and therapeutic applications. Molecules21, 1613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheng, W., Chen, T., Tan, W. & Fan, Z. H. Multivalent DNA nanospheres for enhanced capture of cancer cells in microfluidic devices. ACS Nano7, 7067–7076 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, J. et al. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res.49, 7267–7279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He, J. et al. Recent progress of aptamer‒drug conjugates in cancer therapy. Acta Pharm. Sin. B13, 1358–1370 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen, C. et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl. Acad. Sci. USA105, 15908–15913 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chu, T. et al. Aptamer: toxin conjugates that specifically target prostate tumor cells. Cancer Res.66, 5989–5992 (2006). [DOI] [PubMed] [Google Scholar]
- 55.McNamara, J. et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol.24, 1005–1015 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Ma, Y. et al. Advancing targeted combination chemotherapy in triple negative breast cancer: nucleolin aptamer-mediated controlled drug release. J. Transl. Med.22, 604 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gandotra, R., Wu, H., Kuo, F., Lee, M. & Lee, G. Advancing the diagnosis of periprosthetic joint infections: integrated microfluidic platform for alpha-defensins-specific aptamer selection and its analytical applications. ACS Sens.9, 1775–1784 (2024). [DOI] [PubMed] [Google Scholar]
- 58.Oh, S. et al. Improving aptamer selection efficiency through volume dilution, magnetic concentration, and continuous washing in microfluidic channels. Anal. Chem.83, 6883–6889 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blind, M. & Blank, M. Aptamer selection technology and recent advances. Mol. Ther. Nucleic Acids4, e223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jiehua, Z. & John, R. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov.16, 181–202 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luca, M., Francesca Anna, C., Grazisa, R. & Federica, C. An overview of structural approaches to study therapeutic RNAs. Front. Mol. Biosci.9, 1044126 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paola, A., Soumen, K., Cristian, R.-A. & Gabriel, L.-B. Aptamers: novel therapeutics and potential role in neuro-oncology. Cancers12, 2889 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farzana, Y., Hana, S., Nasir, J., Moon Suk, K. & Sangdun, C. Therapeutic interventions into innate immune diseases by means of aptamers. Pharmaceutics12, 955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Amy D, G., Douglas R, D. & Nebojsa, J. Embracing proteins: structural themes in aptamer-protein complexes. Curr. Opin. Struct. Biol.36, 122–132 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Weihong, T., Michael J, D. & Jianhui, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev.113, 2842–2862 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuanyuan, Y. et al. Molecular selection, modification and development of therapeutic oligonucleotide aptamers. Int J. Mol. Sci.17, 358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chao, L. et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med.21, 288–294 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fabio M, S., Paolo, M. & Carlotta, G. More DNA-aptamers for small drugs: a capture-SELEX coupled with surface plasmon resonance and high-throughput sequencing. ACS Comb. Sci.17, 326–333 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Annamaria, R. & Maria C, D. Small-molecule binding aptamers: selection strategies, characterization, and applications. Front Chem.4, 14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lucy F, Y., Melissa, L., Nataly, K. & Suzie H, P. Aptamers 101: aptamer discovery and in vitro applications in biosensors and separations. Chem. Sci.14, 4961–4978 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shingo, S. SELEX-based DNA aptamer selection: a perspective from the advancement of separation techniques. Anal. Sci.37, 17–26 (2020). [DOI] [PubMed] [Google Scholar]
- 72.Jinpeng, W. et al. Particle display: a quantitative screening method for generating high-affinity aptamers. Angew. Chem. Int. Ed.53, 4796–4801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu, D., Gordon, C. K. L., Shin, J. H., Eisenstein, M. & Soh, H. T. Directed evolution of aptamer discovery technologies. Acc. Chem. Res.55, 685–695 (2022). [DOI] [PubMed] [Google Scholar]
- 74.Yu, Q. et al. Exploring the role of adipokines in exercise-induced inhibition of tumor growth. Sports Med. Health Sci.7, 143–156 (2025). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gordon, C. K. L. et al. Click-particle display for base-modified aptamer discovery. ACS Chem. Biol.14, 2652–2662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dimitriou, R. & Giannoudis, P. V. The genetic profile of bone repair. Clin. Cases Miner. Bone Metab.10, 19–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shunmugam, G. et al. Recombinant human bone morphogenetic protein-2 for treatment of open tibial fractures: a prospective, controlled, randomized study of four hundred and fifty patients. J. Bone Jt. Surg.84, 2123–2134 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Joseph, B. Jr et al. Physiological challenges of bone repair. J. Orthop. Trauma26, 708–711 (2012). [DOI] [PubMed] [Google Scholar]
- 79.Novicoff, W. M. et al. Critical analysis of the evidence for current technologies in bone-healing and repair. J. Bone Jt. Surg.90, 85–91 (2008). [DOI] [PubMed] [Google Scholar]
- 80.Burchardt, H. The biology of bone graft repair. Clin. Orthop. Relat. Res.174, 28–42 (1983). [PubMed] [Google Scholar]
- 81.Roland, B. & Georges, R. Targeting the Wnt/beta-catenin pathway to regulate bone formation in the adult skeleton. Endocrinology148, 2635–2643 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Liu, Q. et al. Advances in the application of bone morphogenetic proteins and their derived peptides in bone defect repair. Composites B262, 110805 (2023). [Google Scholar]
- 83.Wein, M. N. & Kronenberg, H. M. Regulation of bone remodeling by parathyroid hormone. Cold Spring Harb Perspect Med.8, a031237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jilka, R. L. Molecular and cellular mechanisms of the anabolic effect of intermittent PTH. Bone40, 1434–1446 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yongmei, W. et al. IGF-I receptor is required for the anabolic actions of parathyroid hormone on bone. J. Bone Min. Res.22, 1329–1337 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hurley, M. M. et al. Impaired bone anabolic response to parathyroid hormone in Fgf2–/– and Fgf2+/– mice. Biochem. Biophys. Res. Commun.341, 989–994 (2006). [DOI] [PubMed] [Google Scholar]
- 87.Bikle, D. D. et al. Insulin-like growth factor I is required for the anabolic actions of parathyroid hormone on mouse bone. J. Bone Min. Res.17, 1570–1578 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Dobnig, H. & Turner, R. T. Evidence that intermittent treatment with parathyroid hormone increases bone formation in adult rats by activation of bone lining cells. Endocrinology136, 3632–3638 (1995). [DOI] [PubMed] [Google Scholar]
- 89.Bente, L., Serge, F. & David W, D. Bone modeling and remodeling: potential as therapeutic targets for the treatment of osteoporosis. Ther. Adv. Musculoskelet. Dis.8, 225–235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Balani, D. H., Ono, N. & Kronenberg, H. M. Parathyroid hormone regulates fates of murine osteoblast precursors in vivo. J. Clin. Investig.127, 3327–3338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yi, F. et al. Parathyroid hormone directs bone marrow mesenchymal cell fate. Cell Metab.25, 661–672 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jinhu, X. et al. Osteocyte-derived RANKL is a critical mediator of the increased bone resorption caused by dietary calcium deficiency. Bone66, 146–154 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiangwei, W. et al. Inhibition of Sca-1-positive skeletal stem cell recruitment by alendronate blunts the anabolic effects of parathyroid hormone on bone remodeling. Cell Stem Cell7, 571–580 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gaurav, L. et al. BMP-9 induced osteogenic differentiation of mesenchymal stem cells: molecular mechanism and therapeutic potential. Curr. Gene Ther.11, 229–240 (2011). [DOI] [PubMed] [Google Scholar]
- 95.Hongwei, C. et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J. Bone Jt. Surg.85, 1544–1552 (2003). [DOI] [PubMed] [Google Scholar]
- 96.Schoonraad, S. A., Trombold, M. L. & Bryant, S. J. The effects of stably tethered BMP-2 on MC3T3-E1 preosteoblasts encapsulated in a PEG hydrogel. Biomacromolecules22, 1065–1079 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hanada, K., Dennis, J. E. & Caplan, A. I. Stimulatory effects of basic fibroblast growth factor and bone morphogenetic protein-2 on osteogenic differentiation of rat bone marrow-derived mesenchymal stem cells. J. Bone Min. Res.12, 1606–1614 (1997). [DOI] [PubMed] [Google Scholar]
- 98.Naoki, M. et al. Enhancement of osteoblastic differentiation of mesenchymal stromal cells cultured by selective combination of bone morphogenetic protein-2 (BMP-2) and fibroblast growth factor-2 (FGF-2). J. Tissue Eng. Regen. Med.1, 306–313 (2007). [DOI] [PubMed] [Google Scholar]
- 99.Behnoush, K. et al. Regeneration of bone using nanoplex delivery of FGF-2 and BMP-2 genes in diaphyseal long bone radial defects in a diabetic rabbit model. J. Control. Release248, 53–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang, L. L. et al. Bioactive gelatin cryogels with BMP-2 biomimetic peptide and VEGF: a potential scaffold for synergistically induced osteogenesis. Chin. Chem. Lett.33, 1956–1962 (2022). [Google Scholar]
- 101.Winkler, T., Sass, F. A., Duda, G. N. & Schmidt-Bleek, K. A review of biomaterials in bone defect healing, remaining shortcomings and future opportunities for bone tissue engineering: the unsolved challenge. Bone Jt. Res.7, 232–243 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lin, S. et al. Orchestration of energy metabolism and osteogenesis by Mg facilitates low-dose BMP-2-driven regeneration. Bioact. Mater.18, 116–127 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hill, T., Taketo, M., Birchmeier, W. & Hartmann, C. Multiple roles of mesenchymal beta-catenin during murine limb patterning. Development.133, 1219–1229 (2006). [DOI] [PubMed] [Google Scholar]
- 104.Hill, T., Später, D., Taketo, M., Birchmeier, W. & Hartmann, C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell8, 727–738 (2005). [DOI] [PubMed] [Google Scholar]
- 105.Day, T., Guo, X., Garrett-Beal, L. & Yang, Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell8, 739–750 (2005). [DOI] [PubMed] [Google Scholar]
- 106.Bennett, C. et al. Regulation of osteoblastogenesis and bone mass by Wnt10b. Proc. Natl. Acad. Sci. USA102, 3324–3329 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rawadi, G., Vayssière, B., Dunn, F., Baron, R. & Roman-Roman, S. BMP-2 controls alkaline phosphatase expression and osteoblast mineralization by a Wnt autocrine loop. J. Bone Miner. Res.18, 1842–1853 (2003). [DOI] [PubMed] [Google Scholar]
- 108.Simonet, W. et al. Osteoprotegerin: a novel secreted protein involved in the regulation of bone density. Cell89, 309–319 (1997). [DOI] [PubMed] [Google Scholar]
- 109.Eghbali-Fatourechi, G. et al. Role of RANK ligand in mediating increased bone resorption in early postmenopausal women. J. Clin. Investig.111, 1221–1230 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhang, N. et al. Pros and cons of denosumab treatment for osteoporosis and implication for RANKL aptamer therapy. Front. Cell Dev. Biol.8, 325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Holmen, S. et al. Essential role of beta-catenin in postnatal bone acquisition. J. Biol. Chem.280, 21162–21168 (2005). [DOI] [PubMed] [Google Scholar]
- 112.Glass, D. et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev. Cell8, 751–764 (2005). [DOI] [PubMed] [Google Scholar]
- 113.Diarra, D. et al. Dickkopf-1 is a master regulator of joint remodeling. Nat. Med.13, 156–163 (2007). [DOI] [PubMed] [Google Scholar]
- 114.Kolar, P. et al. The early fracture hematoma and its potential role in fracture healing. Tissue Eng. B16, 427–434 (2010). [DOI] [PubMed] [Google Scholar]
- 115.Pape, H. et al. Trauma-induced inflammation and fracture healing. J. Orthop. Trauma24, 522–525 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Grundnes, O. & Reikerås, O. The importance of the hematoma for fracture healing in rats. Acta Orthop. Scand.64, 340–342 (1993). [DOI] [PubMed] [Google Scholar]
- 117.Mizuno, K. et al. The osteogenetic potential of fracture haematoma. Subperiosteal and intramuscular transplantation of the haematoma. J. Bone Jt. Surg.72, 822–829 (1990). [DOI] [PubMed] [Google Scholar]
- 118.Young, R. Cell proliferation and specialization during endochondral osteogenesis in young rats. J. Cell Biol.14, 357–370 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bielby, R., Jones, E. & McGonagle, D. The role of mesenchymal stem cells in maintenance and repair of bone. Injury38, S26–S32 (2007). [DOI] [PubMed] [Google Scholar]
- 120.Bianco, P., Robey, P. & Simmons, P. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell2, 313–319 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Delorme, B., Chateauvieux, S. & Charbord, P. The concept of mesenchymal stem cells. Regen. Med.1, 497–509 (2006). [DOI] [PubMed] [Google Scholar]
- 122.Oe, K. et al. An in vitro study demonstrating that haematomas found at the site of human fractures contain progenitor cells with multilineage capacity. J. Bone Jt. Surg.89, 133–138 (2007). [DOI] [PubMed] [Google Scholar]
- 123.Sun, T. et al. In situ bone regeneration with sequential delivery of aptamer and BMP2 from an ECM-based scaffold fabricated by cryogenic free-form extrusion. Bioact. Mater.6, 4163–4175 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schlickewei, C. et al. Current and future concepts for the treatment of impaired fracture healing. Int. J. Mol. Sci.20, 5805 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Opal, S. Phylogenetic and functional relationships between coagulation and the innate immune response. Crit. Care Med.28, S77–S80 (2000). [DOI] [PubMed] [Google Scholar]
- 126.Pajarinen, J. et al. Mesenchymal stem cell-macrophage crosstalk and bone healing. Biomaterials196, 80–89 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schlundt, C. et al. The multifaceted roles of macrophages in bone regeneration: a story of polarization, activation and time. Acta biomater.133, 46–57 (2021). [DOI] [PubMed] [Google Scholar]
- 128.Li, Z., Wang, Y., Li, S. & Li, Y. Exosomes derived from M2 macrophages facilitate osteogenesis and reduce adipogenesis of BMSCs. Front. Endocrinol.12, 680328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ying, W. et al. MiR-690, an exosomal-derived miRNA from M2-polarized macrophages, improves insulin sensitivity in obese mice. Cell Metab.33, 781–790.e785 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kang, M. et al. Bone regeneration is mediated by macrophage extracellular vesicles. Bone141, 115627 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xiong, Y. et al. M2 Macrophagy-derived exosomal miRNA-5106 induces bone mesenchymal stem cells towards osteoblastic fate by targeting salt-inducible kinase 2 and 3. J. Nanobiotechnology18, 66 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 132.Shou, J. et al. 3WJ RNA nanoparticles-aptamer functionalized exosomes from M2 macrophages target BMSCs to promote the healing of bone fractures. Stem Cells Transl. Med.12, 758–774 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wiklander, O. et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles4, 26316 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kang, M., Jordan, V., Blenkiron, C. & Chamley, L. Biodistribution of extracellular vesicles following administration into animals:a systematic review. J. Extracell. Vesicles10, e12085 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cho, T., Gerstenfeld, L. & Einhorn, T. Differential temporal expression of members of the transforming growth factor beta superfamily during murine fracture healing. J. Bone Miner. Res.: Off. J. Am. Soc. Bone Miner. Res.17, 513–520 (2002). [DOI] [PubMed] [Google Scholar]
- 136.Einhorn, T. The cell and molecular biology of fracture healing. Clin. Orthop. Relat. Res.355, S7–S21 (1998). [DOI] [PubMed] [Google Scholar]
- 137.Oni, O. Early histological and ultrastructural changes in medullary fracture callus. J. bone Jt. Surg. Am. Vol.74, 633–634 (1992). [PubMed] [Google Scholar]
- 138.Capone, A., Marongiu, G. & Sirianni, R. L’osteogenesi riparativa. LO SCALPELLO-OTODI Educ.29, 2–9 (2015). [Google Scholar]
- 139.Dimitriou, R., Tsiridis, E. & Giannoudis, P. Current concepts of molecular aspects of bone healing. Injury36, 1392–1404 (2005). [DOI] [PubMed] [Google Scholar]
- 140.Gerstenfeld, L. et al. Three-dimensional reconstruction of fracture callus morphogenesis. J. Histochem. Cytochem.54, 1215–1228 (2006). [DOI] [PubMed] [Google Scholar]
- 141.Rockey, W. et al. Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling. Nucleic Acid Ther.21, 299–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ye, M. et al. Generating aptamers by cell-SELEX for applications in molecular medicine. Int. J. Mol. Sci.13, 3341–3353 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sefah, K., Shangguan, D., Xiong, X., O'Donoghue, M. & Tan, W. Development of DNA aptamers using cell-SELEX. Nat. Protoc.5, 1169–1185 (2010). [DOI] [PubMed] [Google Scholar]
- 144.Herman, S. et al. Induction of osteoclast-associated receptor, a key osteoclast costimulation molecule, in rheumatoid arthritis. Arthritis Rheum.58, 3041–3050 (2008). [DOI] [PubMed] [Google Scholar]
- 145.Lotinun, S. et al. Osteoclast-specific cathepsin K deletion stimulates S1P-dependent bone formation. J. Clin. Investig.123, 666–681 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zernik, J., Twarog, K. & Upholt, W. Regulation of alkaline phosphatase and alpha 2(I) procollagen synthesis during early intramembranous bone formation in the rat mandible. Differentiation44, 207–215 (1990). [DOI] [PubMed] [Google Scholar]
- 147.Fukushima, N., Hiraoka, K., Shirachi, I., Kojima, M. & Nagata, K. Isolation and characterization of a novel peptide, osteoblast activating peptide (OBAP), associated with osteoblast differentiation and bone formation. Biochem. Biophys. Res. Commun.400, 157–163 (2010). [DOI] [PubMed] [Google Scholar]
- 148.Ishikawa, S. et al. Involvement of FcRgamma in signal transduction of osteoclast-associated receptor (OSCAR). Int. Immunol.16, 1019–1025 (2004). [DOI] [PubMed] [Google Scholar]
- 149.Barnes, G. L., Kostenuik, P. J., Gerstenfeld, L. C. & Einhorn, T. A. Growth factor regulation of fracture repair. J. Bone Miner. Res.14, 1805–1815 (1999). [DOI] [PubMed] [Google Scholar]
- 150.Gerstenfeld, L. C., Cullinane, D. M., Barnes, G. L., Graves, D. T. & Einhorn, T. A. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J. Cell Biochem88, 873–884 (2003). [DOI] [PubMed] [Google Scholar]
- 151.Cai, B. L. et al. Construction of multi-module RNA nanoparticles harboring miRNA, AIE, and CH6 aptamer for bone targeting and bone anabolic therapy. Adv. Funct. Mater.34, 2312260 (2024). [Google Scholar]
- 152.Holstein, J. H. et al. Endostatin inhibits callus remodeling during fracture healing in mice. J. Orthop. Res.31, 1579–1584 (2013). [DOI] [PubMed] [Google Scholar]
- 153.Mark, M., Thomas, H., Lutz, C. & Peter, A. Revascularisation during fracture healing with soft tissue injury. Arch. Orthop. Trauma Surg.128, 1159–1165 (2007). [DOI] [PubMed] [Google Scholar]
- 154.Rowe, N. M. et al. Angiogenesis during mandibular distraction osteogenesis. Ann. Plast. Surg.42, 470–475 (1999). [DOI] [PubMed] [Google Scholar]
- 155.Einhorn, T. A. & Lee, C. A. Bone regeneration: new findings and potential clinical applications. J. Am. Acad. Orthop. Surg.9, 157–165 (2001). [DOI] [PubMed] [Google Scholar]
- 156.Gerber, H. Angiogenesis and bone growth. Trends Cardiovasc. Med.10, 223–228 (2000). [DOI] [PubMed] [Google Scholar]
- 157.Sadegh, D. et al. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): a review. Biosens. Bioelectron.110, 23–37 (2018). [DOI] [PubMed] [Google Scholar]
- 158.Ze, R. et al. GIT1Y321 phosphorylation is required for ERK1/2- and PDGF-dependent VEGF secretion from osteoblasts to promote angiogenesis and bone healing. Int J. Mol. Med.30, 819–825 (2012). [DOI] [PubMed] [Google Scholar]
- 159.Toru, Y. et al. Binding and structural properties of DNA aptamers with VEGF-A-Mimic Activity. Mol. Ther. Nucleic Acids19, 1145–1152 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang, L., Junqi, L. & Qianzhou, J. Inflammasomes in alveolar bone loss. Front. Immunol.12, 691013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bo, Y. & Cun-Yu, W. Osteoporosis and periodontal diseases–an update on their association and mechanistic links. Periodontol 200089, 99–113 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.I, D., S, C. & R, D. P. Ridge preservation: what is it and when should it be considered. Aust. Dent. J.53, 11–21 (2008). [DOI] [PubMed] [Google Scholar]
- 163.Lars, S., Ann, W., Lambros, K. & Thorkild, K. Bone healing and soft tissue contour changes following single-tooth extraction: a clinical and radiographic 12-month prospective study. Int J. Periodontics Restor. Dent.23, 313–323 (2003). [PubMed] [Google Scholar]
- 164.Su, A., Mitchell, D. & Menegaz, R. The effect of tooth loss on craniofacial morphology. FASEB J.35, 03825 (2021). [Google Scholar]
- 165.Christensen, L. V. & Ziebert, G. J. Effects of experimental loss of teeth on the temporomandibular joint. J. Oral. Rehabil.13, 587–598 (1986). [DOI] [PubMed] [Google Scholar]
- 166.E, K., C, H. & R, G. How dentition status and masticatory function affect nutrient intake. J. Am. Dent. Assoc.129, 1261–1269 (1998). [DOI] [PubMed] [Google Scholar]
- 167.Binghan, Z. et al. Notch signaling pathway: architecture, disease, and therapeutics. Signal Transduct. Target Ther.7, 95 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ana, D. K. et al. Notch signalling cascade and proinflammatory mediators in peri-implant lesions with different RANKL/OPG ratios–an observational study. J. Periodontal Res.58, 360–368 (2023). [DOI] [PubMed] [Google Scholar]
- 169.Nadja, N. et al. Notch signaling pathway in apical periodontitis: correlation with bone resorption regulators and proinflammatory cytokines. J. Endod.45, 123–128 (2018). [DOI] [PubMed] [Google Scholar]
- 170.Haruna, N. et al. The Wnt/β-catenin signaling pathway has a healing ability for periapical periodontitis. Sci. Rep.11, 19673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Putri Ayu, J. et al. Overview on postmenopausal osteoporosis and periodontitis: the therapeutic potential of phytoestrogens against alveolar bone loss. Front. Pharm.14, 1120457 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chu-Han, C., Li-Ru, C. & Kuo-Hu, C. Osteoporosis due to hormone imbalance: an overview of the effects of estrogen deficiency and glucocorticoid overuse on bone turnover. Int. J. Mol. Sci.23, 1376 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Fengzhen, L. et al. Treatment of inflammatory bone loss in periodontitis by stem cell-derived exosomes. Acta Biomater.141, 333–343 (2022). [DOI] [PubMed] [Google Scholar]
- 174.Mei-Hua, C. et al. HIF-1α activator DMOG inhibits alveolar bone resorption in murine periodontitis by regulating macrophage polarization. Int. Immunopharmacol.99, 107901 (2021). [DOI] [PubMed] [Google Scholar]
- 175.Zijiao, Z. et al. Pirfenidone inhibits alveolar bone loss in ligature-induced periodontitis by suppressing the NF-κB signaling pathway in mice. Int. J. Mol. Sci.24, 8682 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Xinge, W. et al. Asperuloside prevents peri-implantitis via suppression of NF-κB and ERK1/2 on Rats. Pharmaceuticals15, 1027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Iva, M. et al. The down-regulation of Notch 1 signaling contributes to the severity of bone loss in aggressive periodontitis. J. Periodontol.91, 554–561 (2019). [DOI] [PubMed] [Google Scholar]
- 178.Iva, M. et al. Notch down-regulation and inflammatory cytokines and RANKL overexpression involvement in peri-implant mucositis and peri-implantitis: a cross-sectional study. Clin. Oral. Implants Res.32, 1496–1505 (2021). [DOI] [PubMed] [Google Scholar]
- 179.Purwaningrum, M., Giachelli, C. M., Osathanon, T., Rattanapuchpong, S. & Sawangmake, C. Dissecting specific Wnt components governing osteogenic differentiation potential by human periodontal ligament stem cells through interleukin-6. Sci. Rep.13, 9055 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lihua, L. et al. IL-37 alleviates alveolar bone resorption and inflammatory response through the NF-κB/NLRP3 signaling pathway in male mice with periodontitis. Arch. Oral. Biol.147, 105629 (2023). [DOI] [PubMed] [Google Scholar]
- 181.Jia-He, W., Shu-Juan, Z., Jing, X. & Chen-Chen, Z.Latest findings on NOD-like receptor family pyrin domain containing protein 3 inflammasome and bone and articular diseases. Sichuan Da Xue Xue Bao Yi Xue Ban54, 679–684 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Xiangwei, K. et al. GSK3β is a checkpoint for TNF-α-mediated impaired osteogenic differentiation of mesenchymal stem cells in inflammatory microenvironments. Biochim. Biophys. Acta1830, 5119–5129 (2013). [DOI] [PubMed] [Google Scholar]
- 183.Erik, S. et al. Molecular mechanism of LPS-induced TNF-α biosynthesis in polarized human macrophages. Mol. Immunol.93, 206–215 (2017). [DOI] [PubMed] [Google Scholar]
- 184.T, N. et al. Protein expression and functional difference of membrane-bound and soluble receptor activator of NF-kappaB ligand: modulation of the expression by osteotropic factors and cytokines. Biochem. Biophys. Res. Commun.275, 768–775 (2000). [DOI] [PubMed] [Google Scholar]
- 185.Zhenhua, L. et al. DNA nanostructure-based universal microarray platform for high-efficiency multiplex bioanalysis in biofluids. ACS Appl. Mater. Interfaces6, 17944–17953 (2014). [DOI] [PubMed] [Google Scholar]
- 186.Yanli, W. et al. DNA nanostructure-based Interfacial engineering for PCR-free ultrasensitive electrochemical analysis of microRNA. Sci. Rep.3, 1508 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mi, Z. et al. Effect of tetrahedral DNA nanostructures on proliferation and osteo/odontogenic differentiation of dental pulp stem cells via activation of the notch signaling pathway. Nanomedicine14, 1227–1236 (2018). [DOI] [PubMed] [Google Scholar]
- 188.Mi, Z. et al. Effect of tetrahedral DNA nanostructures on proliferation and osteogenic differentiation of human periodontal ligament stem cells. Cell Prolif.52, e12566 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Qi, Z. et al. Anti-inflammatory and antioxidative effects of tetrahedral dna nanostructures via the modulation of macrophage responses. ACS Appl. Mater. Interfaces10, 3421–3430 (2018). [DOI] [PubMed] [Google Scholar]
- 190.Mi, Z. et al. The protective effect of tetrahedral framework nucleic acids on periodontium under inflammatory conditions. Bioact. Mater.6, 1676–1688 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pinxue, L. et al. Chitosan hydrogel/3D-printed poly(ε-caprolactone) hybrid scaffold containing synovial mesenchymal stem cells for cartilage regeneration based on tetrahedral framework nucleic acid recruitment. Biomaterials278, 121131 (2021). [DOI] [PubMed] [Google Scholar]
- 192.Wenjuan, M. et al. Biomimetic nanoerythrosome-coated aptamer-DNA tetrahedron/maytansine conjugates: pH-Responsive and targeted cytotoxicity for HER2-positive breast cancer. Adv. Mater.34, e2109609 (2022). [DOI] [PubMed] [Google Scholar]
- 193.Wei, W. et al. Antifibrotic effects of tetrahedral framework nucleic acids by inhibiting macrophage polarization and macrophage-myofibroblast transition in bladder remodeling. Adv. Health. Mater.12, e2203076 (2023). [DOI] [PubMed] [Google Scholar]
- 194.Xi, Y. et al. Tetrahedral framework nucleic acids inhibit muscular mitochondria-mediated apoptosis and ameliorate muscle atrophy in sarcopenia. Nano Lett.23, 8816–8826 (2023). [DOI] [PubMed] [Google Scholar]
- 195.Yangxue, Y. et al. A mitochondrial nanoguard modulates redox homeostasis and bioenergy metabolism in diabetic peripheral neuropathy. ACS Nano17, 22334–22354 (2023). [DOI] [PubMed] [Google Scholar]
- 196.Yuxuan, Z. et al. Effects of puerarin-loaded tetrahedral framework nucleic acids on osteonecrosis of the femoral head. Small19, e2302326 (2023). [DOI] [PubMed] [Google Scholar]
- 197.Sophia Fox, A. J., Bedi, A. & Rodeo, S. A. The basic science of articular cartilage: structure, composition, and function. Sports Health1, 461–468 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Buckwalter, J. A., Mankin, H. J. & Grodzinsky, A. J. Articular cartilage and osteoarthritis. Instr. Course Lect.54, 465–480 (2005). [PubMed] [Google Scholar]
- 199.Silvia, L. & Henning, M. Bioinspired scaffolds for osteochondral regeneration. Tissue Eng. A20, 2052–2076 (2014). [DOI] [PubMed] [Google Scholar]
- 200.Makris, E. A., Hadidi, P. & Athanasiou, K. A. The knee meniscus: structure-function, pathophysiology, current repair techniques, and prospects for regeneration. Biomaterials32, 7411–7431 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Fox, A. J., Bedi, A. & Rodeo, S. A. The basic science of human knee menisci: structure, composition, and function. Sports Health4, 340–351 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Arnoczky, S. P. & Warren, R. F. Microvasculature of the human meniscus. Am. J. Sports Med.10, 90–95 (1982). [DOI] [PubMed] [Google Scholar]
- 203.Danzig, L., Resnick, D., Gonsalves, M., Akeson, W. H. Blood supply to the normal and abnormal menisci of the human knee. Clin. Orthop. Relat. Res. 172, 271–276 (1983). [PubMed]
- 204.Davies, D. V. & Edwards, D. A. The blood supply of the synovial membrane and intra-articular structures. Ann. R. Coll. Surg. Engl.2, 142–146 (1948). [PMC free article] [PubMed] [Google Scholar]
- 205.Day, B., Mackenzie, W. G., Shim, S. S. & Leung, G. The vascular and nerve supply of the human meniscus. Arthroscopy1, 58–62 (1985). [DOI] [PubMed] [Google Scholar]
- 206.Scapinelli, R. Studies on the vasculature of the human knee joint. Acta Anat.70, 305–331 (1968). [DOI] [PubMed] [Google Scholar]
- 207.Travascio, F. & Jackson, A. R. The nutrition of the human meniscus: a computational analysis investigating the effect of vascular recession on tissue homeostasis. J. Biomech.61, 151–159 (2017). [DOI] [PubMed] [Google Scholar]
- 208.Vadodaria, K., Kulkarni, A., Santhini, E. & Vasudevan, P. Materials and structures used in meniscus repair and regeneration: a review. Biomedicine9, 2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hao, L. et al. Advanced polymer-based drug delivery strategies for meniscal regeneration. Tissue Eng. B Rev.27, 266–293 (2020). [DOI] [PubMed] [Google Scholar]
- 210.Zong, L. et al. Biomechanically, structurally and functionally meticulously tailored polycaprolactone/silk fibroin scaffold for meniscus regeneration. Theranostics10, 5090–5106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zonghuan, L. & Aixi, Y. Diagnostic value of a PCR-based technique for prosthetic joint infection. J. Clin. Microbiol.52, 2281–2282 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Renke, H. et al. Better choice of the type of specimen used for untargeted metagenomic sequencing in the diagnosis of periprosthetic joint infections. Bone Jt. J.103-B, 923–930 (2021). [DOI] [PubMed] [Google Scholar]
- 213.M, M. et al. Role of universal 16S rRNA gene PCR and sequencing in diagnosis of prosthetic joint infection. J. Clin. Microbiol.50, 583–589 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Xiaohua, L., Xiaobing, J. & Peter X, M. Nanofibrous hollow microspheres self-assembled from star-shaped polymers as injectable cell carriers for knee repair. Nat. Mater.10, 398–406 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Peck, Y., He, P., Chilla, G. S., Poh, C. L. & Wang, D. A. A preclinical evaluation of an autologous living hyaline-like cartilaginous graft for articular cartilage repair: a pilot study. Sci. Rep.5, 16225 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Huey, D. J., Hu, J. C. & Athanasiou, K. A. Unlike bone, cartilage regeneration remains elusive. Science338, 917–921 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xiaoxia, H. et al. A difunctional regeneration scaffold for knee repair based on aptamer-directed cell recruitment. Adv. Mater.29, 1605235 (2017). [DOI] [PubMed] [Google Scholar]
- 218.Andrew D, P., Russell F, W. & Scott A, R. Basic science of articular cartilage and osteoarthritis. Clin. Sports Med.24, 1–12 (2005). [DOI] [PubMed] [Google Scholar]
- 219.Buckwalter, J. A. & Mankin, H. J. Articular cartilage: tissue design and chondrocyte-matrix interactions. Instr. Course Lect.47, 477–486 (1998). [PubMed] [Google Scholar]
- 220.Zhonggang, H. et al. Characterization and target identification of a DNA aptamer that labels pluripotent stem cells. Cell Res.25, 390–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hao, L. et al. Integrated bioactive scaffold with aptamer-targeted stem cell recruitment and growth factor-induced pro-differentiation effects for anisotropic meniscal regeneration. Bioeng. Transl. Med.7, e10302 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Zhong, C. et al. Repairing avascular meniscal lesions by recruiting endogenous targeted cells through bispecific synovial-meniscal aptamers. Am. J. Sports Med.51, 1177–1193 (2023). [DOI] [PubMed] [Google Scholar]
- 223.Forlino, A. & Marini, J. Osteogenesis imperfecta. Lancet387, 1657–1671 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pauli, R. Achondroplasia: a comprehensive clinical review. Orphanet J. Rare Dis.14, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wu, B. et al. A narrative review of diabetic bone disease: characteristics, pathogenesis, and treatment. Front. Endocrinol.13, 1052592 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Terpos, E. et al. Treatment of multiple myeloma-related bone disease: recommendations from the bone working group of the International Myeloma Working Group. Lancet Oncol.22, e119–e130 (2021). [DOI] [PubMed] [Google Scholar]
- 227.Buckwalter, J. & Martin, J. Sports and osteoarthritis. Curr. Opin. Rheumatol.16, 634–639 (2004). [DOI] [PubMed] [Google Scholar]
- 228.Wawrzyniak, A. & Balawender, K. Structural and metabolic changes in bone. Animals12, 1946 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Xiao, W., Li, S., Pacios, S., Wang, Y. & Graves, D. Bone remodeling under pathological conditions. Front. Oral. Biol.18, 17–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Consensus conference: osteoporosis. JAMA. 252, 799-802 (1984). [PubMed]
- 231.Armas, L. & Recker, R. Pathophysiology of osteoporosis: new mechanistic insights. Endocrinol. Metab. Clin. North Am.41, 475–486 (2012). [DOI] [PubMed] [Google Scholar]
- 232.NIH Consensus development panel on osteoporosis prevention, diagnosis, and therapy, March 7-29, 2000: highlights of the conference. South Med. J. 94, 569–573 (2001). [PubMed]
- 233.Lane, N. Epidemiology, etiology, and diagnosis of osteoporosis. Am. J. Obstet. Gynecol.194, S3–S11 (2006). [DOI] [PubMed] [Google Scholar]
- 234.Clynes, M. et al. The epidemiology of osteoporosis. Br. Med. Bull.133, 105–117 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.van Staa, T., Dennison, E., Leufkens, H. & Cooper, C. Epidemiology of fractures in England and Wales. Bone29, 517–522 (2001). [DOI] [PubMed] [Google Scholar]
- 236.Gosset, A., Pouillès, J. & Trémollieres, F. Menopausal hormone therapy for the management of osteoporosis. Best. Pract. Res. Clin. Endocrinol. Metab.35, 101551 (2021). [DOI] [PubMed] [Google Scholar]
- 237.Akhter, M., Lappe, J., Davies, K. & Recker, R. Transmenopausal changes in the trabecular bone structure. Bone41, 111–116 (2007). [DOI] [PubMed] [Google Scholar]
- 238.Cooper, D. et al. Age-dependent change in the 3D structure of cortical porosity at the human femoral midshaft. Bone40, 957–965 (2007). [DOI] [PubMed] [Google Scholar]
- 239.Cardoso, L. et al. Osteocyte apoptosis controls activation of intracortical resorption in response to bone fatigue. J. Bone Miner. Res.24, 597–605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bonewald, L. & Johnson, M. Osteocytes, mechanosensing and Wnt signaling. Bone42, 606–615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Garnero, P., Sornay-Rendu, E., Chapuy, M. & Delmas, P. Increased bone turnover in late postmenopausal women is a major determinant of osteoporosis. J. Bone Miner. Res.11, 337–349 (1996). [DOI] [PubMed] [Google Scholar]
- 242.Riggs, B., Melton, L. & O'Fallon, W. Drug therapy for vertebral fractures in osteoporosis: evidence that decreases in bone turnover and increases in bone mass both determine antifracture efficacy. Bone18, 197S–201S (1996). [DOI] [PubMed] [Google Scholar]
- 243.Boivin, G. et al. The role of mineralization and organic matrix in the microhardness of bone tissue from controls and osteoporotic patients. Bone43, 532–538 (2008). [DOI] [PubMed] [Google Scholar]
- 244.Boivin, G. & Meunier, P. The mineralization of bone tissue: a forgotten dimension in osteoporosis research. Osteoporos. Int.14, S19–S24 (2003). [DOI] [PubMed] [Google Scholar]
- 245.Leblanc, A., Schneider, V., Evans, H., Engelbretson, D. & Krebs, J. Bone mineral loss and recovery after 17 weeks of bed rest. J. Bone Miner. Res.5, 843–850 (1990). [DOI] [PubMed] [Google Scholar]
- 246.Matheson, G. et al. Stress fractures in athletes. A study of 320 cases. Am. J. Sports Med.15, 46–58 (1987). [DOI] [PubMed] [Google Scholar]
- 247.Khan, K. et al. Overuse injuries in classical ballet. Sports Med.19, 341–357 (1995). [DOI] [PubMed] [Google Scholar]
- 248.Herman, B., Cardoso, L., Majeska, R., Jepsen, K. & Schaffler, M. Activation of bone remodeling after fatigue: differential response to linear microcracks and diffuse damage. Bone47, 766–772 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mulcahy, L., Taylor, D., Lee, T. & Duffy, G. RANKL and OPG activity is regulated by injury size in networks of osteocyte-like cells. Bone48, 182–188 (2011). [DOI] [PubMed] [Google Scholar]
- 250.Schaffler, M., Choi, K. & Milgrom, C. Aging and matrix microdamage accumulation in human compact bone. Bone17, 521–525 (1995). [DOI] [PubMed] [Google Scholar]
- 251.Recker, R., Lappe, J., Davies, K. & Heaney, R. Bone remodeling increases substantially in the years after menopause and remains increased in older osteoporosis patients. J. Bone Miner. Res.19, 1628–1633 (2004). [DOI] [PubMed] [Google Scholar]
- 252.Russell, S. & Kahn, C. Endocrine regulation of ageing. Nat. Rev. Mol. Cell Biol.8, 681–691 (2007). [DOI] [PubMed] [Google Scholar]
- 253.Manolagas, S. From estrogen-centric to aging and oxidative stress: a revised perspective of the pathogenesis of osteoporosis. Endocr. Rev.31, 266–300 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Heaney, R. et al. Bone dimensional change with age: interactions of genetic, hormonal, and body size variables. Osteoporos. Int.7, 426–431 (1997). [DOI] [PubMed] [Google Scholar]
- 255.Ahlborg, H., Johnell, O., Turner, C., Rannevik, G. & Karlsson, M. Bone loss and bone size after menopause. N. Engl. J. Med.349, 327–334 (2003). [DOI] [PubMed] [Google Scholar]
- 256.Marie, P. J., Felsenberg, D. & Brandi, M. L. How strontium ranelate, via opposite effects on bone resorption and formation, prevents osteoporosis. Osteoporos. Int.22, 1659–1667 (2010). [DOI] [PubMed] [Google Scholar]
- 257.Häuselmann, H. & Rizzoli, R. A comprehensive review of treatments for postmenopausal osteoporosis. Osteoporos. Int.14, 2–12 (2003). [DOI] [PubMed] [Google Scholar]
- 258.Seeman, E. Reduced bone formation and increased bone resorption: rational targets for the treatment of osteoporosis. Osteoporos. Int.14, S2–S8 (2003). [DOI] [PubMed] [Google Scholar]
- 259.Etminan, M. Long-term use of oral bisphosphonates increases the risk of oesophageal but not gastric or colorectal cancer. Evid.-based Med.16, 28–29 (2011). [DOI] [PubMed] [Google Scholar]
- 260.Martin, T. & Sims, N. Osteoclast-derived activity in the coupling of bone formation to resorption. Trends Mol. Med.11, 76–81 (2005). [DOI] [PubMed] [Google Scholar]
- 261.Yuan-Wei, Z. et al. Diets intervene osteoporosis via gut-bone axis. Gut Microbes16, 2295432 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ye, T., Xinyi, K., Ling, Z. & Xin, X. The associations of gut microbiota, endocrine system and bone metabolism. Front. Microbiol.14, 1124945 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Luigi, B. et al. Nutrition and immune system: from the Mediterranean diet to dietary supplementary through the microbiota. Crit. Rev. Food Sci. Nutr.61, 3066–3090 (2020). [DOI] [PubMed] [Google Scholar]
- 264.Joan, S. et al. High-dose saccharin supplementation does not induce gut microbiota changes or glucose intolerance in healthy humans and mice. Microbiome9, 11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Zhi Qiang, L. et al. A decade of insight: bibliometric analysis of gut microbiota’s role in osteoporosis (2014-2024). Front. Med.11, 1409534 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Calum C, B. & Allan McI, M. Macrophages in intestinal homeostasis and inflammation. Immunol. Rev.260, 102–117 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Yuan-Wei, Z., Yan, W., Xiang-Fei, L., Xiao, C. & Jia-Can, S. Targeting the gut microbiota-related metabolites for osteoporosis: the inextricable connection of gut-bone axis. Ageing Res. Rev.94, 102196 (2024). [DOI] [PubMed] [Google Scholar]
- 268.Kyunghwa, B. et al. TNF-α upregulates sclerostin expression in obese mice fed a high-fat diet. J. Cell Physiol.229, 640–650 (2014). [DOI] [PubMed] [Google Scholar]
- 269.Hideki, K. et al. Osteocyte-related cytokines regulate osteoclast formation and bone resorption. Int. J. Mol. Sci.21, 5169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.N, U. et al. Interleukin (IL)-6 induction of osteoclast differentiation depends on IL-6 receptors expressed on osteoblastic cells but not on osteoclast progenitors. J. Exp. Med.182, 1461–1468 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Shashi, G. et al. Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J. Biol. Chem.289, 8706–8719 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ren, X. M., Gelinas, A. D., von Carlowitz, I., Janjic, N. & Pyle, A. M. Structural basis for IL-1α recognition by a modified DNA aptamer that specifically inhibits IL-1α signaling. Nat. Commun.8, 810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Erik W, O., Nick, J., Yuen Lai, S., Sachdev S, S. & Jean, G. A short DNA aptamer that recognizes TNFα and blocks its activity in vitro. ACS Chem. Biol.8, 170–178 (2012). [DOI] [PubMed] [Google Scholar]
- 274.Sung, H. J. et al. Inhibition of human neutrophil activity by an RNA aptamer bound to interleukin-8. Biomaterials35, 578–589 (2014). [DOI] [PubMed] [Google Scholar]
- 275.Ishiguro, A., Akiyama, T., Adachi, H., Inoue, J. & Nakamura, Y. Therapeutic potential of anti-interleukin-17A aptamer suppression of interleukin-17A signaling and attenuation of autoimmunity in two mouse models. Arthritis Rheum.63, 455–466 (2011). [DOI] [PubMed] [Google Scholar]
- 276.Frasca, D. & Blomberg, B. B. Inflammaging decreases adaptive and innate immune responses in mice and humans. Biogerontology17, 7–19 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Julio, P.-D., Francisco Javier, R.-O., Laura Maria, V.-P. & Angel, G. Evidence of the anti-inflammatory effects of probiotics and synbiotics in intestinal chronic diseases. Nutrients9, 555 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Haslin Madihah, H. & Suzana, M. A review of the preclinical and clinical studies on the role of the gut microbiome in aging and neurodegenerative diseases and its modulation. Front. Cell Neurosci.16, 1007166 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Claes, O. et al. Probiotics protect mice from ovariectomy-induced cortical bone loss. PLoS ONE9, e92368 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Ravinder, N. et al. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci. Rep.8, 12649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sébastien, L. et al. Short-chain fatty acids regulate systemic bone mass and protect from pathological bone loss. Nat. Commun.9, 55 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yong, F. & Oluf, P. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol.19, 55–71 (2020). [DOI] [PubMed] [Google Scholar]
- 283.Savignac, H. M., Tramullas, M., Kiely, B., Dinan, T. G. & Cryan, J. F. Bifidobacteria modulate cognitive processes in an anxious mouse strain. Behav. Brain Res.287, 59–72 (2015). [DOI] [PubMed] [Google Scholar]
- 284.Somayeh, A. N. A. et al. Lactobacilli and bifidobacteria ameliorate memory and learning deficits and oxidative stress in β-amyloid (1-42) injected rats. Appl. Physiol. Nutr. Metab.43, 718–726 (2018). [DOI] [PubMed] [Google Scholar]
- 285.Friedrich, L., Kostja, S., Burkhard, S., Dietmar, F. & Johanna M, G. Probiotic supplementation in patients with Alzheimer’s Dementia–an explorative intervention study. Curr. Alzheimer Res.15, 1106–1113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Xueqin, Y., Dongke, Y., Li, X., Hui, L. & Junrong, D. Probiotics modulate the microbiota-gut-brain axis and improve memory deficits in aged SAMP8 mice. Acta Pharm. Sin. B10, 475–487 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Mingjie, J. et al. Preparation of synbiotic milk powder and its effect on calcium absorption and the bone microstructure in calcium deficient mice. Food Funct.14, 3092–3106 (2023). [DOI] [PubMed] [Google Scholar]
- 288.Bartel, D. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell116, 281–297 (2004). [DOI] [PubMed] [Google Scholar]
- 289.Ambros, V. The functions of animal microRNAs. Nature431, 350–355 (2004). [DOI] [PubMed] [Google Scholar]
- 290.Bartel, D. MicroRNAs: target recognition and regulatory functions. Cell136, 215–233 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tang, P., Xiong, Q., Ge, W. & Zhang, L. The role of microRNAs in osteoclasts and osteoporosis. RNA Biol.11, 1355–1363 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ji, X., Chen, X. & Yu, X. MicroRNAs in osteoclastogenesis and function: potential therapeutic targets for osteoporosis. Int. J. Mol. Sci.17, 349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Meng, F. et al. A potential therapeutic drug for osteoporosis: prospect for osteogenic LncRNAs. Front. Endocrinol.14, 1219433 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lei, S., Papasian, C. & Deng, H. Polymorphisms in predicted miRNA binding sites and osteoporosis. J. Bone Miner. Res.26, 72–78 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Gao, M., Zhang, Z., Sun, J., Li, B. & Li, Y. The roles of circRNA-miRNA-mRNA networks in the development and treatment of osteoporosis. Front. Endocrinol.13, 945310 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Chen, W., Zhang, B. & Chang, X. Emerging roles of circular RNAs in osteoporosis. J. Cell. Mol. Med.25, 9089–9101 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Yang, M. et al. MiR-497∼195 cluster regulates angiogenesis during coupling with osteogenesis by maintaining endothelial Notch and HIF-1α activity. Nat. Commun.8, 16003 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Li, C. et al. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J. Clin. Investig.125, 1509–1522 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Keefe, A. D., Pai, S. & Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov.9, 660 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Xiaoqiu, W. et al. Potential diagnostic and therapeutic applications of oligonucleotide aptamers in breast cancer. Int. J. Mol. Sci.18, 1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Haute, D. V. & Berlin, J. M. Challenges in realizing selectivity for nanoparticle biodistribution and clearance: lessons from gold nanoparticles. Ther. Deliv.8, 763–774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Boomer, R. M. et al. Conjugation to polyethylene glycol polymer promotes aptamer biodistribution to healthy and inflamed tissues. Oligonucleotides15, 183–195 (2005). [DOI] [PubMed] [Google Scholar]
- 303.Healy, J. M. et al. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res.21, 2234–2246 (2005). [DOI] [PubMed] [Google Scholar]
- 304.Bouchard, M. F., Bellinger, D. C., Wright, R. O. & Weisskopf, M. G. Attention-deficit/hyperactivity disorder and urinary metabolites of organophosphate pesticides. Pediatrics125, e1270–e1277 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Jo, V. Y. & Fletcher, C. D. M. WHO classification of soft tissue tumours: an update based on the 2013 (4th) edition. Pathology46, 95–104 (2014). [DOI] [PubMed] [Google Scholar]
- 306.Jundt, G. [Updates to the WHO classification of bone tumours]. Pathologe39, 107–116 (2017). [DOI] [PubMed] [Google Scholar]
- 307.Sbaraglia, M., Bellan, E. & Dei Tos, A. P. The 2020 WHO Classification of Soft Tissue Tumours: news and perspectives. Pathologica113, 70–84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Arndt, C. A. & Crist, W. M. Common musculoskeletal tumors of childhood and adolescence. N. Engl. J. Med.341, 342–352 (1999). [DOI] [PubMed] [Google Scholar]
- 309.Piero, P. Osteosarcoma (osteogenic sarcoma). Orphanet J. Rare Dis.2, 6 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Stiller, C. A., Bielack, S. S., Jundt, G. & Steliarova-Foucher, E. Bone tumours in European children and adolescents, 1978-1997. Report from the Automated Childhood Cancer Information System project. Eur. J. Cancer42, 1978–1997 (2006). [DOI] [PubMed] [Google Scholar]
- 311.Stiller, C. A., Craft, A. W. & Corazziari, I. Survival of children with bone sarcoma in Europe since 1978. Eur. J. Cancer37, 760–766 (2001). [DOI] [PubMed] [Google Scholar]
- 312.Arthur P, S., Richard, L., Keith, R., Joseph B, S. & Michael, W. Osteogenic sarcoma presenting with lung metastasis. Oncologist7, 144–153 (2002). [DOI] [PubMed] [Google Scholar]
- 313.Lamplot, J. D. et al. The current and future therapies for human osteosarcoma. Curr. Cancer Ther. Rev.9, 55–77 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Finkel, M. P., Reilly, C. A. Jr, Biskis, B. O. Pathogenesis of radiation and virus-induced bone tumors. Recent Results Cancer Res. 54, 92–103 (1976). [DOI] [PubMed]
- 315.Swaney J. J. Familial osteogenic sarcoma. Clin. Orthop. Relat. Res. 97, 64–68 (1973). [DOI] [PubMed]
- 316.McIntyre, J. F. et al. Germline mutations of the p53 tumor suppressor gene in children with osteosarcoma. J. Clin. Oncol.12, 925–930 (1994). [DOI] [PubMed] [Google Scholar]
- 317.Tsogtbaatar, K. et al. In vitro selection of DNA aptamers against human osteosarcoma. Investig. N. Drugs40, 172–181 (2021). [DOI] [PubMed] [Google Scholar]
- 318.Wang, L. et al. Generating lung-metastatic osteosarcoma targeting aptamers for in vivo and clinical tissue imaging. Talanta188, 66–73 (2018). [DOI] [PubMed] [Google Scholar]
- 319.Liang, C. et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials147, 68–85 (2017). [DOI] [PubMed] [Google Scholar]
- 320.Yu Z. et al. Epidermal growth factor receptor aptamer‑conjugated polymer‑lipid hybrid nanoparticles enhance salinomycin delivery to osteosarcoma and cancer stem cells. Exp Ther Med15, 1247–1256 (2017). [DOI] [PMC free article] [PubMed]
- 321.Liu, X. Z. et al. Aptamer technology and its applications in bone diseases. Cell Transplant.32, 9636897221144949 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Paydaş, S., Şahin, B., Zorludemir, S. & Hazar, B. All trans retinoic acid as the possible cause of necrotizing vasculitis. Leuk. Res.22, 655–657 (1998). [DOI] [PubMed] [Google Scholar]
- 323.Dong, J. T., Boyd, J. C. & Frierson, H. F. Loss of heterozygosity at 13q14 and 13q21 in high grade, high stage prostate cancer. Prostate49, 166–171 (2001). [DOI] [PubMed] [Google Scholar]
- 324.Hao, Z. et al. Efficient delivery of micro RNA to bone-metastatic prostate tumors by using aptamer-conjugated atelocollagen in vitro and in vivo. Drug Deliv.23, 864–871 (2014). [DOI] [PubMed] [Google Scholar]
- 325.Sherr, C. J. Cancer cell cycles. Science274, 1672–1677 (1996). [DOI] [PubMed] [Google Scholar]
- 326.Dhanasekaran, S. M. et al. Delineation of prognostic biomarkers in prostate cancer. Nature412, 822–826 (2001). [DOI] [PubMed] [Google Scholar]
- 327.Clevers, H. Wnt/β-catenin signaling in development and disease. Cell127, 469–480 (2006). [DOI] [PubMed] [Google Scholar]
- 328.Bonci, D. et al. The miR-15a–miR-16-1 cluster controls prostate cancer by targeting multiple oncogenic activities. Nat. Med.14, 1271–1277 (2008). [DOI] [PubMed] [Google Scholar]
- 329.Eok-Soo, O., Motoharu, S., Martin, G. & Jun, C. Cell adhesion in cancer. Int. J. Cell Biol.2012, 965618 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Bazzoni, G., Martín-Padura, I., Beltràn-Nuñez, A. & Dejana, E. Tumor cell adhesion receptors. J. Surg. Oncol.53, 24–27 (1993). [DOI] [PubMed] [Google Scholar]
- 331.Evan T, K. & Julie, B. Prostate cancer bone metastases promote both osteolytic and osteoblastic activity. J. Cell Biochem.91, 718–729 (2004). [DOI] [PubMed] [Google Scholar]
- 332.Viallard, C. & Larrivée, B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis20, 409–426 (2017). [DOI] [PubMed] [Google Scholar]
- 333.Jun, C. et al. In pursuit of new anti-angiogenic therapies for cancer treatment. Front. Biosci.16, 803–814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Rana, D. et al. Spatiotemporally controlled, aptamers-mediated growth factor release locally manipulates microvasculature formation within engineered tissues. Bioact. Mater.12, 71–84 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Ajona, D. et al. Blockade of the complement C5a/C5aR1 axis impairs lung cancer bone metastasis by CXCL16-mediated effects. Am. J. Respir. Crit. Care Med.197, 1164–1176 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kuo, M. C., Kothari, A. N., Kuo, P. C. & Mi, Z. Cancer stemness in bone marrow micrometastases of human breast cancer. Surgery163, 330–335 (2018). [DOI] [PubMed] [Google Scholar]
- 337.Teramachi, J. et al. Myeloma bone disease: pathogenesis and management in the era of new anti-myeloma agents. J. Bone Miner. Metab.41, 388–403 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Bataille, R. et al. Mechanisms of bone destruction in multiple myeloma: the importance of an unbalanced process in determining the severity of lytic bone disease. J. Clin. Oncol.7, 1909–1914 (1989). [DOI] [PubMed] [Google Scholar]
- 339.Roodman, G. D. Pathogenesis of myeloma bone disease. Leukemia23, 435–441 (2008). [DOI] [PubMed] [Google Scholar]
- 340.Jun Won, L. et al. IL-3 expression by myeloma cells increases both osteoclast formation and growth of myeloma cells. Blood103, 2308–2315 (2003). [DOI] [PubMed] [Google Scholar]
- 341.Nanes, M. S. Tumor necrosis factor-α: molecular and cellular mechanisms in skeletal pathology. Gene321, 1–15 (2003). [DOI] [PubMed] [Google Scholar]
- 342.Kitaura, H. et al. Marrow stromal cells and osteoclast precursors differentially contribute to TNF-α-induced osteoclastogenesis in vivo. J. Immunol.173, 4838–4846 (2004). [DOI] [PubMed] [Google Scholar]
- 343.Masahiro, A. et al. Role for macrophage inflammatory protein (MIP)-1alpha and MIP-1beta in the development of osteolytic lesions in multiple myeloma. Blood100, 2195–2202 (2002). [PubMed] [Google Scholar]
- 344.Colucci, S. et al. Soluble decoy receptor 3 modulates the survival and formation of osteoclasts from multiple myeloma bone disease patients. Leukemia23, 2139–2146 (2009). [DOI] [PubMed] [Google Scholar]
- 345.Ehrlich, L. A. & Roodman, G. D. The role of immune cells and inflammatory cytokines in Paget’s disease and multiple myeloma. Immunol. Rev.208, 252–266 (2005). [DOI] [PubMed] [Google Scholar]
- 346.Uneda, S. et al. Macrophage inflammatory protein‐1 alpha is produced by human multiple myeloma (MM) cells and its expression correlates with bone lesions in patients with MM. Br. J. Haematol.120, 53–55 (2002). [DOI] [PubMed] [Google Scholar]
- 347.Han, J.-H. et al. Macrophage inflammatory protein-1α is an osteoclastogenic factor in myeloma that is independent of receptor activator of nuclear factor κB ligand. Blood97, 3349–3353 (2001). [DOI] [PubMed] [Google Scholar]
- 348.Lentzsch, S. et al. Macrophage inflammatory protein 1-alpha (MIP-1α) triggers migration and signaling cascades mediating survival and proliferation in multiple myeloma (MM) cells. Blood101, 3568–3573 (2003). [DOI] [PubMed] [Google Scholar]
- 349.Vallet, S. et al. A novel role for CCL3 (MIP-1α) in myeloma-induced bone disease via osteocalcin downregulation and inhibition of osteoblast function. Leukemia25, 1174–1181 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.An, G. et al. Osteoclasts promote immune suppressive microenvironment in multiple myeloma: therapeutic implication. Blood128, 1590–1603 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Topalian, S. L., Drake, C. G. & Pardoll, D. M. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell27, 450–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Zaidi, M. Skeletal remodeling in health and disease. Nat. Med.13, 791–801 (2007). [DOI] [PubMed] [Google Scholar]
- 353.Panaroni, C., Yee, A. J. & Raje, N. S. Myeloma and bone disease. Curr. Osteoporos. Rep.15, 483–498 (2017). [DOI] [PubMed] [Google Scholar]
- 354.Baron, R. & Kneissel, M. WNT signaling in bone homeostasis and disease: from human mutations to treatments. Nat. Med.19, 179–192 (2013). [DOI] [PubMed] [Google Scholar]
- 355.Erming, T. et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N. Engl. J. Med349, 2483–2494 (2003). [DOI] [PubMed] [Google Scholar]
- 356.Oshima, T. et al. Myeloma cells suppress bone formation by secreting a soluble Wnt inhibitor, sFRP-2. Blood106, 3160–3165 (2005). [DOI] [PubMed] [Google Scholar]
- 357.Giuliani, N. et al. Production of Wnt inhibitors by myeloma cells: potential effects on canonical Wnt pathway in the bone microenvironment. Cancer Res.67, 7665–7674 (2007). [DOI] [PubMed] [Google Scholar]
- 358.Colucci, S. et al. Myeloma cells suppress osteoblasts through sclerostin secretion. Blood Cancer J.1, e27–e27 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Edwards, C. M. et al. Increasing Wnt signaling in the bone marrow microenvironment inhibits the development of myeloma bone disease and reduces tumor burden in bone in vivo. Blood111, 2833–2842 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Yaccoby, S. et al. Antibody-based inhibition of DKK1 suppresses tumor-induced bone resorption and multiple myeloma growth in vivo. Blood109, 2106–2111 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Fulciniti, M. et al. Anti-DKK1 mAb (BHQ880) as a potential therapeutic agent for multiple myeloma. Blood114, 371–379 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Pozzi, S. et al. In vivo and in vitro effects of a novel anti-Dkk1 neutralizing antibody in multiple myeloma. Bone53, 487–496 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.McDonald, M. M. et al. Inhibiting the osteocyte-specific protein sclerostin increases bone mass and fracture resistance in multiple myeloma. Blood129, 3452–3464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Kang, S. et al. Wnt signaling stimulates osteoblastogenesis of mesenchymal precursors by suppressing CCAAT/enhancer-binding protein α and peroxisome proliferator-activated receptor γ. J. Biol. Chem.282, 14515–14524 (2007). [DOI] [PubMed] [Google Scholar]
- 365.Fairfield, H. et al. The skeletal cell‐derived molecule sclerostin drives bone marrow adipogenesis. J. Cell. Physiol.233, 1156–1167 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Scheller, E. L. et al. Corrigendum: rRegion-specific variation in the properties of skeletal adipocytes reveals regulated and constitutive marrow adipose tissues. Nat. Commun.7, 13775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Landgren, O. et al. Obesity is associated with an increased risk of monoclonal gammopathy of undetermined significance among black and white women. Blood116, 1056–1059 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Islam, R., Altundag, K., Kurt, M., Altundag, O. & Turen, S. Association between obesity and multiple myeloma in postmenopausal women may be attributed to increased aromatization of androgen in adipose tissue. Med. Hypotheses65, 1001–1002 (2005). [DOI] [PubMed] [Google Scholar]
- 369.Wallin, A. & Larsson, S. C. Body mass index and risk of multiple myeloma: a meta-analysis of prospective studies. Eur. J. Cancer47, 1606–1615 (2011). [DOI] [PubMed] [Google Scholar]
- 370.Kamila Anna, Z. et al. Modulation of cell metabolic pathways and oxidative stress signaling contribute to acquired melphalan resistance in multiple myeloma cells. PLoS ONE10, e0119857 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Wang, D., Haile, A. & Jones, L. C. Dexamethasone-induced lipolysis increases the adverse effect of adipocytes on osteoblasts using cells derived from human mesenchymal stem cells. Bone53, 520–530 (2013). [DOI] [PubMed] [Google Scholar]
- 372.Jurczyszyn, A. et al. Plasma fatty acid profile in multiple myeloma patients. Leuk. Res.39, 400–405 (2015). [DOI] [PubMed] [Google Scholar]
- 373.Duque, G., Gimble, J. M., Vidal, C. & Gunaratnam, K. Mechanisms of palmitate-induced lipotoxicity in human osteoblasts. Endocrinology155, 108–116 (2014). [DOI] [PubMed] [Google Scholar]
- 374.Ambrosi, T. H. et al. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell20, 771–784.e776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Pujol-Navarro, N. et al. Multiple myeloma: therapeutic delivery of antibodies and aptamers. Ther. Deliv.12, 705–722 (2021). [DOI] [PubMed] [Google Scholar]
- 376.Soldevilla, M. M., Meraviglia-Crivelli de Caso, D., Menon, A. P. & Pastor, F. Aptamer-iRNAs as therapeutics for cancer treatment. Pharmaceuticals11, 108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Ludwig, H. et al. Olaptesed pegol, an anti-CXCL12/SDF-1 spiegelmer, alone and with bortezomib–dexamethasone in relapsed/refractory multiple myeloma: a Phase IIa Study. Leukemia31, 997–1000 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Ireson, C. R. & Kelland, L. R. Discovery and development of anticancer aptamers. Mol. Cancer Ther.5, 2957–2962 (2006). [DOI] [PubMed] [Google Scholar]
- 379.Vater, A. et al. Hematopoietic stem and progenitor cell mobilization in mice and humans by a first-in-class mirror-image oligonucleotide inhibitor of CXCL12. Clin. Pharmacol. Ther.94, 150–157 (2013). [DOI] [PubMed] [Google Scholar]
- 380.Aravind, A. et al. AS1411 aptamer tagged PLGA‐lecithin‐PEG nanoparticles for tumor cell targeting and drug delivery. Biotechnol. Bioeng.109, 2920–2931 (2012). [DOI] [PubMed] [Google Scholar]
- 381.Teicher, B. A. & Fricker, S. P. CXCL12 (SDF-1)/CXCR4 Pathway in cancer. Clin. Cancer Res.16, 2927–2931 (2010). [DOI] [PubMed] [Google Scholar]
- 382.Balkwill, F. The significance of cancer cell expression of the chemokine receptor CXCR4. Semin. Cancer Biol.14, 171–179 (2004). [DOI] [PubMed] [Google Scholar]
- 383.Domanska, U. M. et al. A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur. J. Cancer49, 219–230 (2013). [DOI] [PubMed] [Google Scholar]
- 384.Sacco, A. et al. Cancer cell dissemination and homing to the bone marrow in a zebrafish model. Cancer Res.76, 463–471 (2016). [DOI] [PubMed] [Google Scholar]
- 385.Roccaro, A. M. et al. SDF-1 inhibition targets the bone marrow niche for cancer therapy. Cell Rep.9, 118–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Seckinger, A. et al. Clinical and prognostic role of annexin A2 in multiple myeloma. Blood120, 1087–1094 (2012). [DOI] [PubMed] [Google Scholar]
- 387.Amundarain, A., Pastor, F., Prósper, F. & Agirre, X. Aptamers, a new therapeutic opportunity for the treatment of multiple myeloma. Cancers14, 5471 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.D’Souza, S. et al. Annexin II interactions with the annexin II receptor enhance multiple myeloma cell adhesion and growth in the bone marrow microenvironment. Blood119, 1888–1896 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Zhou, W. et al. Screening and characterization of an Annexin A2 binding aptamer that inhibits the proliferation of myeloma cells. Biochimie151, 150–158 (2018). [DOI] [PubMed] [Google Scholar]
- 390.Shah, N., Chari, A., Scott, E., Mezzi, K. & Usmani, S. Z. B-cell maturation antigen (BCMA) in multiple myeloma: rationale for targeting and current therapeutic approaches. Leukemia34, 985–1005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Rennert, P. et al. A soluble form of B cell maturation antigen, a receptor for the tumor necrosis factor family member april, inhibits tumor cell growth. J. Exp. Med.192, 1677–1684 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.O'Connor, B. P. et al. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med.199, 91–98 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Catuogno, S. et al. An anti-BCMA RNA aptamer for miRNA intracellular delivery. Mol. Ther.18, 981–990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Mahtouk, K., Tjin, E. P. M., Spaargaren, M. & Pals, S. T. The HGF/MET pathway as target for the treatment of multiple myeloma and B-cell lymphomas. Biochim. Biophys. Acta1806, 208–219 (2010). [DOI] [PubMed] [Google Scholar]
- 395.Zhang, Y. B. et al. Targeting c-met receptor tyrosine kinase by the DNA aptamer SL1 as a potential novel therapeutic option for myeloma. J. Cell. Mol. Med.22, 5978–5990 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Ueki, R. & Sando, S. A DNA aptamer to c-Met inhibits cancer cell migration. Chem. Commun.50, 13131–13134 (2014). [DOI] [PubMed] [Google Scholar]
- 397.Boltz, A. et al. Bi-specific aptamers mediating tumor cell lysis. J. Biol. Chem.286, 21896–21905 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.van de Donk, N. W. C. J., Richardson, P. G. & Malavasi, F. CD38 antibodies in multiple myeloma: back to the future. Blood131, 13–29 (2018). [DOI] [PubMed] [Google Scholar]
- 399.Wen, J., Tao, W., Hao, S., Iyer, S. P. & Zu, Y. A unique aptamer-drug conjugate for targeted therapy of multiple myeloma. Leukemia30, 987–991 (2015). [DOI] [PubMed] [Google Scholar]
- 400.Neogi, T. The epidemiology and impact of pain in osteoarthritis. Osteoarthr. Cartil.21, 1145–1153 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Ewa M, R. & Nigel K, A. Strategies for the prevention of knee osteoarthritis. Nat. Rev. Rheumatol.12, 92–101 (2015). [DOI] [PubMed] [Google Scholar]
- 402.Martel-Pelletier, J. et al. Osteoarthritis. Nat. Rev. Dis. Prim.2, 16072 (2016). [DOI] [PubMed] [Google Scholar]
- 403.Glyn-Jones, S. et al. Osteoarthritis. Lancet386, 376–387 (2015). [DOI] [PubMed] [Google Scholar]
- 404.Neogi, T. & Zhang, Y. Epidemiology of osteoarthritis. Rheum. Dis. Clin. North Am.39, 1–19 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Lawrence, R. C. et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States: Part II. Arthritis Rheum.58, 26–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Christiansen, B. A. et al. Non-invasive mouse models of post-traumatic osteoarthritis. Osteoarthr. Cartil.23, 1627–1638 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Kuyinu, E. L., Narayanan, G., Nair, L. S. & Laurencin, C. T. Animal models of osteoarthritis: classification, update, and measurement of outcomes. J. Orthop. Surg. Res.11, 19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Kraus, V. B., Blanco, F. J., Englund, M., Karsdal, M. A. & Lohmander, L. S. Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthr. Cartil.23, 1233–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Sharma, L. & Solomon, C. G. Osteoarthritis of the Knee. N. Engl. J. Med.384, 51–59 (2021). [DOI] [PubMed] [Google Scholar]
- 410.Ji, M.-L. et al. Precise targeting of miR-141/200c cluster in chondrocytes attenuates osteoarthritis development. Ann. Rheum. Dis.80, 356–366 (2021). [DOI] [PubMed] [Google Scholar]
- 411.Smith, M. D. The normal synovium. Open Rheumatol. J.5, 100–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.de Sousa, E. B., Casado, P. L., Moura Neto, V., Duarte, M. E. & Aguiar, D. P. Synovial fluid and synovial membrane mesenchymal stem cells: latest discoveries and therapeutic perspectives. Stem Cell Res. Ther.5, 112 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Jay, G. D., Britt, D. E. & Cha, C. J. Lubricin is a product of megakaryocyte stimulating factor gene expression by human synovial fibroblasts. J. Rheumatol.27, 594–600 (2000). [PubMed] [Google Scholar]
- 414.Jay, G. D. & Waller, K. A. The biology of lubricin: near frictionless joint motion. Matrix Biol.39, 17–24 (2014). [DOI] [PubMed] [Google Scholar]
- 415.Scanzello, C. R. & Goldring, S. R. The role of synovitis in osteoarthritis pathogenesis. Bone51, 249–257 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Sanchez-Lopez, E., Coras, R., Torres, A., Lane, N. E. & Guma, M. Synovial inflammation in osteoarthritis progression. Nat. Rev. Rheumatol.18, 258–275 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Ni, S. et al. The involvement of follistatin-like protein 1 in osteoarthritis by elevating NF-κB-mediated inflammatory cytokines and enhancing fibroblast like synoviocyte proliferation. Arthritis Res. Ther.17, 91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Kapoor, M., Martel-Pelletier, J., Lajeunesse, D., Pelletier, J.-P. & Fahmi, H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat. Rev. Rheumatol.7, 33–42 (2010). [DOI] [PubMed] [Google Scholar]
- 419.Pap, T., Dankbar, B., Wehmeyer, C., Korb-Pap, A. & Sherwood, J. Synovial fibroblasts and articular tissue remodelling: role and mechanisms. Semin. Cell Dev. Biol.101, 140–145 (2020). [DOI] [PubMed] [Google Scholar]
- 420.Chen, X. et al. Specific clearance of senescent synoviocytes suppresses the development of osteoarthritis based on aptamer-functionalized targeted drug delivery system. Adv. Funct. Mater.32, 202109460 (2022). [Google Scholar]
- 421.Jeon, O. H. et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat. Med.23, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Zhao, R. et al. Inflammatory factors are crucial for the pathogenesis of post-traumatic osteoarthritis confirmed by a novel porcine model: “Idealized” anterior cruciate ligament reconstruction” and gait analysis. Int. Immunopharmacol.99, 107905 (2021). [DOI] [PubMed] [Google Scholar]
- 423.Wojdasiewicz, P., Poniatowski, Ł. A. & Szukiewicz, D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediat. Inflamm.2014, 1–19 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Marcu, K. B., Otero, M., Olivotto, E., Borzi, R. M. & Goldring, M. B. NF-kappaB signaling: multiple angles to target OA. Curr. Drug Targets11, 599–613 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Shakibaei, M., Schulze-Tanzil, G., John, T. & Mobasheri, A. Curcumin protects human chondrocytes from IL-1β-induced inhibition of collagen type II and β1-integrin expression and activation of caspase-3: an immunomorphological study. Ann. Anat.187, 487–497 (2005). [DOI] [PubMed] [Google Scholar]
- 426.Stöve, J., Huch, K., Günther, K.-P. & Scharf, H.-P. Interleukin-1β induces different gene expression of stromelysin, aggrecan and tumor-necrosis-factor-stimulated gene 6 in human osteoarthritic chondrocytes in vitro. Pathobiology68, 144–149 (2000). [DOI] [PubMed] [Google Scholar]
- 427.Mengshol, J. A., Vincenti, M. P., Coon, C. I., Barchowsky, A. & Brinckerhoff, C. E. Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-jun N-terminal kinase, and nuclear factor κB: Differential regulation of collagenase 1 and collagenase 3. Arthritis Rheum.43, 801–811 (2000). [DOI] [PubMed] [Google Scholar]
- 428.Vincenti, M. P. & Brinckerhoff, C. E. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res. Ther.4, 157–164 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Meszaros, E. & Malemud, C. J. Prospects for treating osteoarthritis: enzyme-protein interactions regulating matrix metalloproteinase activity. Ther. Adv. Chronic Dis.3, 219–229 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Verma, P. & Dalal, K. ADAMTS-4 and ADAMTS-5: key enzymes in osteoarthritis. J. Cell. Biochem.112, 3507–3514 (2011). [DOI] [PubMed] [Google Scholar]
- 431.Dinarello, C. A. Overview of the interleukin-1 family of ligands and receptors. Semin. Immunol.25, 389–393 (2013). [DOI] [PubMed] [Google Scholar]
- 432.Saha, N. et al. Interleukin-1β-converting enzyme/caspase-1 in human osteoarthritic tissues: localization and role in the maturation of interleukin-1β and interleukin-18. Arthritis Rheum.42, 1577–1587 (1999). [DOI] [PubMed] [Google Scholar]
- 433.Olee, T., Hashimoto, S., Quach, J. & Lotz, M. IL-18 is produced by articular chondrocytes and induces proinflammatory and catabolic responses. J. Immunol.162, 1096–1100 (1999). [PubMed] [Google Scholar]
- 434.B, M. et al. Interferon-gamma induces expression of interleukin-18 binding protein in fibroblast-like synoviocytes. Rheumatol.42, 442–445 (2003). [DOI] [PubMed] [Google Scholar]
- 435.Udagawa, N. et al. Interleukin-18 (Interferon-γ–inducing factor) is produced by osteoblasts and acts via granulocyte/macrophage colony-stimulating factor and not via interferon-γ to inhibit osteoclast formation. J. Exp. Med.185, 1005–1012 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Cheng-zhong, P. et al. Concentration of IL-18 and PGE2 in synovial fluid in patients with osteoarthritis and its significance. Zhong Nan Da Xue Xue Bao Yi Xue Ban31, 862–865 (2007). [PubMed] [Google Scholar]
- 437.Wang, Y. et al. Correlation between plasma, synovial fluid and articular cartilage Interleukin-18 with radiographic severity in 33 patients with osteoarthritis of the knee. Clin. Exp. Med.14, 297–304 (2013). [DOI] [PubMed] [Google Scholar]
- 438.Denoble, A. E. et al. Uric acid is a danger signal of increasing risk for osteoarthritis through inflammasome activation. Proc. Natl. Acad. Sci. USA108, 2088–2093 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Dai, S. M., Shan, Z. Z., Nishioka, K. & Yudoh, K. Implication of interleukin 18 in production of matrix metalloproteinases in articular chondrocytes in arthritis: direct effect on chondrocytes may not be pivotal. Ann. Rheum. Dis.64, 735–742 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Joosten, L. A. B. et al. Interleukin-18 promotes joint inflammation and induces interleukin-1-driven cartilage destruction. Am. J. Pathol.165, 959–967 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Inoue, H. et al. High levels of serum IL-18 promote cartilage loss through suppression of aggrecan synthesis. Bone42, 1102–1110 (2008). [DOI] [PubMed] [Google Scholar]
- 442.John, T., Kohl, B., Mobasheri, A., Ertel, W. & Shakibaei, M. Interleukin-18 induces apoptosis in human articular chondrocytes. Histol. Histopathol.22, 469–482 (2007). [DOI] [PubMed] [Google Scholar]
- 443.Futani, H. et al. Relation between interleukin-18 and PGE2 in synovial fluid of osteoarthritis: a potential therapeutic target of cartilage degradation. J. Immunother.25, S61–S64 (2002). [DOI] [PubMed] [Google Scholar]
- 444.Cho, M. et al. Interleukin-18 induces the production of vascular endothelial growth factor (VEGF) in rheumatoid arthritis synovial fibroblasts via AP-1-dependent pathways. Immunol. Lett.103, 159–166 (2006). [DOI] [PubMed] [Google Scholar]
- 445.Yong, L., Jian-ming, J., De-hong, Y., Feng-long, W. & Zhong-xuan, M. Determination of the concentrations of interleukin-18 and other cytokines in the synovial fluid in patients with osteoarthritis. Nan Fang Yi Ke Da Xue Xue Bao29, 729–731 (2009). [PubMed] [Google Scholar]
- 446.Feng-long, W., Jian-ming, J., Fei, W., Zhao-zong, F. & Zhao-fei, Z. [Expressions of interleukin 18 and prostaglandin E2 and their correlation in the synoviocytes of patients with osteoarthritis]. Nan Fang. Yi Ke Da Xue Xue Bao30, 731–733 (2010). [PubMed] [Google Scholar]
- 447.Saklatvala, J. Tumour necrosis factor α stimulates resorption and inhibits synthesis of proteoglycan in cartilage. Nature322, 547–549 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Séguin, C. A. & Bernier, S. M. TNFα suppresses link protein and type II collagen expression in chondrocytes: Role of MEK1/2 and NF‐κB signaling pathways. J. Cell. Physiol.197, 356–369 (2003). [DOI] [PubMed] [Google Scholar]
- 449.Henderson, B. & Pettipher, E. R. Arthritogenic actions of recombinant IL-1 and tumour necrosis factor alpha in the rabbit: evidence for synergistic interactions between cytokines in vivo. Clin. Exp. Immunol.75, 306–310 (1989). [PMC free article] [PubMed] [Google Scholar]
- 450.Koshy, P. J. T. et al. The modulation of matrix metalloproteinase and ADAM gene expression in human chondrocytes by interleukin‐1 and oncostatin M: a time‐course study using real‐time quantitative reverse transcription–polymerase chain reaction. Arthritis Rheum.46, 961–967 (2002). [DOI] [PubMed] [Google Scholar]
- 451.Ye, Z. et al. c-Jun N-terminal kinase–c-Jun pathway transactivates Bim to promote osteoarthritis. Can. J. Physiol. Pharmacol.92, 132–139 (2014). [DOI] [PubMed] [Google Scholar]
- 452.Héraud, F., Héraud, A. & Harmand, M. F. Apoptosis in normal and osteoarthritic human articular cartilage. Ann. Rheum. Dis.59, 959–965 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.López-Armada, M. J. et al. Mitochondrial activity is modulated by TNFα and IL-1β in normal human chondrocyte cells. Osteoarthr. Cartil.14, 1011–1022 (2006). [DOI] [PubMed] [Google Scholar]
- 454.López-Armada, M. J. et al. Cytokines, tumor necrosis factor-α and interleukin-1β, differentially regulate apoptosis in osteoarthritis cultured human chondrocytes. Osteoarthr. Cartil.14, 660–669 (2006). [DOI] [PubMed] [Google Scholar]
- 455.Guerne, P. A., Carson, D. A. & Lotz, M. IL-6 production by human articular chondrocytes. Modulation of its synthesis by cytokines, growth factors, and hormones in vitro. J. Immunol.144, 499–505 (1990). [PubMed] [Google Scholar]
- 456.Lotz, M., Terkeltaub, R. & Villiger, P. M. Cartilage and joint inflammation. Regulation of IL-8 expression by human articular chondrocytes. J. Immunol.148, 466–473 (1992). [PubMed] [Google Scholar]
- 457.Alaaeddine, N., Olee, T., Hashimoto, S., Creighton-Achermann, L. & Lotz, M. Production of the chemokine RANTES by articular chondrocytes and role in cartilage degradation. Arthritis Rheum.44, 1633–1643 (2001). [DOI] [PubMed] [Google Scholar]
- 458.Honorati, M. C., Cattini, L. & Facchini, A. VEGF production by osteoarthritic chondrocytes cultured in micromass and stimulated by IL-17 and TNF-α. Connect. Tissue Res.48, 239–245 (2009). [DOI] [PubMed] [Google Scholar]
- 459.Steeve, K. T., Marc, P., Sandrine, T., Dominique, H. & Yannick, F. IL-6, RANKL, TNF-alpha/IL-1: interrelations in bone resorption pathophysiology. Cytokine Growth Factor Rev.15, 49–60 (2004). [DOI] [PubMed] [Google Scholar]
- 460.Sakao, K. et al. Osteoblasts derived from osteophytes produce interleukin-6, interleukin-8, and matrix metalloproteinase-13 in osteoarthritis. J. Bone Miner. Metab.27, 412–423 (2009). [DOI] [PubMed] [Google Scholar]
- 461.Honsawek, S. et al. Association of the IL-6 -174G/C gene polymorphism with knee osteoarthritis in a Thai population. Genet. Mol. Res.10, 1674–1680 (2011). [DOI] [PubMed] [Google Scholar]
- 462.E, P. et al. Interleukin-6 gene polymorphism and risk of osteoarthritis of the hip: a case-control study. Osteoarthr. Cartil.13, 1025–1028 (2005). [DOI] [PubMed] [Google Scholar]
- 463.Benoît, P. et al. Interleukin-6 (IL-6) and/or soluble IL-6 receptor down-regulation of human type II collagen gene expression in articular chondrocytes requires a decrease of Sp1.Sp3 ratio and of the binding activity of both factors to the COL2A1 promoter. J. Biol. Chem.283, 4850–4865 (2007). [DOI] [PubMed] [Google Scholar]
- 464.Rowan, A. D. et al. Synergistic effects of glycoprotein 130 binding cytokines in combination with interleukin-1 on cartilage collagen breakdown. Arthritis Rheum.44, 1620–1632 (2001). [DOI] [PubMed] [Google Scholar]
- 465.Cawston, T. E. et al. The role of oncostatin M in animal and human connective tissue collagen turnover and its localization within the rheumatoid joint. Arthritis Rheum.41, 1760–1771 (1998). [DOI] [PubMed] [Google Scholar]
- 466.de Hooge, A. S. K. et al. Male IL-6 gene knock out mice developed more advanced osteoarthritis upon aging. Osteoarthr. Cartil.13, 66–73 (2005). [DOI] [PubMed] [Google Scholar]
- 467.Kunkel, S. L., Lukacs, N. W. & Strieter, R. M. The role of interleukin‐8 in the infectious process. Ann. N. Y. Acad. Sci.730, 134–143 (2006). [DOI] [PubMed] [Google Scholar]
- 468.Rolfe, M. W. et al. Pulmonary fibroblast expression of interleukin-8: a model for alveolar macrophage-derived cytokine networking. Am. J. Respir. Cell Mol. Biol.5, 493–501 (1991). [DOI] [PubMed] [Google Scholar]
- 469.Standiford, T. J. et al. Interleukin-8 gene expression by a pulmonary epithelial cell line. A model for cytokine networks in the lung. J. Clin. Investig.86, 1945–1953 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Korn, T., Bettelli, E., Oukka, M. & Kuchroo, V. K. IL-17 and Th17 Cells. Annu. Rev. Immunol.27, 485–517 (2009). [DOI] [PubMed] [Google Scholar]
- 471.Pawłowska, J. et al. Different distribution of CD4 and CD8 T cells in synovial membrane and peripheral blood of rheumatoid arthritis and osteoarthritis patients. Folia Histochem. Cytobiol.47, 627–632 (2010). [DOI] [PubMed] [Google Scholar]
- 472.Ishii, H. et al. Characterization of infiltrating T cells and Th1/Th2-type cytokines in the synovium of patients with osteoarthritis. Osteoarthr. Cartil.10, 277–281 (2002). [DOI] [PubMed] [Google Scholar]
- 473.Alsalameh, S. et al. Cellular immune response toward human articular chondrocytes. T cell reactivities against chondrocyte and fibroblast membranes in destructive joint diseases. Arthritis Rheum.33, 1477–1486 (2005). [DOI] [PubMed] [Google Scholar]
- 474.Lubberts, E., Joosten, L. A., Van De Loo, F. A., Van Den Gersselaar, L. A. & Van Den Berg, W. B. Reduction of interleukin-17-induced inhibition of chondrocyte proteoglycan synthesis in intact murine articular cartilage by interleukin-4. Arthritis Rheum.43, 1300–1306 (2000). [DOI] [PubMed] [Google Scholar]
- 475.Martel-Pelletier, J., Mineau, F. O., Jovanovic, D., Di Battista, J. A. & Pelletier, J.-P. Mitogen-activated protein kinase and nuclear factor kappaB together regulate interleukin-17-induced nitric oxide production in human osteoarthritic chondrocytes: possible role of transactivating factor mitogen-activated protein kinase-activated protein kinase (MAPKAPK). Arthritis Rheum.42, 2399–2409 (1999). [DOI] [PubMed] [Google Scholar]
- 476.Mohamed, B. et al. Interleukin 17 (IL-17) induces collagenase-3 production in human osteoarthritic chondrocytes via AP-1 dependent activation: differential activation of AP-1 members by IL-17 and IL-1beta. J. Rheumatol.29, 1262–1272 (2002). [PubMed] [Google Scholar]
- 477.LeGrand, A. et al. Interleukin-1, tumor necrosis factor, and interleukin-17 synergistically up-regulate nitric oxide and prostaglandin E2 production in explants of human osteoarthritic knee menisci. Arthritis Rheum.44, 2078–2083 (2001). [DOI] [PubMed] [Google Scholar]
- 478.Honorati, M. C., Bovara, M., Cattini, L., Piacentini, A. & Facchini, A. Contribution of interleukin 17 to human cartilage degradation and synovial inflammation in osteoarthritis. Osteoarthr. Cartil.10, 799–807 (2002). [DOI] [PubMed] [Google Scholar]
- 479.Attur, M. G., Patel, R. N., Abramson, S. B. & Amin, A. R. Interleukin‐17 up‐regulation of nitric oxide production in human osteoarthritis cartilage. Arthritis Rheum.40, 1050–1053 (2005). [DOI] [PubMed] [Google Scholar]
- 480.Honorati, M. C., Neri, S., Cattini, L. & Facchini, A. Interleukin-17, a regulator of angiogenic factor release by synovial fibroblasts. Osteoarthr. Cartil.14, 345–352 (2006). [DOI] [PubMed] [Google Scholar]
- 481.Han, L. et al. Association of IL-17A and IL-17F single nucleotide polymorphisms with susceptibility to osteoarthritis in a Korean population. Gene533, 119–122 (2014). [DOI] [PubMed] [Google Scholar]
- 482.Adachi, H. et al. Antagonistic RNA aptamer specific to a heterodimeric form of human interleukin-17A/F. Biochimie93, 1081–1088 (2011). [DOI] [PubMed] [Google Scholar]
- 483.Baslund, B. et al. Targeting interleukin‐15 in patients with rheumatoid arthritis: a proof‐of‐concept study. Arthritis Rheum.52, 2686–2692 (2005). [DOI] [PubMed] [Google Scholar]
- 484.McInnes, I. B. et al. The role of interleukin–15 in T–cell migration and activation in rheumatoid arthritis. Nat. Med.2, 175–182 (1996). [DOI] [PubMed] [Google Scholar]
- 485.Waldmann, T. A. & Tagaya, Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in Nk cell differentiation and host response to intracellular pathogens. Annu. Rev. Immunol.17, 19–49 (1999). [DOI] [PubMed] [Google Scholar]
- 486.Scanzello, C. R. et al. Local cytokine profiles in knee osteoarthritis: elevated synovial fluid interleukin-15 differentiates early from end-stage disease. Osteoarthr. Cartil.17, 1040–1048 (2009). [DOI] [PubMed] [Google Scholar]
- 487.Marini, J. C. et al. Osteogenesis imperfecta. Nat. Rev. Dis. Prim.3, 17052 (2017). [DOI] [PubMed] [Google Scholar]
- 488.Neri Morales, C. et al. Osteogenesis imperfecta: a case series and literature review. Cureus15, e33864 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Bini, L. et al. Intracellular and extracellular markers of lethality in osteogenesis imperfecta: a quantitative proteomic approach. Int. J. Mol. Sci.22, 429 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Marini, J. C. et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat.28, 209–221 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Sillence, D. O., Senn, A. & Danks, D. M. Genetic heterogeneity in osteogenesis imperfecta. J. Med. Genet.16, 101–116 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Valadares, E. R., Carneiro, T. B., Santos, P. M., Oliveira, A. C. & Zabel, B. What is new in genetics and osteogenesis imperfecta classification? J. Pediatr.90, 536–541 (2014). [DOI] [PubMed] [Google Scholar]
- 493.Dogba, M. J., Rauch, F., Douglas, E. & Bedos, C. Impact of three genetic musculoskeletal diseases: a comparative synthesis of achondroplasia, Duchenne muscular dystrophy and osteogenesis imperfecta. Health Qual. Life Outcomes12, 151 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Botor, M. et al. Osteogenesis imperfecta: current and prospective therapies. Biomolecules11, 1493 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Perosky, J. E. et al. Single dose of bisphosphonate preserves gains in bone mass following cessation of sclerostin antibody in Brtl/+ osteogenesis imperfecta model. Bone93, 79–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Holdsworth, G., Roberts, S. J. & Ke, H. Z. Novel actions of sclerostin on bone. J. Mol. Endocrinol.62, R167–R185 (2019). [DOI] [PubMed] [Google Scholar]
- 497.Wijenayaka, A. R. et al. Sclerostin stimulates osteocyte support of osteoclast activity by a RANKL-dependent pathway. PLoS ONE6, e25900 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.van Bezooijen, R. L., ten Dijke, P., Papapoulos, S. E. & Löwik, C. W. SOST/sclerostin, an osteocyte-derived negative regulator of bone formation. Cytokine Growth Factor Rev.16, 319–327 (2005). [DOI] [PubMed] [Google Scholar]
- 499.Poole, K. E. S. et al. Sclerostin is a delayed secreted product of osteocytes that inhibits bone formation. FASEB J.19, 1842–1844 (2005). [DOI] [PubMed] [Google Scholar]
- 500.Scheiber, A. L. et al. Sclerostin antibody-induced changes in bone mass are site specific in developing crania. J. Bone Miner. Res.34, 2301–2310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Cosman, F. et al. Romosozumab treatment in postmenopausal women with osteoporosis. N. Engl. J. Med.375, 1532–1543 (2016). [DOI] [PubMed] [Google Scholar]
- 502.Neer, R. M. et al. Effect of parathyroid hormone (1-34) on fractures and bone mineral density in postmenopausal women with osteoporosis. N. Engl. J. Med.344, 1434–1441 (2001). [DOI] [PubMed] [Google Scholar]
- 503.Moester, M. J. C., Papapoulos, S. E., Löwik, C. W. G. M. & van Bezooijen, R. L. Sclerostin: current knowledge and future perspectives. Calcif. Tissue Int.87, 99–107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Vahle, J. L. et al. Skeletal changes in rats given daily subcutaneous injections of recombinant human parathyroid hormone (1-34) for 2 years and relevance to human safety. Toxicol. Pathol.30, 312–321 (2002). [DOI] [PubMed] [Google Scholar]
- 505.de Vlaming, A. et al. Atrioventricular valve development: new perspectives on an old theme. Differentiation84, 103–116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Folkestad, L. et al. Cardiovascular disease in patients with osteogenesis imperfecta–a nationwide, register-based cohort study. Int. J. Cardiol.225, 250–257 (2016). [DOI] [PubMed] [Google Scholar]
- 507.Wang, L. Y. et al. Therapeutic aptamer targeting sclerostin loop3 for promoting bone formation without increasing cardiovascular risk in osteogenesis imperfecta mice. Theranostics12, 5645–5674 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 508.Gubu, A. et al. Unique quinoline orientations shape the modified aptamer to sclerostin for enhanced binding affinity and bone anabolic potential. Mol. Ther. Nucleic Acids35, 102146 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Sarkar, I. N. et al. Selecting essential information for biosurveillance—a multi-criteria decision analysis. PLoS ONE9, e86601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Summerfield, S., Barfield, M., Spooner, N. & White, S. From patient to tube: the importance of physiologically relevant quantitative bioanalytical assays. Bioanalysis8, 2595–2604 (2016). [DOI] [PubMed] [Google Scholar]
- 511.Poveda-Rogers, C. & Morrissette, J. J. D. Greater expectations: meeting clinical needs through broad and rapid genomic testing. Clin. Chem. Lab. Med.61, 654–661 (2023). [DOI] [PubMed] [Google Scholar]
- 512.Zhang, Z. et al. Aptamer-based fluorescence polarization assay for separation-free exosome quantification. Nanoscale11, 10106–10113 (2019). [DOI] [PubMed] [Google Scholar]
- 513.Zhang, X. Q. et al. Strain H37Rv electrochemical sensor mediated by aptamer and AuNPs-DNA. ACS Sens.4, 849–855 (2019). [DOI] [PubMed] [Google Scholar]
- 514.Yang, S. et al. A label-free fluorescent biosensor based on specific aptamer-templated silver nanoclusters for the detection of tetracycline. J. Nanobiotechnol.21, 22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Zheng, J. et al. Design of aptamer-based sensing platform using triple-helix molecular switch. Anal. Chem.83, 6586–6592 (2011). [DOI] [PubMed] [Google Scholar]
- 516.Saad, M. & Faucher, S. P. Aptamers and aptamer-coupled biosensors to detect water-borne pathogens. Front. Microbiol.12, 643797 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Sun, D. P., Lu, J., Zhang, L. Y. & Chen, Z. G. Aptamer-based electrochemical cytosensors for tumor cell detection in cancer diagnosis: a review. Anal. Chim. Acta1082, 1–17 (2019). [DOI] [PubMed] [Google Scholar]
- 518.Shin, W. R. et al. Aptamer-based pathogen monitoring for Salmonella enterica ser. typhimurium. J. Biomed. Nanotechnol.14, 1992–2002 (2018). [DOI] [PubMed] [Google Scholar]
- 519.Chen, X. Y., Ma, Y., Xie, Y. Q. & Pu, J. Aptamer-based applications for cardiovascular disease. Front. Bioeng. Biotechnol.10, 1002285 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Li, H. et al. Fluorescence assay for detecting four organophosphorus pesticides using fluorescently labeled aptamer. Sensors22, 5712 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Cruz-Aguado, J. A. & Penner, G. Determination of ochratoxin a with a DNA aptamer. J. Agric. Food Chem.56, 10456–10461 (2008). [DOI] [PubMed] [Google Scholar]
- 522.Qiao, Q. Q. et al. Aptamer-based fluorescence quenching approach for detection of aflatoxin M1 in milk. Front. Chem.9, 653869 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Wang, L. J., Zhou, H., Hu, H. X., Wang, Q. & Chen, X. G. Regulation mechanism of ssDNA aptamer in nanozymes and application of nanozyme-based aptasensors in food safety. Foods11, 544 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Qi, Y. et al. Practical aptamer-based assay of heavy metal mercury ion in contaminated environmental samples: convenience and sensitivity. Anal. Bioanal. Chem.412, 439–448 (2019). [DOI] [PubMed] [Google Scholar]
- 525.Ge, K., Hu, Y. L., Zheng, Y. J., Jiang, P. C. & Li, G. K. Aptamer/derivatization-based surface-enhanced Raman scattering membrane assembly for selective analysis of melamine and formaldehyde in migration of melamine kitchenware. Talanta235, 122743 (2021). [DOI] [PubMed] [Google Scholar]
- 526.Yi, J. C. et al. The research of aptamer biosensor technologies for detection of microorganism. Appl. Microbiol. Biotechnol.104, 9877–9890 (2020). [DOI] [PubMed] [Google Scholar]
- 527.Chauhan, N. et al. Net-shaped DNA nanostructures designed for rapid/sensitive detection and potential inhibition of the SARS-CoV-2 virus. J. Am. Chem. Soc.145, 20214–20228 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Basu, A. et al. Performance of Abbott ID now COVID-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City Academic Institution. J. Clin. Microbiol.58, e01136–01120 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Lu, J. et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. eBioMedicine59, 102960 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Litke, J. L. & Jaffrey, S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol.37, 667–675 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature495, 333–338 (2013). [DOI] [PubMed] [Google Scholar]
- 532.Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One7, e30733 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J.30, 4414–4422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Wang, Y. & Wang, Z. F. Efficient backsplicing produces translatable circular mRNAs. Rna21, 172–179 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Li, H. M., Ma, X. L. & Li, H. G. Intriguing circles: conflicts and controversies in circular RNA research. WIREs RNA10, e1538 (2019). [DOI] [PubMed] [Google Scholar]
- 536.Karoline K, E., Thomas B, H. & Jørgen, K. Insights into circular RNA biology. RNA Biol.14, 1035–1045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA19, 141–157 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Bai, Y. et al. Research progress on circular RNA vaccines. Front. Immunol.13, 1091797 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Zhang, Z. et al. Advances in engineering circular RNA vaccines. Pathogens13, 692 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Choi, S.-W. & Nam, J.-W. Optimal design of synthetic circular RNAs. Exp. Mol. Med.56, 1281–1292 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 541.Cui, J. et al. A precise and efficient circular RNA synthesis system based on a ribozyme derived from Tetrahymena thermophila. Nucleic Acids Res.51, e78–e78 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Beadudry, D. & Perreault, J.-P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res.23, 3064–3066 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 543.Lee, K. H. et al. Efficient circular RNA engineering by end-to-end self-targeting and splicing reaction using Tetrahymena group I intron ribozyme. Mol. Ther.33, 587–598 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Wang, W. et al. Regulatory circular RNAs in viral diseases: applications in diagnosis and therapy. RNA Biol.20, 847–858 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun.9, 2629 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell74, 508–520.e504 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol.41, 262–272 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.