Abstract
Nuclear transplantation is one of the very few ways by which the genetic content and capacity for nuclear reprogramming can be assessed in individual cells of differentiated somatic tissues. No more than 6% of the cells of differentiated tissues have thus far been shown to have nuclei that can be reprogrammed to elicit the formation of unrelated cell types. In Amphibia, about 25% of such nuclear transfers form morphologically abnormal partial blastulae that die within 24 h. We have investigated the genetic content and capacity for reprogramming of those nuclei that generate partial blastulae, using as donors the intestinal epithelium cells of feeding Xenopus larvae. We have analyzed single nuclear transplant embryos obtained directly from intestinal tissue, thereby avoiding any genetic or epigenetic changes that might accumulate during cell culture. The expression of the intestine-specific gene intestinal fatty acid binding protein is extinguished by at least 104 times, within a few hours of nuclear transplantation. At the same time several genes that are normally expressed only in early embryos are very strongly activated in nuclear transplant embryos, but to an unregulated extent. Remarkably, cells from intestine-derived partial blastulae, when grafted to normal host embryos, contribute to several host tissues and participate in the normal 100-fold increase in axial muscle over several months. Thus, cells of defective cloned embryos unable to survive for more than 1 day can be reprogrammed to participate in new directions of differentiation and to maintain indefinite growth, despite the abnormal expression of early genes.
The earliest successful nuclear transplantation experiments were designed to show whether the genetic content of vertebrate somatic cells undergoes stable changes during cell differentiation (1). Following early indications from work on Rana pipiens that this might be so (2), it was found in Xenopus laevis, and later in Rana, that even nuclei from differentiated tissues could, when transplanted to enucleated eggs, reveal their toti- or multipotency (3–5). However, it was found in all amphibian nuclear transfer experiments that the efficiency of obtaining normal development, as a percentage of total transfers, became progressively reduced as more differentiated cells were used (2, 3). The same result has been obtained with the more recent nuclear transfer experiments with mammals (6–8).
In Amphibia, nuclei from the intestinal epithelial cells of feeding Xenopus larvae yielded fertile adult frogs in less than 1% of transfers (9) and feeding tadpoles in 1.5% (3). Nuclei from adult frog skin cells gave heart beat tadpoles in 3% of transfers, and this proportion was increased to 5.3% by use of serial nuclear transfers (4). In sheep and mice, offspring were obtained from 0.4% and 2.8% of nuclear transfers from cultured mammary gland (6) and ovarian cumulus cells (7), respectively. Even the highest cloning efficiencies in mammals have not exceeded 6% of total transfers yielding live births (7, 10–12). This result is also true when cells of early nuclear transfer embryos are differentiated in vitro or injected into host blastocysts (13). Cell fusion experiments have also demonstrated a reprogramming of gene activity (14–16), but even when hybrid cells, obtained by fusing somatic cells with embryonic germ and embryonic stem cells, are transferred to host embryos, the proportion of total somatic nuclei shown to undergo reprogramming is less than 6% (15, 17). Therefore, it has not yet been demonstrated that more than 6% of differentiated somatic cell nuclei are capable of being reprogrammed to elicit the formation of wholly unrelated cell types.
The 94% or more of somatic cell nuclear transfers that do not undergo nuclear reprogramming either fail to initiate cleavage or become abnormal and die at different stages of development, both in Amphibia (2) and mammals (7). In the case of tadpole intestinal epithelium, which is representative of differentiated cells, about 50% of nuclear transfers fail to initiate cleavage of recipient eggs and about 25% undergo partial cleavage and die within 24 h (3). Here we have investigated the nuclear reprogramming and developmental capacity of cells in these partially cleaved embryos. We find a high degree of pluripotentiality in these somatic nuclei, which represent at least 16% of the cells of differentiated tissue. Although these nuclei undergo, after transplantation, a rapid and extensive change in gene expression, this is often quantitatively aberrant, and yet our results demonstrate that their capacity to direct subsequent cell differentiation and indefinite growth is remarkably normal.
Materials and Methods
Donor Cells.
Stage-47 (18) tadpoles were killed with tricaine methanesulphonate. The intestinal tract was removed, and the central wide-diameter part, about 1 mm in length, was separated and used as a source of donor cells. This part of the intestine was opened longitudinally, food removed, and dissociated in calcium and magnesium-deficient modified Barth saline (19) including 0.2% BSA and 1 mM EDTA, at a pH of 8.2. The first cells to be released are those of the intestinal epithelium, which strongly express the marker intestinal fatty acid binding protein (IFABP) (20–22).
Nuclear Transplantation.
This procedure was carried out as described (23).
Grafts.
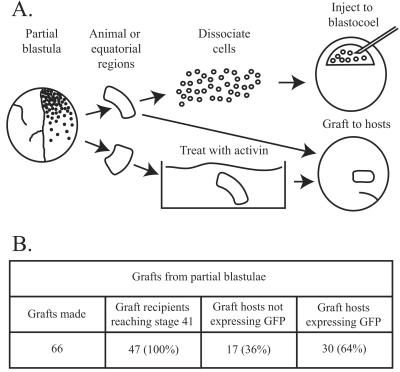
These were carried out in 1× modified Barth saline (MBS) medium at stage 10½. To facilitate the healing and retention of grafts, host embryos were cultured in this medium for about 2 h. This salt concentration inhibits gastrulation, compared with the 0.1× MBS preferred by embryos at this stage, and this probably accounts for the loss of some grafts and host embryos at this stage (Fig. 5B).
Figure 5.
Nuclear transplant embryo development. (A) Experimental design. (B) Differentiation of grafts. In 6 experimental series, a total of 66 host embryos received grafts, and 47 of these reached the normal swimming tadpole stage 41.
In Situ Hybridization.
This procedure was carried out as described (24).
Reverse Transcription (RT)-PCR Analysis.
This procedure was carried out as described (25). To calculate the number of transcripts, the PCR product was quantified as a function of cycle number with a PhosphorImager and compared with a dilution series of a plasmid DNA of known concentration. Cells were counted with a haemocytometer.
Results
Donor Cells and Nuclear Transfer Success.
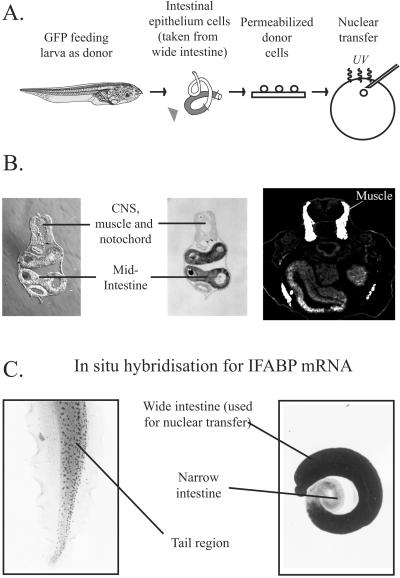
The first stage of our experimental procedure involves transplanting nuclei from the intestine of feeding Xenopus larvae to enucleated eggs (Fig. 1A). As a genetic marker to follow the fate of cells derived from nuclear transplant embryos, we have used donor tadpoles carrying a single genomic green fluorescent protein (GFP) insertion. This GFP marker is under the control of a cytomegalovirus promoter and is thus expressed in all Xenopus cells (26). Donor cells were prepared from the epithelium of the mid-intestinal tract of stage-47 tadpoles (18) that had commenced feeding. These epithelial cells have a striated epithelium and stain with the intestinal markers endodermin (27) and IFABP (20–22) but not with the muscle marker 12-101 (28) (Fig. 1B). We performed in situ hybridization for IFABP mRNA. This procedure confirmed that IFABP is expressed uniformly in the wide mid-intestinal region that we used to obtain donor epithelial cells but not in the narrow intestine region, nor in any of the tissues in the part of the tail that was used as a control (Fig. 1C). The light staining observed in the tail region is attributable to endogenous alkaline phosphatase activity and not the presence of IFABP mRNA (20).
Figure 1.
Nuclear transfer experiment and donor cell characterization. (A) Design of nuclear transfer experiment. Arrowhead indicates region of intestine used for nuclear transfer. (B) Transverse sections of a stage-47 tadpole. (Left) Nomarski view; (Center) section in situ to show IFABP mRNA in mid-intestine; (Right) 12-101 antibody staining of muscle. (C) Whole-mount in situ hybridization for IFABP mRNA. Tail (control) (Left); intestine (Right). The dark spots in the center of the tail are pigment cells.
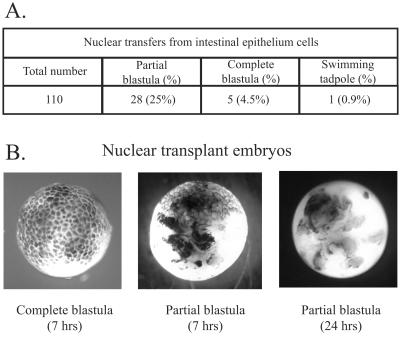
Recipient unfertilized eggs were UV-enucleated (29) and were genetically wild type (not GFP transgenic). The results of our nuclear transfer experiments are summarized in Fig. 2A and are similar to those previously obtained with intestinal epithelium cells (3). Complete cleavage of recipient eggs is obtained in only 5% of total nuclear transfers, and only a few of these, about 1% of total transfers, become morphologically normal feeding tadpoles (Fig. 2A). In contrast, about 25% of total nuclear transfers form partially cleaved blastulae. About half of each partial blastula consists of normal-sized cells, the other half being cleaved very irregularly or not at all (Fig. 2B). These partially cleaved nuclear transplant blastulae die within 24 h of nuclear transfer (Fig. 2B).
Figure 2.
Nuclear transfer experiment results. (A) Nuclear transfer results. (B) Photographs of whole embryos.
Extinction of Intestine-Specific Genes.
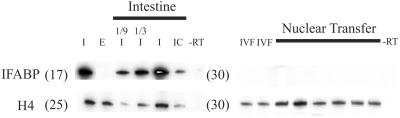
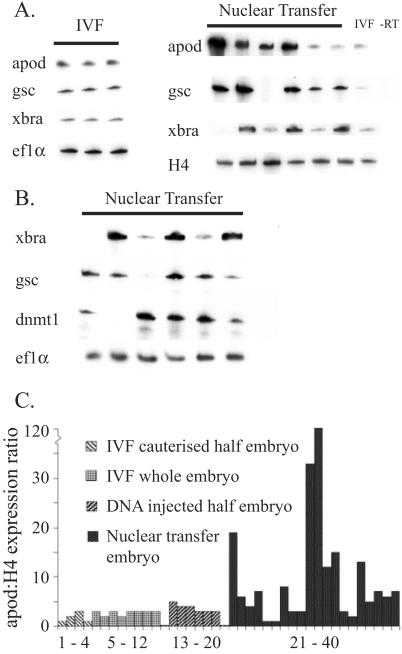
To investigate the reprogramming of gene expression, we have used RT-PCR to analyze mRNA transcripts in the cellular part of partial nuclear transplant blastulae at the late-blastula or early-gastrula stages. IFABP is expressed strongly in the epithelial cells of the mid-intestine of Xenopus tadpoles from the prefeeding stage 43 to the end of larval feeding at metamorphosis (20–22) and can be readily seen in 5 × 104 dissociated epithelium cells of the kind used for nuclear transfer (Fig. 3). This level of expression works out at 104 molecules of IFABP mRNA per intestinal epithelium cell, as determined from RT-PCR quantitation of purified IFABP mRNA. Even with a much greater level of PCR amplification, IFABP is completely undetectable in single partial nuclear transplant blastulae at stage 9, each containing 10,000–15,000 cells (Fig. 3), just as this marker is also undetectable in half-embryos reared from fertilized eggs (see IVF lanes in Fig. 3). The sensitivity of detection by RT-PCR is such that there must have been less than one molecule of IFABP mRNA per partial blastula cell. Therefore, the transplantation of nuclei to eggs causes rapid extinction of genes expressed in specialized cells.
Figure 3.
Extinction of intestine-specific genes. RT-PCR analysis using primers for IFABP and, on the same samples, for histone H4. This analysis demonstrates extinction of intestine-specific IFABP expression after nuclear transfer; E, eye; I (1, 1/3, 1/9), whole or fractions of one stage-47 tadpole mid-intestine; IC, 5 × 104 intestinal epithelial cells; IVF, blastula from in vitro fertilization; nuclear transfer, single partial blastulae. Numbers in brackets represent cycles of PCR amplification. Strong IFABP expression is seen in intestine cells with only 17 cycles but no expression is observed in nuclear transfer embryos, even after 30 cycles. RT, omission of reverse transcriptase.
Activation of Early Embryo Genes.
We have also used RT-PCR to provide a quantitatively sensitive assay for early zygotic gene expression. Single whole blastulae grown from fertilized eggs show consistent and quantitatively similar levels of expression of gsc (dorsal), Xbra and Apod (pan-equatorial), and Ef1α (maternal) (Fig. 4A). Nearly all half-blastulae obtained by nuclear transfer express these early zygotic genes, thereby showing that they have undergone gene reprogramming within a few hours of nuclear transfer (Fig. 4A). However, the extent of gene activation is not normal and most half-embryos, each derived from a single transplanted nucleus, show large but variable overexpression of early zygotic genes (Fig. 4 A and B and summarized in C).
Figure 4.
Activation of early embryo genes. (A) Activation of early zygotic gene expression in single in vitro fertilization blastula (IVF) and in single partial nuclear transplant blastulae. (B) Another series of experiments in which part of each embryo was analyzed by RT-PCR (as shown). The other part of the same embryo was used to graft to hosts, as described in the text. dnmt1, DNA methyltransferase. (C) Summary of expression of the “zygotic gene Apod: histone H4” ratio in single, whole, or partial blastulae analyzed by RT-PCR. Apod, antipodean (40); gsc, goosecoid (41); Xbra, Xenopus brachyury (42).
To verify that the cause of the abnormal gene expression was not caused by the abnormal cleavage of the cloned partial embryos, we have analyzed abnormally cleaved noncloned partial blastulae. These partial blastulae were created either by cautery or by injecting an excess of DNA into one blastomere at the two-cell stage. In none of these experimentally induced partial blastulae do we see abnormally high levels of gene expression (Fig. 4C). The few cases in which partial nuclear transfer blastulae fail to express zygotic genes (Fig. 4 A and B) are probably attributable to delayed division of some cells before the normal stage of zygotic gene expression. The extinction and reactivation of genes has been described before in amphibian nuclear transplant embryos, but only in embryos that undergo normal cleavage (30–32). Mammalian experiments have demonstrated abnormal imprinted (33) and nonimprinted (34) gene expression in cloned embryos but these embryos were derived from cultured donor cells, and culturing cells could be a possible cause of aberrant gene expression (23, 33). In the experiments described here, we have analyzed single nuclear transplant embryos obtained directly from intestinal tissue, thereby avoiding any genetic or epigenetic changes that might accumulate during cell culture.
Grafts to Wild-Type Hosts.
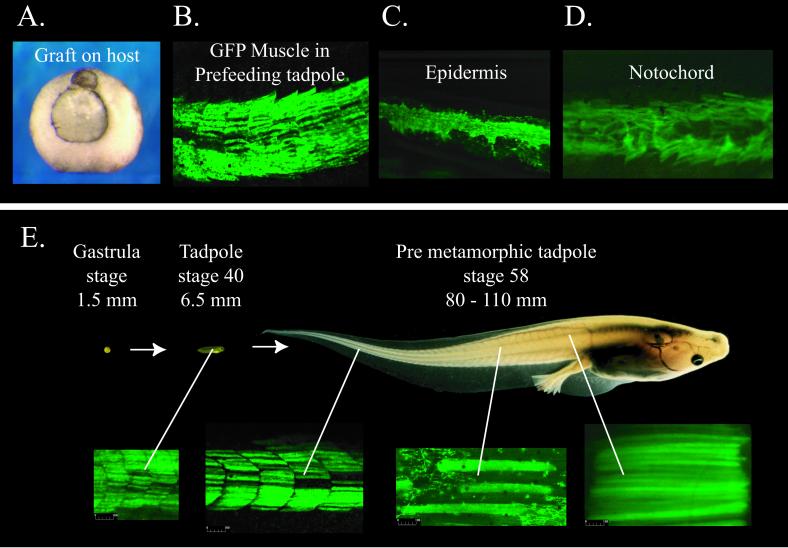
To test the developmental capacity of cells from partial nuclear transplant blastulae, we have transferred some of these cells to wild-type (non-GFP) host embryos. We have used two cell transfer regimes (Fig. 5A). In one we injected dissociated cells from nuclear transplant blastulae into the blastocoel of recipient blastulae (35). In the other we grafted portions of donor partial blastulae to host embryos at the early gastrula stage. Within this second regime, we either grafted material directly or we treated parts of the animal cap blastulae with activin to direct these cells toward a mesodermal fate (36, 37) and then grafted these pieces of tissue to host embryos. Both regimes gave similar results, although the yield was higher with grafts than with cell injections. The results of grafting experiments are summarized in Fig. 5B, and a grafted embryo is illustrated in Fig. 6A. About one-third of host embryos that received grafts failed to gastrulate normally and did not reach stage 41 (see Materials and Methods). Of the 47 graft hosts that became surviving tadpoles, 17 (36%) did not contain GFP cells, either because the grafted cells were inviable or possibly because grafts were lost soon after they were made. Of the 47 graft hosts, 25 (53%) contained GFP cells integrated into muscle (Fig. 6B), as expected of grafts made to the lateral equatorial region of the hosts, and 5 (11%) integrated GFP cells into several different tissues (including muscle, notochord, and epidermis). Overall, 30 of 47 surviving tadpoles (64%) expressed GFP. In all of the GFP-expressing grafts, GFP cells were found in muscle tissue and were found to be the same size as, and spatially intermixed with, host muscle cells. The observation that 11% of the surviving tadpoles had integrated GFP cells into multiple tissues, including muscle, notochord, and/or epidermis, demonstrated that a range of cell types can be obtained (Fig. 6 B–D).
Figure 6.
Growth and differentiation of muscle. (A) Mid-gastrula with graft. (B–D) Differentiated cells derived from grafts of GFP-partial nuclear transplant blastula tissue into wild-type hosts. These fluorescent photographs do not show host cells. (E) Growth of a gastrula that received a graft of cells from a GFP-partial nuclear transplant embryo. The fluorescent photografts of axial muscle show the great enlargement of muscle cells during tadpole growth. Embryos and tadpoles are shown at the same relative size (diameter and length are shown in millimeters).
In another series of experiments, we divided the cellular portion of partial blastulae into two parts. One part of each embryo was grafted to a host embryo, and the other part of the same embryo was used for RT-PCR analysis of gene expression. We again saw abnormal amounts of Eomes, gsc, and apod expression in cells of the same embryos from which grafts contributed to muscle (Fig. 4B). We conclude that most partial nuclear transplant blastulae contain cells that can differentiate, after grafting to host embryos, into cell types unrelated to the original donor tissue (intestinal epithelium). This conclusion applies to at least 16% of the cells of the original intestinal epithelium, that is, to 64% (Fig. 5B) of the 25% of the total nuclear transfers that become partial blastulae.
Growth of Grafted Cells.
We next asked whether reprogrammed intestine nuclei in partial embryos due to die in 24 h can participate in the massive growth that takes place as a hatched prefeeding 3-day-old tadpole progresses to metamorphosis, a process that takes up to 100 days according to temperature, feeding, and other conditions, and entails a 100-fold increase in weight. We see in Fig. 6E that GFP muscle cells continue to populate axial muscle of tadpoles up to the stage of metamorphosis, after which axial muscle is reabsorbed as tadpoles change into frogs. GFP muscle cells also increase in size in accord with host muscle (Fig. 6E). The abundance of GFP muscle cells is similar in tadpoles that reach metamorphosis and in 3-day-old prefeeding tadpoles, and there is no obvious loss of GFP muscle cells during growth. By staining these cells with the muscle-specific antibody 12-101 (28) (data not shown), we confirmed that the GFP cells that integrated into the host muscle were in fact muscle. In conclusion, we see that both the differentiation and growth of muscle cells derived from transplanted intestine nuclei take place normally and indistinguishably from muscle in host larvae reared from fertilized eggs. It is remarkable that normal differentiation and this massive growth for several months can be obtained from cells of embryos that were destined to die within 24 h.
Discussion
Three conclusions can be drawn from our results. First, we have gained information about the genetic content of differentiated cells. Much of our knowledge of the genetic composition of differentiated somatic cells comes from analyzing samples of several million cells. Only nuclear transfer (1, 2, 4, 6, 7, 10–12) and cell fusion (14–17) experiments provide direct genetic information about single somatic cells. Nearly all previous nuclear transfer work has paid attention to embryos that cleave normally. Although our earlier experiments with larval intestine nuclei included the use of partially cleaved embryos for serial transfer, these extended the proportion of total transfers that reached a feeding tadpole stage only from 1.5% to 5.3% (3). By sampling all partially cleaved nuclear transplant embryos, as we have done here, we can conclude that at least 16% (64% of 25%; see Figs. 5B and 2A, respectively) of differentiated intestine cells have functional genes for pathways of differentiation wholly unrelated to their own.
Our second conclusion is that transplanted nuclei eliciting only abnormal development are nevertheless extensively reprogrammed, as revealed by the extinction and activation of gene expression. It is important to appreciate that this degree of gene reprogramming takes place within 6 h at 23°C from the time when nuclei are transplanted, and without a cell culture stage as is sometimes used in mammalian experiments. Unexpectedly, we found a very large and misregulated overexpression of induced early zygotic genes in single nuclear transplant embryos, compared with normal or partial embryos derived from fertilized eggs. The observed misregulation of gene expression could be connected with aberrant methylation or demethylation (38).
Third, we see a remarkable capacity for muscle growth and differentiation by transplanted intestinal nuclei, even though such nuclei supported only very defective development of embryos and expressed early zygotic genes incorrectly. Cell differentiation seems surprisingly tolerant of abnormal early gene expression, as has also been observed in some mouse nuclear transfer experiments (39). It may be that cells from nonviable cloned human embryos, which could not survive independently, may be useful for research and therapeutic purposes.
Acknowledgments
We thank Karen Butler for help with the in situ hybridization, N. Marsh-Armstrong and D. D. Brown (Carnegie Institution of Washington, Baltimore) for the generous donation of a transgenic cytomegalovirus-GFP X. laevis, and R. Jaenisch and I. Wilmut for comments on the manuscript. We acknowledge support from the Biotechnology and Biological Sciences Research Council, Cancer Research Campaign, the Manifold Trust, and the Swedish Foundation for International Cooperation in Research and Higher Education (STINT).
Abbreviations
- IFABP
intestinal fatty acid binding protein
- GFP
green fluorescent protein
- RT
reverse transcription
References
- 1.Briggs R, King T J. Proc Natl Acad Sci USA. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Briggs R, King T J. J Morphol. 1957;100:269–312. [Google Scholar]
- 3.Gurdon J B. J Embryol Exp Morph. 1962;10:622–640. [PubMed] [Google Scholar]
- 4.Gurdon J B, Laskey R A, Reeves O R. J Embryol Exp Morph. 1975;34:93–112. [PubMed] [Google Scholar]
- 5.Di Berardino M A, Hoffner N. Science. 1983;219:862–864. doi: 10.1126/science.6600520. [DOI] [PubMed] [Google Scholar]
- 6.Wilmut I, Schnieke A E, McWhir J, Kind A J, Campbell K H. Nature (London) 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 7.Wakayama T, Perry A C, Zuccotti M, Johnson K R, Yanagimachi R. Nature (London) 1998;394:369–374. doi: 10.1038/28615. [DOI] [PubMed] [Google Scholar]
- 8.Polejaeva I A, Chen S H, Vaught T D, Page R L, Mullins J, Ball S, Dai Y, Boone J, Walker S, Ayares D L, Colman A, Campbell K H. Nature (London) 2000;407:86–90. doi: 10.1038/35024082. [DOI] [PubMed] [Google Scholar]
- 9.Gurdon J B, Uehlinger V. Nature (London) 1966;210:1240–1241. doi: 10.1038/2101240a0. [DOI] [PubMed] [Google Scholar]
- 10.Kato Y, Tani T, Sotomaru Y, Kurokawa K, Kato J, Doguchi H, Yasue H, Tsunoda Y. Science. 1998;282:2095–2098. doi: 10.1126/science.282.5396.2095. [DOI] [PubMed] [Google Scholar]
- 11.Kato Y, Tani T, Tsunoda Y. J Reprod Fertil. 2000;120:231–237. [PubMed] [Google Scholar]
- 12.Baguisi A, Overstrom E W. Theriogenology. 2000;53:209. doi: 10.1016/S0093-691X(00)00253-3. [DOI] [PubMed] [Google Scholar]
- 13.Wakayama T, Tabar V, Rodriguez I, Perry A C, Studer L, Mombaerts P. Science. 2001;292:740–743. doi: 10.1126/science.1059399. [DOI] [PubMed] [Google Scholar]
- 14.Blau H M, Pavlath G K, Hardeman E C, Chiu C P, Silberstein L, Webster S G, Miller S C, Webster C. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 15.Tada M, Tada T, Lefebvre L, Barton S C, Surani M A. EMBO J. 1997;16:6510–6520. doi: 10.1093/emboj/16.21.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blau H M, Blakely B T. Semin Cell Dev Biol. 1999;10:267–272. doi: 10.1006/scdb.1999.0311. [DOI] [PubMed] [Google Scholar]
- 17.Matveeva N M, Shilov A G, Kaftanovskaya E M, Maximovsky L P, Zhelezova A I, Golubitsa A N, Bayborodin S I, Fokina M M, Serov O L. Mol Reprod Dev. 1998;50:128–38. doi: 10.1002/(SICI)1098-2795(199806)50:2<128::AID-MRD2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 18.Nieuwkoop P D, Faber J. Normal Table of Xenopus Laevis (Daudin) Amsterdam: North–Holland; 1975. [Google Scholar]
- 19.Gurdon J B. Methods Cell Biol. 1977;16:125–139. doi: 10.1016/s0091-679x(08)60096-5. [DOI] [PubMed] [Google Scholar]
- 20.Shi Y B, Hayes W P. Dev Biol. 1994;161:48–58. doi: 10.1006/dbio.1994.1006. [DOI] [PubMed] [Google Scholar]
- 21.Chalmers A D, Slack J M. Dev Dyn. 1998;212:509–521. doi: 10.1002/(SICI)1097-0177(199808)212:4<509::AID-AJA4>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Chalmers A D, Slack J M, Beck C W. Mech Dev. 2000;96:125–128. doi: 10.1016/s0925-4773(00)00379-8. [DOI] [PubMed] [Google Scholar]
- 23.Chan A P, Gurdon J B. Int J Dev Biol. 1996;40:441–451. [PubMed] [Google Scholar]
- 24.Butler K, Zorn A M, Gurdon J B. Methods. 2001;23:303–312. doi: 10.1006/meth.2000.1142. [DOI] [PubMed] [Google Scholar]
- 25.Wilson P A, Melton D A. Curr Biol. 1994;4:676–686. doi: 10.1016/s0960-9822(00)00152-4. [DOI] [PubMed] [Google Scholar]
- 26.Marsh-Armstrong N, Huang H, Berry D L, Brown D D. Proc Natl Acad Sci USA. 1999;96:14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasai Y, Lu B, Piccolo S, De Robertis E M. EMBO J. 1996;15:4547–4555. [PMC free article] [PubMed] [Google Scholar]
- 28.Kintner C R, Brockes J P. Nature (London) 1984;308:67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- 29.Gurdon J B. Methods Cell Biol. 1991;36:299–309. doi: 10.1016/s0091-679x(08)60284-8. [DOI] [PubMed] [Google Scholar]
- 30.Gurdon J B, Brown D D. J Mol Biol. 1965;12:27–33. doi: 10.1016/s0022-2836(65)80279-0. [DOI] [PubMed] [Google Scholar]
- 31.Wakefield L, Gurdon J B. EMBO J. 1983;2:1613–1619. doi: 10.1002/j.1460-2075.1983.tb01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurdon J B, Brennan S, Fairman S, Mohun T J. Cell. 1984;38:691–700. doi: 10.1016/0092-8674(84)90264-2. [DOI] [PubMed] [Google Scholar]
- 33.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout W M, 3rd, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 34.Daniels R, Hall V, Trounson A O. Biol Reprod. 2000;63:1034–1040. doi: 10.1095/biolreprod63.4.1034. [DOI] [PubMed] [Google Scholar]
- 35.Heasman J, Wylie C C, Hausen P, Smith J C. Cell. 1984;37:185–194. doi: 10.1016/0092-8674(84)90314-3. [DOI] [PubMed] [Google Scholar]
- 36.Green J B, New H V, Smith J C. Cell. 1992;71:731–739. doi: 10.1016/0092-8674(92)90550-v. [DOI] [PubMed] [Google Scholar]
- 37.Dyson S, Gurdon J B. Cell. 1998;93:557–568. doi: 10.1016/s0092-8674(00)81185-x. [DOI] [PubMed] [Google Scholar]
- 38.Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Proc Natl Acad Sci USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rideout W M, 3rd, Eggan K, Jaenisch R. Science. 2001;293:1093–1098. doi: 10.1126/science.1063206. [DOI] [PubMed] [Google Scholar]
- 40.Stennard F, Carnac G, Gurdon J B. Development (Cambridge, UK) 1996;122:4179–4188. doi: 10.1242/dev.122.12.4179. [DOI] [PubMed] [Google Scholar]
- 41.Cho K W, Blumberg B, Steinbeisser H, De Robertis E M. Cell. 1991;67:1111–1120. doi: 10.1016/0092-8674(91)90288-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith J C, Price B M, Green J B, Weigel D, Herrmann B G. Cell. 1991;67:79–87. doi: 10.1016/0092-8674(91)90573-h. [DOI] [PubMed] [Google Scholar]