Abstract
Eusocial insects are characterized by reproductive division of labor, cooperative brood care, and the presence of a sterile worker caste. It is generally accepted that caste determination, including the differentiation of females into sterile workers and reproductive queens, is determined by environmental factors. In contrast, we find that in the red harvester ant, Pogonomyrmex barbatus, an individual's genotype at a particular microsatellite locus predicts its caste. We propose that this microsatellite locus is in tight linkage disequilibrium with at least one locus that plays an important role in caste determination. We call this the caste locus. We hypothesize that the system of caste determination we observe segregates the population into two distinct genetic lineages, each of which has distinct alleles at the microsatellite locus and also has distinct alleles, we propose, at caste. Workers are the offspring of parents from different lineages, and are thus heterozygous at caste, whereas queens are the offspring of parents from the same lineage, and are, therefore, homozygous at caste. This mode of caste determination has important consequences for the evolution of multiple mating by females and for control of the sex ratio and reproductive allocation in social insect colonies.
Studies of caste determination in the social Hymenoptera (ants, bees, and wasps) have shown that the development of a female larva into a sterile worker or a reproductive queen depends on environmental factors (1–3). There are only two known cases in which caste determination is not purely environmental (4). In both cases (5–10), a genetic factor confers the potential for a female larva to develop into a queen, but the caste of the larva ultimately depends on environmental factors such as nutrition (11, 12), incubation temperature (13), egg size and queen age (14), as well as queen influence (15, 16). It has been argued that environmental caste determination is essential to the evolution of eusociality (17).
Pogonomyrmex barbatus is a perennial ant species that reproduces by sending winged reproductives (males and virgin queens) to an annual mating flight. After mating, males die, whereas newly mated queens leave the mating aggregation to found new colonies. P. barbatus is a monogynous, polyandrous species; each ant colony is founded by a single queen that has mated with one or more males. A queen lives for 15–20 years (18), using sperm from her original mating to produce all of the colony's workers and reproductives. No additional queens are adopted during the lifetime of the colony.
Methods
Study Population.
All ant samples were collected from a population of P. barbatus colonies that has been part of an ongoing demographic and behavioral study for the past 20 years (19). Workers were collected from 40 known colonies of the study population. Of the 59 queens we genotyped, 43 were collected from a total of 19 known colonies before the onset of the mating flight, and 16 were taken from the mating aggregation. Of the 62 males we genotyped, 1 individual was taken from each of 16 known colonies, and 46 males were collected from the aggregation. The colonies of origin are not known for reproductives taken from the mating aggregation. Two congeners of P. barbatus are present in the vicinity of the study population. Pogonomyrmex maricopa and Pogonomyrmex rugosus colonies can be found nearby, but not immediately around or within the boundaries of the study site. P. rugosus is rarely found at the P. barbatus mating aggregation, whereas P. maricopa is often present (unpublished observations). There is no evidence of hybridization between P. barbatus and P. maricopa.
Microsatellite Amplification.
As part of a larger, multilocus analysis of population structure in the red harvester ant, we used primers for microsatellite locus Myrt3, originally developed for Myrmica tahoensis (20), to cross-amplify this locus in P. barbatus. We also genotyped workers from a subset of the 40 colonies included in this study at ten microsatellite loci, Pb 1–Pb 10, developed specifically for P. barbatus (21). DNA was extracted from ant heads by crushing the sample with a Teflon pestle, adding 150 μl of 10% Chelex solution (Bio-Rad), and incubating at 95°C for 20 min. The samples were centrifuged for 2 min, and the supernatant containing the DNA was removed. Two microliters of each sample were used in 15 μl of PCRs containing 0.5 units of Taq DNA Polymerase (Qiagen, Chatsworth, CA), 0.8 mM dNTPs, 1× Q-solution (Qiagen), 1× PCR buffer (Qiagen), and 0.5 μM of each primer, where the reverse primer was fluorescently labeled with 6-FAM (Operon). The PCR cycle consisted of 95°C for 5 min, followed by 25 cycles of 95°C for 30 s, 48°C for 30 s, 72°C for 30 s, followed by 72°C for 7 min.
Microsatellite Analysis.
PCR products were run on 5% Sequagel (National Diagnostics) acrylamide gels on an ABI 377 automated sequencer (Applied Biosystems). Gels were analyzed with GENESCAN V.3.1.2. software (Perkin-Elmer). Myrt3 is an imperfect microsatellite locus composed of several arrays of dinucleotide repeats. This locus has five alleles in P. barbatus, labeled 2, 4, 6, 10, and 12. The numerical difference between the allele labels is the number of basepairs by which the alleles differ in size.
Statistical Analyses.
Expected heterozygosities and significance levels were estimated by implementing a randomized resampling procedure that takes into account the colony structure of the sample of workers. The effective number of mates, me, was calculated for each colony from the average relatedness among sisters of the same matriline, rs, by using the equation rs = 0.25 + 0.5(1/me) (22). Average within-colony relatedness of workers was calculated for each colony in RELATEDNESS V.5.0.8 (23) by using data from microsatellite loci Pb 1–Pb 10 and weighting individuals equally.
Results and Discussion
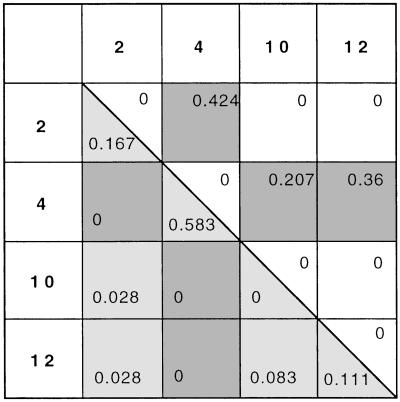
All 563 workers we genotyped were heterozygous at locus Myrt3. Moreover, we found that 555 of the 563 workers were heterozygous for allele 4. Their genotype was 4,X, where X is 2, 10, or 12 but never 4 or 6 (Fig. 1). The only exception was found in a single colony, where 8 of 15 workers genotyped were 6,X instead of 4,X (Table 3, which is published as supporting information on the PNAS web site, www.pnas.org).
Figure 1.
Genotype frequencies among queens and workers. Light-shaded half of the genotype matrix (lower left) contains genotype frequencies among queens. Because individuals collected from the same colony do not represent independent samples, we used one queen per colony to calculate genotype frequencies. Queens collected from the mating aggregation were considered independent samples. Genotype frequencies are based on data from 36 queens. The upper right half of the genotype matrix contains genotype frequencies among workers. Genotype frequencies were calculated from colonies from which we had equal numbers of samples (15 workers from each of 28 colonies). Dark-shaded cells correspond to heterozygotes for allele 4. Note that all workers, but no queens, are heterozygous for allele 4. For clarity of presentation, the rare allele 6, which was found in eight workers but no queens (see text), is not included in the table.
To investigate further the transmission of allele 4, we genotyped P. barbatus reproductives from the study population. All queens were either 4,4 homozygotes, or X,X homozygotes or heterozygotes (Fig. 1; Table 4, which is published as supporting information on the PNAS web site). Males, which develop from unfertilized eggs and are thus haploid, were either 4 or X (Table 1). Allele 6 was not observed among any of the reproductives. Thus, with the exception of the eight workers that carried allele 6, all workers had one copy of allele 4, whereas queens had either two or zero copies of allele 4. A G test of association shows a highly significant association between genotype and caste: G = 104, d.f. = 2, P < 10−22 (Table 2).
Table 1.
Genotype frequencies among males
| Genotype | Frequency |
|---|---|
| 2 | 0.290 |
| 4 | 0.387 |
| 10 | 0.097 |
| 12 | 0.226 |
Genotype frequencies were calculated from 62 males (one male from each of 16 colonies, and 46 males from the mating aggregation).
Table 2.
Test of association using G test
| Genotype | Queens | Workers |
|---|---|---|
| 4,4 | 21 | 0 |
| 4,X | 0 | 39 |
| X,X | 15 | 0 |
Due to nonindependence of individuals sampled from the same colony, we used one ant per colony for this test. We used data from the 36 queens used to calculate genotype frequencies for Fig. 1. For workers, we included one individual from each colony. The single colony with workers of genotype 6,X (see text) was excluded, leaving a total of 39 colonies (G = 104, d.f. = 2, P < 10−22).
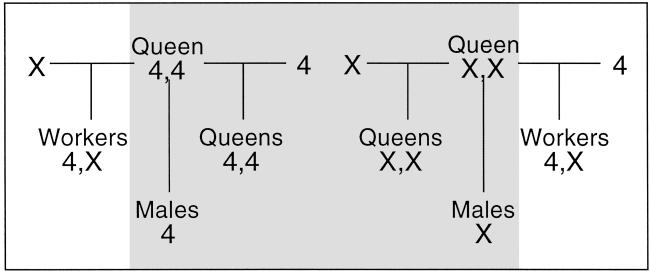
Examination of the mating structure shown in Fig. 2 suggests that this population of P. barbatus is effectively segregated into two distinct lineages. We call these lineage 4 and lineage X. Female reproductives result from the mating of a queen with a male from the same lineage. Workers result from the mating of a queen with a male from the other lineage. Therefore, workers are the only individuals in which the genomes from both lineages are present. However, if workers do not reproduce, there will be no genetic exchange between lineage 4 and lineage X. The two lineages thus would be evolving separately.
Figure 2.
Diagrams of possible matings. Shaded area represents single matings from which workers will not develop.
Our data show that the genotype of an individual at Myrt 3 is predictive of its caste. We postulate the existence of at least one locus (a gene or set of genes) that plays a central role in caste determination. We call this the caste locus, and propose that alleles 4 and 6 of Myrt3 are in strong positive linkage disequilibrium with an important allele at caste. All individuals that are heterozygous for this allele at caste develop into workers. Individuals that possess two copies or no copies of this allele become queens.
Myrt3 is not necessarily close to the caste locus in the genome, as the linkage disequilibrium between the two loci is probably because of the segregation of the population into two lineages. If the two lineages are evolving independently, one would expect to find genome-wide differences between them. Genetic drift should operate at neutral loci throughout the genome to gradually create distinct allele pools in the two lineages. If the frequencies of particular alleles at a given locus differ between the two lineages, one would expect increased levels of heterozygosity among workers. In fact, 4 of 10 loci developed specifically for P. barbatus show significantly higher levels of heterozygosity in workers than would be expected under the null hypothesis of population-wide random mating (randomization test; Pb 7, P = 0.002; Pb 8, P < 0.001; Pb 9, P = 0.001; Pb 10, P < 0.001). Two of these loci, Pb 8 and Pb 9, exhibit 100% heterozygosity. Because of the large number of alleles at these loci (17 and 25, respectively), a pattern as clear as that observed at Myrt3 does not emerge. It will be necessary to genotype reproductives at these loci to determine whether the alleles present in the workers segregate into two distinct lineages. (Although it is not unusual to observe lower-than-expected levels of heterozygosity because of the presence of null alleles (24), significantly increased heterozygosity is not generally artifactual. Two of the 10 loci show low heterozygosity: Pb 4, P = 0.002; Pb 5, P < 0.001).
Our data do not establish whether caste is determined at the time of fertilization. The possibility remains that individuals of any genotype initiate development into either caste, but are culled according to genotype, removing 4,X queens as well as 4,4 and X,X workers at some point before they emerge from their natal colony. The elimination of 4,X queens and 4,4 or X,X workers could be because of intrinsic developmental causes, or because of extrinsic factors such as the execution of these individuals by members of the same colony, as in the behavioral response of the red fire ant, Solenopsis invicta, toward conspecifics of a certain genotype (25).
Consideration of the possible matings that can take place among P. barbatus reproductives (Fig. 2) reveals several striking implications of the mode of caste determination we have found. First, the system of caste determination described here may produce a strong selective pressure in favor of polyandry. Several hypotheses for the evolution of polyandry have been advanced (26, 27), but few have unequivocal, empirical support (28). Under the system of caste determination described here, a queen whose mates were all from a single lineage could produce either workers or female reproductives, but never both. Queens that produce only reproductives (Fig. 2, shaded area) may not be able to establish colonies if workers are necessary to raise the brood. If p1 is the frequency of 4,4 among queens, and p2 is the frequency of 4 among males, the expected frequency of a single mating that will not result in workers is p1p2 + (1−p1)(1−p2). The expected frequency of n independent matings that will not result in workers is p1p2n + (1−p1)(1−p2)n. The observed genotype frequencies among the reproductives (Fig. 1 and Table 1) suggest that under random mating, 48% of singly mated queens, 24% of doubly mated queens, and 13% of triply mated queens would produce only reproductives and thus fail to establish colonies. Furthermore, a queen whose matings allow her to produce workers (Fig. 2, nonshaded area) will not be able to produce female reproductives unless her mates were from different lineages. If her mates were from the same lineage, all of her reproductives will be male. We do indeed observe multiple mating by P. barbatus queens. All 23 colonies for which workers were genotyped at Pb 1–Pb 10 have four or more alleles at least at one locus, which excludes the possibility that the queen mated only once. The average effective mate number, me, which takes into account the relative contributions of each male, is 3.34 for the study population.
A further consequence of the type of caste determination we describe here is that it prevents workers of singly mated queens from maximizing their own inclusive fitness by controlling the sex ratio among the reproductives. The split sex ratio hypothesis states that workers of a singly mated queen will increase their inclusive fitness by producing a female-biased sex ratio (29). This hypothesis has been corroborated by empirical studies (30). Under the type of caste determination we present here, workers of a singly mated queen will never have the opportunity to bias the sex ratio in their favor, because such queens have exclusively males among their reproductives.
The mode of caste determination we find in P. barbatus also imposes constraints on reproductive allocation, the division of resources between colony maintenance and reproduction. A genetic system of caste determination will constrain the ability of a colony to adjust its ratio of workers to reproductives according to its changing needs in different seasons and stages of life. Because the lifespan of a worker does not exceed 1 year (31), a colony must produce workers throughout its lifetime. P. barbatus colonies do not produce reproductives until the colony attains a size of 10,000–12,000 workers, which occurs at about 5 yrs of age (32, 33). Furthermore, once colonies have reached reproductive maturity, adult reproductives are apparently produced seasonally in preparation for the mating flight, as in other Pogonomyrmex species (34). Under the mode of caste determination described here, a colony's ability to adjust its investment in workers vs. reproductives is severely constrained. Although workers could control reproductive allocation by selectively feeding or neglecting certain larvae, they will have to make use of the initial ratio of workers to reproductives provided by the queen, which depends on the genotypes and number of males with which she mated. As a result of a colony's adjustment of this initial ratio, some eggs will not complete their development. Consequently, it may be costly for colonies to adjust the ratio of workers to reproductives.
Although very little is known about the genetic variation for traits relevant to social evolution, most theories about the evolution of eusociality are based on genetic models (35). Empirical research is now beginning to uncover genetic patterns underlying social systems. Recent work on the red fire ant, S. invicta, has identified a gene that may affect workers' abilities to recognize queens and regulate their numbers (36). Our study of queen–worker dimorphism in the red harvester ant, P. barbatus, reveals a genetic basis for differences between the two castes. We hypothesize that the mode of caste determination segregates the population of P. barbatus into two distinct lineages that are evolving without genetic exchange. Yet the lineages remain codependent, as the establishment of colonies depends on the viability of workers. Not only must the genomes of the two lineages remain compatible with one another, but recognition ability and reproductive compatibility between the lineages must be maintained as well. Thus, although the lineages are following independent evolutionary trajectories, they are highly constrained in their degree of divergence. The unveiling of such genetic underpinnings of social systems may provide additional insight into the evolution and maintenance of eusociality.
Supplementary Material
Acknowledgments
We thank Marc Feldman, Aaron E. Hirsh, Krista K. Ingram, Dmitri A. Petrov, and Guy Sella for helpful discussion and comments on the manuscript. We also thank two anonymous referees whose comments helped us improve the manuscript. We are especially indebted to Aaron E. Hirsh for developing the randomization test used in this study. This work was funded by grants from the Center for Evolutionary Studies at Stanford University, Sigma-Xi, and the American Museum of Natural History (to V.P.V.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Michener C D. The Social Behavior of the Bees. Cambridge, MA: Harvard Univ. Press; 1974. [Google Scholar]
- 2.Wilson E O. The Insect Societies. Cambridge, MA: Harvard Univ. Press; 1971. [Google Scholar]
- 3.Craig R, Crozier R H. Isozyme Bull. 1978;11:66–67. [Google Scholar]
- 4.Crozier R H. Annu Rev Entomol. 1977;22:263–288. [Google Scholar]
- 5.Buschinger A, Winter U. Insectes Soc. 1975;22:333–362. [Google Scholar]
- 6.Buschinger A. Naturwissenschaften. 1975;62:239–240. [Google Scholar]
- 7.Buschinger A, Winter U. Insectes Soc. 1978;25:63–78. [Google Scholar]
- 8.Buschinger A. Insectes Soc. 1978;25:163–172. [Google Scholar]
- 9.Buschinger A. Naturwissenschaftliche Rundschau. 1992;45:85–92. [Google Scholar]
- 10.Kerr W E. Genetics. 1950;35:143–152. doi: 10.1093/genetics/35.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brian M V. Insectes Soc. 1956;3:369–394. [Google Scholar]
- 12.Wesson L G J. Psyche. 1940;47:105–111. [Google Scholar]
- 13.Gösswald K, Bier K. Insectes Soc. 1957;4:335–348. [Google Scholar]
- 14.Brian M V, Hibble J. Insectes Soc. 1964;11:223–238. [Google Scholar]
- 15.Brian M V, Hibble J. Insectes Soc. 1963;10:71–81. [Google Scholar]
- 16.Gösswald K, Bier K. Insectes Soc. 1954;1:305–318. [Google Scholar]
- 17.Bourke A F G, Franks N R. Social Evolution in Ants. Princeton: Princeton Univ. Press; 1995. [Google Scholar]
- 18.Gordon D M. Am Nat. 1991;138:379–411. [Google Scholar]
- 19.Gordon D M, Kulig A W. Ecology. 1996;77:2393–2409. [Google Scholar]
- 20.Evans J D. Mol Ecol. 1993;2:393–397. doi: 10.1111/j.1365-294x.1993.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 21. Volny, V. P. & Gordon, D. M. (2002) Mol. Ecol. Notes, in press.
- 22.Pamilo P. Heredity. 1993;70:472–480. [Google Scholar]
- 23.Goodnight K F, Queller D C. RELATEDNESS V.5.0.8. Houston: Goodnight Software; 2001. [Google Scholar]
- 24.Brookfield J F Y. Mol Ecol. 1996;5:453–455. doi: 10.1111/j.1365-294x.1996.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 25.Keller L, Ross K G. Nature (London) 1998;394:573–575. [Google Scholar]
- 26.Cole B J. Behav Ecol Sociobiol. 1983;18:165–173. [Google Scholar]
- 27.Crozier R H, Page R E. Behav Ecol Sociobiol. 1985;18:105–115. [Google Scholar]
- 28.Strassmann J. Insectes Soc. 2001;48:1–13. [Google Scholar]
- 29.Boomsma J J, Grafen A. Evolution (Lawrence, Kans) 1990;44:1026–1034. doi: 10.1111/j.1558-5646.1990.tb03823.x. [DOI] [PubMed] [Google Scholar]
- 30.Sundström L, Chapuisat M, Keller L. Science. 1996;274:993–995. doi: 10.1126/science.274.5289.993. [DOI] [PubMed] [Google Scholar]
- 31.Gordon D M, Hölldobler B. Psyche. 1987;94:341–346. [Google Scholar]
- 32.Gordon D M. Anim Behav. 1995;49:649–659. [Google Scholar]
- 33.Gordon D M. Behav Ecol Sociobiol. 1992;31:417–427. doi: 10.1007/s00265-015-2045-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacKay W P. Psyche. 1981;88:25–73. [Google Scholar]
- 35.Crozier R H, Pamilo P. Evolution of Social Insect Colonies: Sex Allocation and Kin Selection. Oxford: Oxford Univ. Press; 1996. [Google Scholar]
- 36.Krieger M J B, Ross K G. Science. 2002;295:328–332. doi: 10.1126/science.1065247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.