Abstract
In recent years a great deal of biochemical and genetic research has focused on the C-terminal domain (CTD) of the largest subunit (RPB1) of DNA-dependent RNA polymerase II. This strongly conserved domain of tandemly repeated heptapeptides has been linked functionally to important steps in the initiation and processing of mRNA transcripts in both animals and fungi. Although they are absolutely required for viability in these organisms, C-terminal tandem repeats do not occur in RPB1 sequences from diverse eukaryotic taxa. Here we present phylogenetic analyses of RPB1 sequences showing that canonical CTD heptads are strongly conserved in only a subset of eukaryotic groups, all apparently descended from a single common ancestor. Moreover, eukaryotic groups in which the most complex patterns of ontogenetic development occur are descended from this CTD-containing ancestor. Consistent with the results of genetic and biochemical investigations of CTD function, these analyses suggest that the enhanced control over RNA polymerase II transcription conveyed by acquired CTD/protein interactions was an important step in the evolution of intricate patterns of gene expression that are a hallmark of large, developmentally complex eukaryotic organisms.
Keywords: development‖RPB1‖transcription
Each of the core subunits present in all cellular DNA-dependent RNA polymerase (pol) enzymes shares a common evolutionary origin (1, 2). The largest of these subunits contains eight highly conserved domains, designated regions A through H (2), which are common to all prokaryotic and eukaryotic homologues (3). Unlike all other members of this protein family, however, the largest subunit of eukaryotic RNA pol II has an additional C-terminal domain (CTD), comprising a varied number of tandemly repeated heptapeptides with the consensus sequence Tyr-1–Ser-2–Pro-3–Thr-4–Ser-5–Pro-6–Ser-7 (4). In animals and yeast, where the mechanics of transcription are understood most clearly, the CTD has become a focal point of investigations into the interactions between RNA pol II and a variety of transcription-related proteins.
Reversible phosphorylation of the CTD regulates the cycling of RNA pol II between a hypophosphorylated (IIO) form, which is competent to enter the preinitiation complex, and a hyperphosphorylated (IIA) form capable of processive transcript elongation (5). Throughout this cycle the CTD binds essential transcription-related proteins that help to regulate gene expression (6–9), promote efficient elongation (10), and effectively couple transcription to pre-mRNA processing (11–15). So central is its role in these interactions that the CTD has been called, by one reviewer, “the tail that wags the dog” of RNA pol II (16).
Tandemly repeated CTD heptads occur in all RNA pol II largest subunits isolated to date from animals, fungi, and green plants. Given the importance of the CTD for pol II function, it is not surprising that it has been conserved so strongly during the evolution of these groups. A typical CTD also is present in certain protists; however, canonical heptad repeats do not occur in RPB1 sequences isolated from a diverse array of eukaryotes (17). For example, in the amitochondriate soil amoeba, Mastigamoeba invertens, there are well-ordered heptads, but with a nearly invariant consensus sequence that is different from that of the typical CTD (18). In other protists, such as parasitic trypanosomids (19), there is no recognizable CTD whatsoever. One of the most interesting cases is that of the red algae; both canonical and noncanonical heptads are present in RPB1 from Glaucosphaera vacuolata, a unicellular alga that appears to represent one of the earliest diverging rhodophyte evolutionary branches. In contrast, heptad repeats are absent from three more developmentally complex red algae that have been examined (17).
Thus far, the significance of the evolutionary variation found in RPB1 C-terminal sequences among diverse eukaryotes has not been investigated rigorously. As a step in that investigation, we examine the phylogenetic distribution of the CTD among eukaryotic organisms. Our analyses indicate that there is a distinct group of eukaryotes, designated here as the CTD-clade, in which repetitive heptads are under functional constraints that result in powerful stabilizing selection on CTD structure. We propose that only in this group of organisms was the CTD so thoroughly integrated into the pol II transcription cycle that it became absolutely essential for viability and, therefore, could not be lost. If true, this notion suggests that the full complement of CTD functions found in animals and fungi represents an evolutionary modification of the mRNA synthetic machinery, one that led to more intricate patterns of gene expression and a greater potential for ontogenetic developmental complexity.
Methods
A data set of 25 RPB1 sequences that we have isolated previously or retrieved from GenBank was assembled, along with three archaean largest subunits for use as an outgroup. Archaean and RPB1 inferred amino acid sequences, encompassing regions A through H (C-terminal sequences were not used in phylogenetic reconstruction), first were aligned separately with clustal x (20), and unique insertions in individual sequences, as well as regions that could not be aligned with confidence, were removed from each subalignment. The two sets of sequences then were aligned with each other in clustal x and adjusted through visual inspection. All regions of further ambiguity between the subalignments were removed, leaving 1,102 positions for phylogenetic reconstruction.
The maximum-likelihood tree for this alignment was determined by using puzzle (21) under a mixed model for variation among sites, with one category for invariable sites and a four-category discrete approximation to a gamma-distribution, and the JTT weighting matrix for probability of change among amino acids. An initial quartet-puzzling run resolved a number of major eukaryotic groups clearly but left a polytomy among key taxa. Further analyses were performed in paup (22), to produce all possible branching combinations among the well-resolved quartet-puzzling clades. The individual tree with the highest likelihood from among the resulting 210 trees was determined in puzzle under the model and parameters described above. In addition, a most parsimonious tree was recovered by using protpars in PHYLIP 3.573 (23), which weights changes among amino acids based on the number of nucleotide replacements needed for a given substitution. One thousand protpars bootstrap replicates were performed.
To further assess the strength of support for hypotheses relating to CTD evolution, Bayesian phylogenetic inference was performed by using metropolis-coupled Markov chain Monte Carlo analysis (24). Four simultaneous Markov chains were run under a gamma approximation for rate variation among sites and a JTT substitution matrix. Chains were begun with random a priori trees; subsequently trees were sampled from the posterior probability distribution every 10 generations. The burn-in period required for likelihoods to converge on stable values was determined empirically to be approximately 10,000 generations; an additional 50,000 generations were run and the first 20,000 were excluded from analyses of the posterior probability distribution. Thus, a total of 4,000 trees were examined to determine Bayesian support values.
In addition, several a priori alternative hypotheses of eukaryotic evolution were compared with the CTD-clade hypothesis by using Kishino–Hasegawa and Templeton tests (25, 26). First, we tested the hypothesis of a single origin of a repetitive CTD with no subsequent losses. In this case, trees were constrained to require all organisms with well-ordered tandem heptads, regardless of the consensus sequence present, to form a clade. In addition, because several recent studies have suggested that red algae may be the sister group to green plants (27, 28), the best tree constraining that relationship also was analyzed. Kishino–Hasegawa tests were performed both by using the parameters discussed above as well as under the assumption of two possible rates among sites, one variable and one invariable. Although only small differences in significance levels were obtained in the two sets of analyses, the results reported are from two rate tests because gamma estimates appeared to be biased slightly toward whatever topology was used as a guide tree for estimating rate categories.
Results
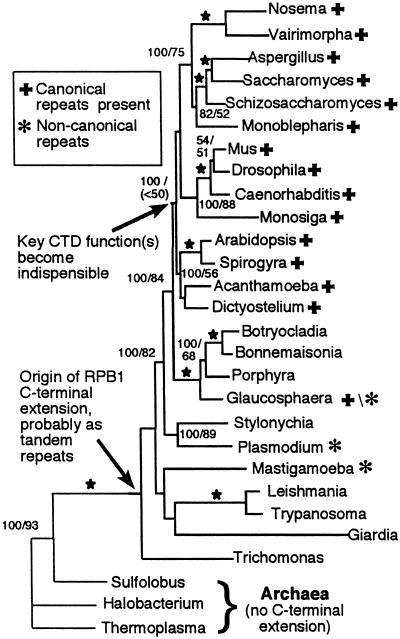
Largely congruent trees were recovered from parsimony, maximum-likelihood, and Bayesian analyses of aligned RPB1 regions A-H (Fig. 1). The only differences in these trees occurred in relationships among the most deeply branching sequences. We have demonstrated that this cluster of deep-branching sequences is subject to topological distortions known as long-branch artifacts (17); therefore, branching order among these taxa should be viewed with caution in any case. All of the relationships found among members of the CTD-clade, however, as well as the two nodes and intra-clade relationships immediately ancestral to it, were recovered regardless of the phylogenetic inference method used.
Figure 1.
Best tree and branch lengths recovered from maximum-likelihood analyses. Support values for each node from Bayesian inference and parsimony bootstrap analyses are shown either above or to the right of a given internode. In each case, Bayesian values are on the left of the slash with parsimony bootstrap values following. A star designates where both values are 100%. No values are given below 50% or for nodes that were not recovered in all three analyses. GenBank accessions used: Arabidopsis thaliana, P31635; Spirogyra sp., U90210; Bonnemaisonia hamifera, U90209; Botryocladia uvariodes, AF315819; Glaucosphaera vacuolata, AF315820; Porphyra yezoensis, U90208; Caenorhabditis elegans, P16356; Drosophila melanogaster, P04052; Monosiga brevicalis, AF315821; Mus musculus, NM_009089; Monoblepharis macandra, AF31582.2; Aspergillus oryzae, AB017184; Saccharomyces cerevisiae, P04050; Schizosaccharomyces pombe, P36594; Vairimorpha necatrix, AF060234; Nosema locustae, AF061288; Plasmodium falciparum, P14248; Stylonychia mytilus, AF315819; Acanthamoeba castellanii, U90211; Dictyostelium discoideum, AF58710/S52651; Mastigamoeba invertens, AF083338; Trichomonas vaginalis, U20501; Trypanosoma brucei, P17545; Leishmania donovani, AF126254; Halobacterium sp. NRC-1, AE005138; Sulfolobus acidocaldarius, X14818; Thermoplasma acidophilum, AL445064. The Giardia lamblia sequence was provided by H.-P. Klenk, Epidaurus, Bernried, Germany.
Previously, we proposed that the CTD originated only once in the course of eukaryotic evolution and, based on a survey of RNA pol largest subunit sequences available, that its full complement of functions was codified only after the evolutionary divergence of red algae (29). Based on the sequences available at the time, however, it was not possible to assess whether tandem heptad repeats originated only once, in a CTD-clade ancestor, or whether repeats at one time were present in the other eukaryotic groups but were lost during subsequent evolutionary diversification. Therefore, we sought to investigate this question explicitly by using an expanded data set of RPB1 genes, from a more diverse array of eukaryotes, to compare the RPB1 C-terminal structure in a given organism with its inferred evolutionary position on the RPB1 tree.
The occurrence of tandemly repeated heptapeptides is not distributed randomly among major eukaryotic taxa (Fig. 1). Although a well-defined CTD is absent from over a third of the RPB1 sequences in our analysis, there is a clearly defined clade in which tandem heptads always are present. With some internal variation, the heptads in members of this clade share the consensus CTD sequence. This CTD-clade is composed of animals, plants, fungi, and several related protistan groups. The consensus for the first six CTD heptad positions (Y-S-P-T-S-P) is conserved in all members of this clade; the most exceptional case is the fungus, Aspergillus oryzae, in which approximately half of the Tyr-1 residues have been substituted by Phe (30).
In contrast with this strong evolutionary conservation, only two sequences outside the CTD-clade contain an appreciable number of regularly repeated heptads: those are from Mastigamoeba and Glaucosphaera. Moreover, in neither instance does a consensus of the heptads conform to the typical CTD sequence (17, 18). It should be noted that RPB1 from Plasmodium falciparum contains a short stretch of tandemly repeated and noncanonical heptads; however, examinations both of related taxa and codon usage (29, 31) indicate that these repeats are of very recent origin within P. falciparum or an immediate ancestor. Therefore, they are not likely to be evolutionary homologues of the repeats found in the CTD-clade.
Support for the CTD-clade hypothesis is highly significant in Bayesian phylogenetic inference; it is recovered in 100% of the 4,000 trees sampled from the metropolis-coupled Markov chain Monte Carlo analysis posterior probability distribution (Fig. 1). Support for the CTD clade also is strong in Kishino–Hasegawa and Templeton tests when compared with reasonable alternative a priori evolutionary hypotheses. For example, constraining all taxa with C-terminal repeats as a clade, regardless of the consensus sequence present, is rejected with significance (Table 1). Thus, a single origin of all taxa containing tandem heptad repeats is inconsistent with this larger RPB1 data set. This finding, in turn, suggests that tandem heptads have been lost from a number of lineages outside the CTD-clade (see Discussion).
Table 1.
Results of testing specific a priori alternatives to CTD-clade hypothesis
| Constraint | Kishino–Hasegawa
|
Templeton
|
||||
|---|---|---|---|---|---|---|
| Δlog L | s | P | ΔSteps | s | P | |
| (R,P) | 31.73 | 18.24 | 0.08 | 43.0 | 20.721 | 0.04 |
| (M,CTD-clade) | 90.31 | 24.73 | 0.0001 | 110.0 | 27.544 | 0.0001 |
R, red algae; P, green plants; M, Mastigamoeba.
Despite its strong support from Bayesian inference, the CTD-clade hypothesis is not upheld with significance in Shimodaira–Hasegawa tests (32) when challenged by multiple alternative topologies. Comparative likelihood scores of trees with the 210 possible combinations of major, well-resolved RPB1 clades indicate that support for the CTD-clade is eroded in two ways. Trees in which microsporidians do not associate with fungi, but rather branch nearer to the base of the RPB1 tree, represent an alternative cluster of topologies with likelihoods that are not significantly different from the favored tree. This finding is consistent with a general problem of long-branch attraction of microsporidians toward the bases of broad-scale molecular trees (33) and may account for the weak bootstrap support for the CTD-clade in parsimony analyses. Unlike Bayesian inference, parsimony algorithms do not account for rate variation among sites, nor do they incorporate equally realistic models of substitution among amino acids. Indeed, removal of the microsporidians from RPB1 analyses increases parsimony bootstrap support for the CTD-clade by 30%.
In addition to problems posed by microsporidian sequences, trees that place red algae as the sister group to either of the two major subgroups within the CTD-clade (Fig. 1) are not significantly worse than the favored tree topology. This finding raises an important issue of a potential conflict between the CTD-clade hypothesis and several recent phylogenetic studies that suggest red algae and green plants share a common ancestor (27, 28). The evolutionary position of red algae has been debated for over a century and remains unresolved (17). Nevertheless, we tested the specific a priori hypothesis of a red algal/green plant relationship against the CTD-clade hypothesis. In addition to the highly significant support for independent origins of red algae and green plants in Bayesian inference, best trees constraining a green-red clade are rejected at P = 0.04 and 0.08 in Templeton and Kishino–Hasegawa tests, respectively (Table 1). Thus, although alternative evolutionary hypothesis cannot always be rejected with significance, the most parsimonious and consistent explanation for the observed RPB1 phylogeny is the CTD-clade hypothesis.
Discussion
The analyses presented here suggest that thorough integration of the CTD into the RNA pol II transcription cycle resulted in important changes in the mechanics of eukaryotic mRNA synthesis. The distinct phylogenetic bifurcation between eukaryotic groups in which tandem heptads invariably are present, and those in which they are mostly absent, supports the proposition that an essential function or set of functions was locked into the pol II transcription machinery in the ancestor of the CTD-clade. Such a change in function would account for the clear differences in stabilizing selection on CTD structure between members of the CTD-clade and other eukaryotes.
The CTD-Clade Hypothesis and Eukaryotic Phylogeny.
Recently, large, concatenated molecular data sets constructed from multiple genes have been used in an attempt to resolve eukaryotic relationships more clearly (27, 34). Based on these studies, and the cumulative results of many individual molecular phylogenetic investigations, the CTD-clade hypothesis is consistent with most reasonably well-supported relationships among eukaryotes. There is one possible exception to this generalization; that is a putative sister relationship between green plants and red algae that has been recovered in certain broad-scale phylogenetic investigations (27, 28). Based on these analyses, it has been suggested that the results of RPB1 phylogenies, which consistently show independent origins for red algae and green plants, may be a phylogenetic artifact because of biases among RPB1 sequences.
To address these issues, we recently performed an extensive investigation of a number of key molecular data sets, including RPB1, with respect to the relative branching positions of green plants and red algae (17). These analyses indicate that the separate origins of red algae and green plants recovered in RPB1 trees are not caused by attraction of rhodophyte sequences toward the more divergent sequences near the base of the tree. In fact, an overall evaluation of the biases that do exist in the RPB1 data set, as well as an empirical assessment of long-branch attraction among RPB1 sequences, reveals that phylogenetic artifacts act to reduce support for the divergent position of red algae, rather than to cause it. At present, relationships among many major eukaryotic taxa, including green plants and red algae remain unclear, but there does not appear to be convincing evidence from molecular phylogenetic studies that is in conflict with the CTD-clade hypothesis (17).
Origin of Heptad Repeats?
Unlike largest subunits from RNA pol I, RNA pol III, and prokaryotic pols, all RPB1 genes isolated to date have an appreciable C-terminal extension beyond the last universally conserved H domain (Fig. 2). Moreover, repeated heptads, albeit with variant consensus sequences, occur in several taxa outside the CTD-clade (Fig. 1). A number of these variant sequences can complement CTD function in yeast, as long as a regular tandem heptad structure is maintained (ref. 35 and unpublished data). Even in deeply branching organisms with no discernable CTD, the RPB1 C terminus usually is enriched in Tyr, Ser, Thr, and Pro residues (31). Together, these observations suggest that the RPB1 C-terminal extension originated as a tandem array of heptapeptides sequences, the same or similar to the CTD consensus, that subsequently degenerated in a number of eukaryotic lineages.
Figure 2.
Alignment of last 38 H region residues from prokaryotic and eukaryotic RNA polymerase largest subunits. The number of amino acids that occur distal to the extremely conserved GTG motif are indicated after each sequence. If tandem heptads are present, the number is in bold. Absolutely conserved residues are highlighted in black and those with only conservative substitutions in gray. Abbreviated designations are: Bacteria—Ta, Thermoplasma acidophilum and Ec, E. coli; Archaea—Hh, Halobacterium and Sa, Sulfolobus; Pol I—Y1, yeast and M1, mouse; Pol III—Y3, yeast and H3, human; Pol II—Y2, yeast; M2, mouse; A2, Arabidopsis; B2, Bonnemaisonia; L2, Leishmania; G2, Giardia.
A possible clue to the origin of C-terminal heptads may be the importance of the CTD for efficient pre-mRNA splicing. In mammalian, yeast, and plant systems the CTD targets splicing factors to the site of transcription, thereby dramatically increasing splicing efficiency (36, 37). This colocalization of mRNA synthesizing and splicing machinery should increase in importance as the density of introns grows within a given genome. In general, eukaryotes outside the CTD-clade are depauperate of introns (38); however, our survey of RPB1 genes suggests that there may be exceptions to this generalization. For example, unique among the most basal RPB1 sequences, the gene from Mastigamoeba is interrupted by five introns. If this intron density reflects the general condition of the Mastigamoeba genome, it would be unusually high for a deeper branching eukaryote (38). If the initial selective advantage of a tandem heptad domain was to increase the efficiency of splicing, it is not be surprising to find that the domain has degenerated in those lineages from which most introns were lost.
The details of mRNA transcription and processing have not been clarified for most organisms outside the CTD-clade. As more genes are isolated from diverse eukaryotes, it will be interesting to see whether an apparent correlation between intron density and conservation of heptad structure persists, or whether other functional constraints emerge as more likely explanations for the presence or absence of heptads in deep-branching eukaryotes. Regardless of the ultimate explanation for the origin of tandem heptads, the extremely low intron density in yeast and microsporidians make it clear that the CTD's role in splicing cannot be solely responsible for the intense stabilizing selection on tandem heptad structure in members of the CTD-clade.
A New Functional Role for Heptads?
A CTD-clade is recovered consistently in RPB1-based phylogenetic trees; as new sequences have been isolated from a wide variety of eukaryotes, the phylogenetic bifurcation between those containing, and those missing a canonical CTD has remained intact (refs. 17 and 18, Fig. 1). Although this node distinctly groups CTD containing taxa, it does not appear to correlate with other obvious organismal characteristics. Multicellular, unicellular, parasitic, and free-living forms occur both inside and outside the CTD-clade. Most significant is the presence of the CTD in the Microsporidia, a group of amitochondriate intracellular parasites once believed to be among the earliest branches of eukaryotic evolution but now considered to be close relatives of fungi (33). Microsporidians have the most severely reduced molecular machinery among eukaryotes; for example, they have lost all but the most essential portions of their major ribosomal RNA subunits, including regions that are otherwise present even in prokaryotic homologues (39). Yet canonical heptad repeats are retained in both of the microsporidians examined (33). In contrast, other amitochondriate and highly derived parasitic taxa, which lie outside the CTD-clade, have no semblance of a CTD whatsoever (e.g., ref. 19).
Our hypothesis of extreme stabilizing selection in members of the CTD-clade is in agreement with evidence from in vivo genetic investigations that show the CTD to be essential for viability in both complex multicellular metazoans, as well as more developmentally simple unicellular fungi (40–42). Thus, if heptapeptide repeats were present in the ancestors of most extant eukaryotic taxa, they must not have evolved under the same strict functional constraints in lineages outside the CTD-clade. This finding suggests that tandem heptads acquired a new function, or set of functions, in the ancestor of the CTD-clade that transformed Y-S-P-T-S-P-S repeats into an essential component of RNA pol II.
Constraint Within the CTD-Clade.
As discussed above, no animal, plant, or fungal RPB1 sequence found to date lacks a CTD. Although substitutions that deviate from the canonical sequence have been commonplace within individual heptads throughout CTD evolution (4), the overall repetitive structure of the CTD has been highly conserved in these groups. Even when large numbers of individual substitutions have accumulated (2, 4, 30), the global structure of tandemly repeated heptads is retained. For example, in Drosophila melanogaster only two of 44 heptads present have the canonical CTD consensus sequence, whereas approximately 75% lie in the tandemly repeated register (2). Again, these evolutionary observations are in accord with mutational analyses of CTD function, which indicate that maintenance of a tandem heptad structure is required for CTD function. In the yeast CTD-based transcription system, various individual differences including some of those found in Mastigamoeba and red algal C-terminal sequences are not lethal as long as heptads are maintained in tandem repeats (35, 43); however, cells are inviable when the tandem register is disrupted (unpublished data). Thus, genetic and biochemical analyses of CTD-based pol II transcription in yeast and animals are entirely consistent with our evolutionary hypothesis that strong stabilizing selection on tandem heptad structure uniquely characterize members of the CTD-clade.
Why a tandemly repeated CTD is essential in these organisms remains unclear. Alteration of a regular physical structure certainly could interfere with the binding of one more of the CTD-related proteins required for the efficient initiation, elongation, and processing of mRNA transcripts. Disruption of the heptad register also might inhibit efficient phosphorylation of the CTD, which is essential for both transcript elongation and the binding of processing-related proteins. Multiple kinases phosphorylate various CTD residues (44) and the pattern of phosphorylation is complex, often involving other components of the pol II transcription apparatus (5). If these kinases operate in a cooperative or hierarchical manner, then maintenance of the correct spacing between Tyr and/or Ser-Pro residues may be essential for substrate recognition in each sequential step of CTD phosphorylation (45).
The circular dichroism of tandem heptad fragments indicates that at least eight repeats are necessary to achieve a secondary structure resembling that of the full-length mouse CTD (46). It is interesting that this also is the minimum number of heptads needed to convey viability in yeast (43). Thus, truncation of the tandem repeats, as well as disruption of their regular structure, both have a dramatic impact on CTD conformation and, therefore, on the ability of CTD heptapeptides to bind other proteins properly. Based on the complexity of CTD-protein interactions throughout the pol II transcription cycle, it seems likely that multiple factors play a role in the strong selection on tandem structure in members of the CTD-clade.
The CTD and the Evolution of Eukaryotic Complexity.
Whichever of their myriad of functions originally turned tandem heptads into an indispensable component of RNA pol II, their importance in the subsequent elaboration of gene regulation is clear. Both biochemical and genetic assays have demonstrated that the CTD plays a central role during the entire transcription cycle by regulating pol II activity through reversible phosphorylation, acting as a platform for assembling the transcription complex, and participating directly in certain processing reactions (36). Our phylogenetic analyses suggest that this full complement of essential functions first coalesced in the common ancestor of the CTD-clade.
The surprisingly small number of genes found in the human genome (47, 48) illustrates the importance of evolutionary advances in the control of gene expression. Developmentally complex organisms do not appear to be distinguished so much by their total number of genes, as by the number of ways these genes can be expressed and controlled. Green plants and metazoans are the only eukaryotic groups whose members are primarily multicellular and are known to have developmental programs tightly controlled by regulated expression of homeotic genes. In addition, nuclear mRNA synthesis in animals, plants, and fungi requires multiple protein–RNA interactions to successfully cap, splice, polyadenylate, and cleave a completed message. What is more, these various steps in mRNA synthesis are found to be interdependent and are accomplished in a coordinated manner by holoenzyme complexes consisting of the pol II core enzyme and scores of other proteins (5–16).
In animals and yeast, the CTD is essential for regulating these processes at a number of levels. The hypophosphorylated CTD binds the mediator that transduces control signals to the pol II/promoter complex (6). The CTD also recruits SR (serine-arginine) proteins and other splicing factors to the elongating message (37, 49), thereby mediating alternative splicing of exon junctions to produce different tissue-specific or developmentally specific products from the same gene. Approximately 40% of genes in the human genome appear to be subject to such alternative splicing, resulting in a more than 3-fold increase in complexity of gene products over gene content (50). Thus, full-scale integration of RPB1 C-terminal heptads into the pol II transcription cycle, and the dramatic increase in flexibility they confer upon gene regulation, was likely one of the prerequisites for the evolution of large, multicellular organisms with true tissue differentiation and complex patterns of development.
Broader examinations are needed, both of the evolutionary history of the CTD and the types of physiological interactions that constrain its structure. The combined evidence to date, however, suggests that the coalescence of CTD function, as characterized in animals, plants, and fungi, dramatically enhanced the capacity to regulate gene expression in one particular group of eukaryotes and helped pave the way for an explosion of diversity in its descendents.
Abbreviations
- CTD
C-terminal domain
- pol
polymerase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Langer D, Hain J, Thuriaux P, Zillig W. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 3.Pühler G, Leffers H, Gropp F, Palm P, Klenk H-P, Lottspeich F, Garret R A, Zillig W. Proc Natl Acad Sci USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corden J L. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 5.Riedl T, Egly J M. Gene Expression. 2000;9:3–13. doi: 10.3727/000000001783992704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim Y-J, Björklund S, Li Y, Sayre M H, Kornberg R D. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 7.Nonet M L, Young R A. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laio S-M, Taylor I C A, Kingston R E, Young R A. Genes Dev. 1991;5:2431–2440. doi: 10.1101/gad.5.12b.2431. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto S, Watanabe Y, van der Spek P J, Watanabe T, Fujimoto H, Hanaoka F, Ohkuma Y. Mol Cell Biol. 2001;21:1–15. doi: 10.1128/MCB.21.1.1-15.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M G, Gustafsson C M, Erdjument-Bromage J, Tempst P, Svejstrup J Q. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 11.Maldonado E, Shiekhatter R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. Nature (London) 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 12.Cho E-J, Takagi T, Moore C R, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer P, Pesce C G, Baralle F E, Kornblihtt A R. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCrackin S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 15.Ho C K, Sriskanda V, McCrackin S, Bentley D, Schwer B, Shuman S. J Biol Chem. 1998;273:9577–9585. doi: 10.1074/jbc.273.16.9577. [DOI] [PubMed] [Google Scholar]
- 16.Steinmetz E J. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 17.Stiller J W, Riley J, Hall B D. J Mol Evol. 2001;52:527–539. doi: 10.1007/s002390010183. [DOI] [PubMed] [Google Scholar]
- 18.Stiller J W, Duffield E C S, Hall B D. Proc Natl Acad Sci USA. 1998;95:11769–11774. doi: 10.1073/pnas.95.20.11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evers R, Hammer A, Köck J, Waldemar J, Borst P, Mémet S, Cornelissen A W C A. Cell. 1989;56:585–597. doi: 10.1016/0092-8674(89)90581-3. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 22.Swofford D L. paup. Sunderland, MA: Sinauer; 1997. , Version 4.0b3a. [Google Scholar]
- 23.Felsenstein J. Cladistics. 1989;5:164–165. [Google Scholar]
- 24.Huelsenbeck J P, Ronquist F. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 25.Kishino H, Hasegawa M. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 26.Templeton A R. Evolution. 1983;37:221–244. doi: 10.1111/j.1558-5646.1983.tb05533.x. [DOI] [PubMed] [Google Scholar]
- 27.Moreira D, Le Guyader H, Philippe H. Nature (London) 2000;405:69–72. doi: 10.1038/35011054. [DOI] [PubMed] [Google Scholar]
- 28.Burger G, Saint-Louis D, Gray M W, Lang B F. Plant Cell. 1999;11:1675–1694. doi: 10.1105/tpc.11.9.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stiller J W, Hall B D. J Phycol. 1998;34:857–864. [Google Scholar]
- 30.Nakajima K, Chang Y C, Suzuki T, Jigami Y, Machida M. Biosci Biotechnol Biochem. 2000;64:641–646. doi: 10.1271/bbb.64.641. [DOI] [PubMed] [Google Scholar]
- 31.Giesecke H, Barale J C, Langsley G, Cornelissen A W. Biochem Biophys Res Commun. 1991;180:1350–1355. doi: 10.1016/s0006-291x(05)81344-0. [DOI] [PubMed] [Google Scholar]
- 32.Shimodaira H, Hasegawa M. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 33.Hirt R P, Logsdon J M, Healy B, Dorey M W, Doolittle W F, Embley T M. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldauf S L, Roger A J, Wenk-Siefert I, Doolittle W F. Science. 2000;290:972–977. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 35.Stiller J W, McConaughy B L, Hall B D. Yeast. 2000;16:57–64. doi: 10.1002/(SICI)1097-0061(20000115)16:1<57::AID-YEA509>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 36.Hirose Y, Manley J L. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 37.Cordon J L, Patturajan M. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 38.Logsdon J M. Curr Opin Genet Dev. 1998;8:637–648. doi: 10.1016/s0959-437x(98)80031-2. [DOI] [PubMed] [Google Scholar]
- 39.Peyretaillade E, Biderre C, Peyret P, Duffieux F, Metenier G, Gouy M, Michot B, Vivares C P. Nucleic Acids Res. 1998;26:3513–3520. doi: 10.1093/nar/26.15.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nonet M, Sweetser D, Young R A. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 41.Bartolomei M S, Halden N F, Cullen C R, Corden J L. Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zehring W A, Lee J M, Weeks J R, Jokerst R S, Greenleaf A L. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West M L, Corden J L. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trigon S, Serizawa H, Weliky Conaway J, Conaway R C, Jackson S P, Morange M. J Biol Chem. 1998;273:6769–6775. doi: 10.1074/jbc.273.12.6769. [DOI] [PubMed] [Google Scholar]
- 45.Roach P J. J Biol Chem. 1991;262:14042–14048. [PubMed] [Google Scholar]
- 46.Bienkiewicz E A, Moon Woody A-Y, Woody R W. J Mol Biol. 2000;297:119–133. doi: 10.1006/jmbi.2000.3545. [DOI] [PubMed] [Google Scholar]
- 47.Genome International Sequencing Consortium. Nature (London) 2001;409:860–921. [Google Scholar]
- 48.Venter J C, Adams M D, Myers E W, Li P W, Mural R J, Sutton G G, Smith H O, Yandell M, Evans C A, Holt R A, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 49.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]