Abstract
Control of gene expression requires cis-acting regulatory DNA sequences. Historically these sequences have been difficult to identify. Conserved noncoding sequences (CNSs) have recently been identified in mammalian genes through cross-species genomic DNA comparisons, and some have been shown to be regulatory sequences. Using sequence alignment algorithms, we compared genomic noncoding DNA sequences of the liguleless1 (lg1) genes in two grasses, maize and rice, and found several CNSs in lg1. These CNSs are present in multiple grass species that represent phylogenetically disparate lineages. Six other maize/rice genes were compared and five contained CNSs. Based on nucleotide substitution rates, these CNSs exist because they have biological functions. Our analysis suggests that grass CNSs are smaller and far less frequent than those identified in mammalian genes and that mammalian gene regulation may be more complex than that of grasses. CNSs make excellent pan-grass PCR-based genetic mapping tools. They should be useful as characters in phylogenetic studies and as monitors of gene regulatory complexity.
The elucidation of gene regulatory networks depends on identification of cis-acting elements and the products that bind them. Cross-species genomic DNA comparisons offer a powerful method for finding cis regulatory sequences (1). Rice and maize are two domesticated species in the Poaceae, the grass family of flowering plants, which includes wheat, barley, sugarcane, and other cultivated staples. This economically important family has about 10,000 species that have been grouped together based on shared morphological traits and nucleotide sequences. Phylogenies built on these data suggest that grasses are monophyletic, with an ancestral genome existing about 50 million years ago (MYA) for almost all of the common grasses, and perhaps 70 MYA if the basal grass genera are included (2). Many grass genes are similar enough at a nucleotide level to permit comparative grass gene mapping by cross species Southern blot hybridization (3). Current data support the hypothesis that all grasses have approximately the same genes in roughly the same order (4) although this order may need to be deduced by a process called consolidation (5). If so, then the morphological and physiological diversity of grasses is essentially allelic diversity distributed phylogenetically. Patterson and coworkers (6) have shown that domestication of several grasses has focused on selection for allelic variations at the same ancestral genes. Thus, it is of particular importance and interest to identify and phylogenetically analyze those potential regulatory sequences responsible for the allelic diversity. There are now enough orthologous genomic DNA sequences in grasses, notably between maize and rice, to begin to address the question of grass gene regulatory diversity.
Regulatory elements have historically been difficult to identify because, unlike exons, they are not constrained by the codon syntax necessary for use in translation. Traditionally, cis-acting regulatory elements have been identified through biochemical and molecular genetic experimental approaches. Regulatory sequences are generally small, e.g., 6–8 nt long (7–10). A recent advance in identifying mammalian cis regulatory elements was made by leveraging the power of genomic DNA comparisons based on the premise that these functionally important regulatory regions will be conserved between species (1). Comparisons between a syntenous human and mouse genomic DNA segment identified conserved noncoding sequences (CNSs) that were experimentally shown to regulate the expression of several nearby genes (11). CNSs correspond to regions of endonuclease sensitivity in human/mouse comparisons (12) as well as other previously defined regulatory elements (10, 13, 14). A study of 502 human genes showed that regulatory sequences are enriched in CNSs compared with nonconserved noncoding regions (15). Most identified CNSs have no experimentally defined function (16). There are now several web-based tools that can be used to identify CNSs (17, 18) and functional RNAs (19). Very few examples of CNSs have been reported in plants (20, 21).
In this article we identify multiple CNSs in grass genes and show they are conserved from a common ancestor over the 50 million years to maize and also over the 50 million years to rice. In addition, we compared higher plant CNS frequency and length distributions with mammalian CNS data. To the extent that CNS frequency and length is a measure of gene complexity, the grass genes we analyzed are far less complex than mammalian genes.
Materials and Methods
Identification of CNSs.
DNA sequences were processed by using Perl scripts to automate CNS discovery. Coding regions of maize and rice genes were identified by comparing genomic sequences to predicted protein translations by using the National Center for Biotechnology Information (NCBI)'s blastx (22). The coding regions were masked out for further comparisons. The two masked genomic sequences were then compared by using NCBI's BL2SEQ (23), with the following parameters: for finding stringent CNSs (Fig. 1A and Table 1) word size = 7, gap penalties, existence = 5, extension = 2; for finding long CNSs (Fig. 1B) word size = 7, gap penalties, existence = 2, extension = 1. The positions of CNSs relative to intron/exon structure were visualized by using Perl scripts to parse the output of both the blastx and BL2SEQ results.
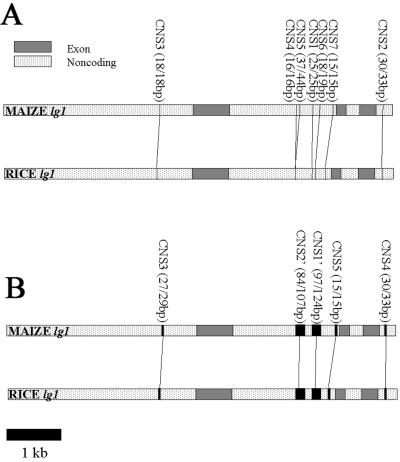
Figure 1.
lg1 CNSs conserved between maize and rice. Graphic display of CNS locations and sizes found when comparing 7,142 bp of maize sequence at lg1 with 7,100 bp of orthologous rice sequence. (A) blast parameters adjusted to favor shorter, more stringent CNSs. (B) blast parameters adjusted to favor longer, less exact matches.
Table 1.
Summary of CNS frequencies and size distributions in seven rice/maize gene pairs
| Gene name | 15–20 bp | 21–30 bp | 31–60 bp | 61–99 bp | >100 bp | Size of compared region, bp |
|---|---|---|---|---|---|---|
| lg1 | 4 | 1 | 2 | 0 | 0 | 7,142 |
| 1g3/osh6 | 1 | 0 | 1 | 0 | 0 | 2,300 |
| Chalcone synthase | 1 | 0 | 0 | 0 | 0 | 4,000 |
| wx1 | 0 | 0 | 0 | 0 | 0 | 4,800 |
| adh1 | 2 | 0 | 0 | 0 | 0 | 6,000 |
| sh2 | 0 | 1 | 0 | 0 | 0 | 7,320 |
| H+ ATPase | 1 | 0 | 0 | 0 | 0 | 6,725 |
| Six gene mammalian region | 26 | 29 | 29 | 10 | 12 | 51,187 |
Maize and rice genomic DNA sequences were analyzed for the presence of CNSs. The genes are listed with the frequency of CNSs of the various size classes indicated. The last column lists the size of the smaller of the two sequences being compared.
Sequence Information.
The GenBank accession number for the rice liguleless1 (lg1) containing bacterial artificial chromosome is AL442117. A total of 7,142 bp of maize lg1 was sequenced from a genomic DNA clone (AF451895). The lg1 intron 1 grasses GenBank accession numbers are: Setaria (AF451892), Arundo (AF451893), and bamboo (AF451894). The GenBank accession numbers for the additional six maize and rice genes used to obtain the data of Table 1 are: adh1 maize (X04049) and rice (AF172282); chalcone synthase maize (X60204.1) and rice (AB058397.1); maize lg3, missing intron3 (AF479591, AF479592) and rice osh6 (AP002881); sh2 maize (M81603) and rice (AF101045); wx1 maize (X03935) and rice (AF141955); H+ ATPase maize (AF480431) and rice (AL442117). The mouse bacterial artificial chromosome is AC002397. The human region is 12p13, U47924.
PCR and Southern Blots.
The lg1 CNS3:exon 1 PCR products were amplified by using primer pair CNS3–2 (gctatcaccmctcaccaactccaattaa; m = a or c) and lg1–77 (cgctggagaggtcggcctt) with 94°C 30 sec, 60°C 30 sec, 72°C 1 min cycling conditions for 35 cycles. The PCR products were hybridized with a combined maize and rice lg1 exon 1-derived probe that did not include the highly conserved SBP domain. The maize exon 1 probe was amplified from a maize lg1 cDNA clone by using primers ccgaattcatggagcagcagcaggagagc and tggccgtacgtgtagcctcctctggg. The rice exon 1 probe was amplified from Nipponbare genomic DNA by using primers atgatgaacgttccatccgcc and tcggcgggaagtaggtccggta. Both amplification conditions were the same as used to amplify the CNS3–2:lg1–77 PCR products. Hybridization and washes were done at 65°C, and the blot was washed in 0.2× SSC/0.2% SDS. lg1 intron 1 from various grasses was PCR-amplified in two overlapping fragments by using an exon 1-derived primer (lg1–77 Rev, aaggccgacctctccagcg) with a CNS 1 primer (tcagctgcaaaactccaagctgtct), and a CNS 4 primer (gatcaagactttgtacagcc) with an exon 2-derived primer (tcttrgcatcgtcgaactcatc; r = a or g). Conditions for Lg1–77Rev:CNS 1–2R were: 97°C 2 min, 30 cycles of 94°C 30 sec, 62°C 1 min, 72°C 2 min. Conditions for CNS 4-lg1–79 were: 97°C 2 min, 30 cycles of 94°C 30 sec, 54°C 30 sec, 72°C 1 min. The PCR products were cloned by using Invitrogen's pTOPO cloning system and sequenced at the University of California-Berkeley DNA sequencing facility. The fragments were assembled by using dnastar seqman ii software. When necessary, strand-specific primers were used to fill in any gaps in the sequences.
Plant Materials.
We used the following sources of grasses for our analyses. Maize is Zea mays L. inbred B73 (gift of Pioneer Hi-Bred Seed International, Des Moines, IA, UT90323-C3OC) and rice is Oryza sativa L. cv. Nipponbare, a gift from Torrey Mesa Research Institute, Syngenta. The bamboo, Bambusa multiplex (Lour.) Raeusch. cv. golden goddess, was purchased from Magic Gardens, Berkeley, CA, and the chloridoid, Muhlenbergia porteri Scribn. Ex Beal, is accession no. 0079538, Royal Botanical Gardens at Kew, Surrey, UK. The Setaria viridis var. Pachystachys was collected from Hagi City, Japan. Arundo donax L. tissue was collected from the University of California-Berkeley Botanical Garden. Sorghum bicolor (L.) Moench cv. Tx430 seeds were a gift from Peggy Lemeaux, University of California-Berkeley.
Results
CNSs Exist in lg1.
lg1 encodes a member of the squamosa promoter binding protein (SBP) family. It is required to specify the ligule, an epidermal organ that exists along a line that bisects the grass leaf into sheath and blade (24–27). Because grasses share the homologous ligule structure, we reasoned that lg1 gene regulation may also be evolutionarily conserved. To assess the conserved regulatory sequences we identified an unannotated rice bacterial artificial chromosome that contains the rice lg1 orthologous gene. This sequence maps to rice chromosome 4L (data not shown), which is the syntenous region to maize chromosome 2S where lg1 resides (26). Within the coding region a comparison of the predicted maize and rice proteins shows them to be 70% similar in amino acid sequence overall and identical in the SBP domain (data not shown). We identified CNSs based on three criteria: conservation of sequence, conservation of position relative to the intron/exon structure of the gene, and a size greater than or equal to 15 bp. For reference, any 15-bp stretch is expected to exist about once (1/4)15, in the entire ≈400-megabasepair rice genome. Therefore, we used 15 identical bp conserved between maize and rice (i.e., 15/15) as our cut-off, but we recognize that shorter sequences are also important.
A comparison of 7,142 bp of maize genomic DNA sequence containing the three exons of lg1 (and about 5.9 kb of noncoding sequence) and 7,100 bp of rice genomic sequence identified seven CNSs that met our stringent criteria (see Materials and Methods). The same CNSs were identified when the entire rice bacterial artificial chromosome sequence (≈50 kb) was used in the comparison (data not shown). Fig. 1A shows lg1 “stringent CNSs” listed as CNSs 1–7 in the order of their significance. These seven CNSs identified in Fig. 1A are too small to fit the definition used by most researchers working with mammalian systems, >100 bp in length and >70% identity (11). However, if we relax the gap and extension penalties to those used by Levy and coworkers (15), we display the lg1 CNSs in a different way (Fig. 1B). Here, we find two long CNSs, CNS1′ (97/124) and CNS2′ (84/107), in intron 1 replacing the CNS4-CNS5-CNS1-CNS6 region of Fig. 1A. Both of these less stringent CNSs meet the >100-bp, >70% identity criteria often used in mammalian studies.
lg1 CNSs Are Conserved Among Many Grasses.
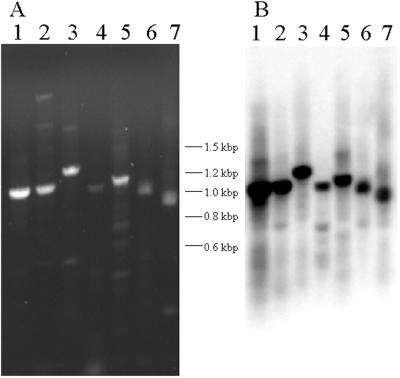
To test whether the CNSs identified between maize and rice are conserved in other grasses we devised a PCR-based assay. CNS3 (Fig. 1A) was used to design a PCR primer (CNS3–2) facing downstream toward exon 1. A second primer (lg1–77) was chosen within exon 1 directed upstream toward the promoter and CNS3. In addition to maize and rice, this primer pair amplified a fragment from five more grass species (Fig. 2). In all, seven tribes and five different subfamilies (panicoids, chloridoids, bambusoids, oryzoids, and arundinoids) are represented (ref. 28 and Grass Phylogeny Working Group at www.ftg.fiu.edu/grass/gpwg). Fig. 2A shows an ethidium bromide-stained gel with one predominant amplification product from each species. Fig. 2B shows a Southern blot of this gel hybridized with a combined maize and rice lg1-specific exon 1 probe. In each case the predominant PCR product is homologous to the lg1 exon probe. Note the lg1 hybridizing sequences are polymorphic in length between the various grasses, which is expected because they contain noncoding sequences.
Figure 2.
lg1 CNS3 is conserved in the grasses. CNS3 was used as a PCR primer site, paired with a lg1 exon 1 site, to amplify the promoter, 5′ untranslated region and first exon regions of lg1 from various grasses. (A) An ethidium bromide-stained gel of the PCR products. The gel was transferred onto a nylon membrane and hybridized with a combined lg1 exon 1 region from maize and rice. (B) The autoradiogram after hybridization. Lane 1, rice (Oryza sativa); lane 2, weedy foxtail (Setaria viridis); lane 3, sorghum (Sorghum bicolor); lane 4, maize (Zea mays); lane 5, Muhlenbergia porteri; lane 6, Arundo donax; lane 7, bamboo (Bambusa multiplex).
To further examine the conservation of CNSs in lg1 we PCR-amplified intron 1 from three other grasses (a bamboo, a Setaria, and an Arundo) and compared them with maize and rice by using clustalx to generate a five-way sequence alignment (29). These five sequences represent five tribes and four subfamilies of grasses. The results are displayed in Table 2. Full intron sequences are shown in Fig. 3, which is published as supporting information on the PNAS web site, www.pnas.org. All five intron 1 stringent CNSs are highly conserved among all these grasses in both sequence and position. Note that when the bamboo, Setaria, and Arundo sequences were added to the original rice and maize comparison, the CNSs became more refined.
Table 2.
lg1 intron 1 CNSs are present in widely divergent grasses
| Species | CNS4 | CNS5 | CNS1 | CNS6 | CNS7 |
|---|---|---|---|---|---|
| Maize | CAA-GACTTTGTACAGCC | ACGTACGTACGTACGTACGAGCAAAAA-TTGCAAATGCAAGCAAC | TCAGCTGCAAAACTCCAAGCTGTCT | ATTGGTCCCCGGAATGTTC | GTATTTCATGTGTAT |
| Rice | CAAAGACTTTGTACAGCC | ACGTACATATGTACGTACCAGCACAAG-TTGCATTTGCAAGCAAC | TCAGCTGCAAAACTCCAAGCTGTCT | ATTGGTCCCCAGAATGTTC | GTATTTCATGTGTAT |
| Setaria | CAA-GACTTTGTACAGCC | ---CGTGCACGTATGTACGAGCAAAAAATTCCAATTGCAAGCAAC | TTAGCTGCAAGACTCCAAGCTGTCT | GTTGGTCCC-GGAATGTTC | GTCTTTGCTGTTAAT |
| Arundo | CAA-GACTTTGTACAGCC | ---CATGTACGTACGTACCAGCAAAAG-TTGCAATTGCGAGCGAC | TCAGCTGCAAGAATCCAAGCTGTCT | ATTGGTCCCCGGAATGTTC | GTATTTCATGTGTAT |
| Bamboo | CAA-GACTTTGTACAGCC | --------ACGTACGTACCAGCACAAG-TTGCGATTGCAAGCAAC | TCAGCTGCAAAACTCCAAGCTGTCT | ATTGGTCCCCAGAATGTTC | GTATTTCATGTGTAT |
| Consensus | *** ************** | * *** **** **** ** ** * *** *** ** | * ******** * ************ | ******** ******** | ** *** *** ** |
lg1 intron1 sequences from Arundo donax, Setaria viridis, a bamboo, maize, and rice were aligned by using ClustalX 1.81. CNSs 4, 5, 1, 6, and 7 are shown. Asterisks represent sequence identity among all five species.
CNSs Exist in Other Grass Genes.
To establish that the results we obtained at lg1 were general properties of grass genes, we searched for stringent CNSs in six additional maize/rice gene pairs. The genes we have chosen have unambiguous, experimental exon annotation. Table 1 summarizes the number of stringent CNSs found, sorted by length; six of these genes contained one or more CNSs with a significance greater than or equal to a 15-bp exact match. None of these sequences contained stringent CNSs more than 60 bp in length. Researchers working in mammalian systems have identified larger CNSs (11, 15), suggesting that mammalian and higher plant gene CNSs may, on average, differ. To compare our results directly to mammalian analyses, we used our stringent algorithm to identify the CNSs in a previously annotated 52-kb region of human and mouse genomic DNA containing six genes (30). Our analysis is in complete concordance with that of other mammalian CNS researchers as we identified 12 large (>100 bp) CNSs and 94 smaller CNSs in these mammalian genes (Table 1).
Discussion
Using cross-species genomic DNA sequence comparisons, we have identified CNSs in several grass genes. Comparison of maize and rice genomic sequences at seven loci identified CNSs in six genes. We extended this finding by showing that the lg1 CNSs are highly conserved among all grasses analyzed. These potential regulatory elements will help to elucidate networks of gene regulation.
Our results would be trivial if sequence conservation over 50 million years of divergent evolution was actually nothing more than neutral carryover of the ancestral sequence. We assume that the process of DNA replication generates errors at some measurable rate, and that no region is a mutational “cold spot.” This rate, in the absence of selection, has been estimated by Gaut and coworkers (31) to be ≈7 × 10−9 substitution/bp per yr in the grasses. Our own measure of synonymous substitutions/bp per yr at lg1 is 5.8 × 10−9 using their methods. The chance of mutation at any one position between maize and rice and their shared ancestor is about 0.35 (7 × 10−9 mutations/neutral bp per yr × 50 × 106 yr). Thus, the chance of carryover/bp is 0.65 (1–0.35), and carryover for both maize and rice is 0.65maize × 0.65rice = 0.42. The chance that a 15-bp maize/rice CNS at a fixed position could have been passed down from an ancestor without selection is (0.42)15, which is infinitesimal. Because we measure our CNSs in multiple grasses representing deeply diverging branches of the grass phylogeny, we have extended the effective amount of time during which evolution might have randomized the sequences. Because the CNSs are shared among all these grasses, they are not a chance result of the maize vs. rice comparison. The conservation of lg1 CNSs (as shown in Table 2 and Fig. 2) argues for their function.
The question of what a functional CNS unit is becomes particularly interesting because mammals certainly have more long CNSs than plants (Table 1). This observation raises the question of whether a long CNS functions as a unit, is a collection of shorter functional units, or is both. One hypothetical function for long CNSs is as a template for the assembly of regulatory protein complexes that may not associate on their own. Our data suggest that regulatory complexity differences between mammals and grasses might be reflected in the complexity of the genes themselves. Human and mouse are estimated to have diverged from a common ancestor living in the late Cretaceous period (65–98.9 million years ago) (32), and a median of many rodent/human genes have a synonymous (neutral) substitution frequency of 4 × 10−9 mutations/yr (33). Thus, mammalian and grass CNSs may be compared with one another.
Even if it takes years to evaluate the functional importance of CNSs, their utility is immediate. CNSs could make valuable phylogenetic characters. Moreover, if one is studying genes that are known to be regulated differently in different grasses, the phylogenetic analysis of CNSs could illuminate mechanisms of regulatory evolution even without exact knowledge of which CNS binds to which protein(s).
Comparative gene mapping in grasses has been done by using conserved exons as hybridization probes (4). Current gene-specific PCR-based mapping tools, like SSR primers, are useful within one or a few closely related species. CNS-derived mapping tools should work for all species within a family and perhaps related families. With or without exon sequence, CNSs could be used to prepare PCR-based gene-specific amplification products that are designed to detect polymorphisms. We successfully used primers complementary to CNSs 1 and 4 along with exon-derived primers to amplify lg1 intron 1, demonstrating that CNSs 1 and 4 are useful pan-grass primer sites. The other CNS primer we tested, CNS3, also specifically amplified lg1 from all of the grasses tested (Fig. 2). The identification of CNSs in many orthologous gene pairs should quickly lead to the development of multiple PCR markers and high-density linkage maps.
No lg1 CNSs exist intact anywhere in the rice genome outside of lg1 itself; this conclusion results from conducting short sequence searches of the complete Torrey Mesa Research Institute Nipponbare Japonica rice genome. However, we were able to detect partial identity to lg1 stringent CNSs in rice and other organisms. For example, CNS1 (25/25 bp) exists as 17/17 bp in a rice gene (GenBank accession no. AF488772) similar to a hypothetical Arabidoposis gene. However, as we lack the sequence of a maize ortholog of this rice gene, we cannot assay whether this 17-bp sequence is itself a CNS, and therefore meaningful. As argued convincingly by Hardison and coworkers (34), a sequenced mouse genome is necessary to annotate the human genome. Without an annotation sequence, finding human exons has proven problematic (35). With the rice genome now completed, this same scenario is now true in the grasses (36). A second sequenced grass genome is needed for identification and annotation of conserved sequences, including coding and regulatory components of genes. As another example, of the seven lg1 CNSs, only one is obviously over-represented in GenBank. A component of CNS5, acgtacgtacgtacgtacga (Table 2) is found in 12 of the 17 top hits (19/19 bp) in different genes of various grasses. The other five top hits were animal, and surprisingly, none were from Arabidopsis. We suspected a grass family transposon sequence, but there were no transposon terminal repeats or insertion site duplications nearby. With a second fully sequenced grass genome, testing the hypothesis that this sequence is conserved and thus biologically important would be possible.
Beyond the obvious utility of CNS PCR primer sites, it is premature to speculate on what proportion of CNS distribution is mutable during evolution, or what percent is locked in as part of generic, cis-acting machinery. Measuring CNS patterns and complexity at phylogenetic branch points and in polyploid clades should be particularly useful to sort out these two broad categories of CNS function.
Supplementary Material
Acknowledgments
We thank Nancy Nelson, Damon Lisch, Randall Tyers, Paula McSteen, and Justine Walsh for critically reading this article and Zoya Akulova-Barlow for providing vouchered grasses. This work was supported by grants from the National Institutes of Health and the Plant and Microbial Biology-Syngenta Collaborative Research Program. D.M.B. was supported by National Institutes of Health Postdoctoral Fellowship F32 GM 19107. N.J.K. was supported by a Graduate Research Fellowship from the National Science Foundation.
Abbreviation
- CNS
conserved noncoding sequence
Footnotes
References
- 1.Hardison R C. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- 2.Kellogg E A. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gale M D, Devos K M. Proc Natl Acad Sci USA. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Devos K M, Gale M D. Plant Cell. 2000;12:637–646. doi: 10.1105/tpc.12.5.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freeling M. Plant Physiol. 2001;125:1191–1197. doi: 10.1104/pp.125.3.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paterson A H, Lin Y-R, Li Z, Schertz K F, Doebley J F, Pinson S R M, Liu S-C, Stansel J W, Irvine J E. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 7.Tjian R. Sci Am. 1995;272:54–61. doi: 10.1038/scientificamerican0295-54. [DOI] [PubMed] [Google Scholar]
- 8.Ficket J W, Hatzigeorgiou A G. Genome Res. 1997;7:861–878. doi: 10.1101/gr.7.9.861. [DOI] [PubMed] [Google Scholar]
- 9.Bucher P. Curr Opin Struct Biol. 1999;9:400–407. doi: 10.1016/S0959-440X(99)80054-2. [DOI] [PubMed] [Google Scholar]
- 10.Sumiyama K, Kim C-B, Ruddle F H. Genomics. 2001;71:260–262. doi: 10.1006/geno.2000.6422. [DOI] [PubMed] [Google Scholar]
- 11.Loots G G, Locksley R M, Blankespoor C M, Wang Z E, Miller W, Rubin E M, Frazer K A. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 12.Gottgens B, Gilbert J G R, Barton L M, Grafham D, Rogers J, Bentley D R, Green A R. Genome Res. 2001;11:87–97. doi: 10.1101/gr.153001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flint J, Tufarelli C, Peden J, Clark K, Daniels R J, Hardison R, Miller W, Philipsen S, Tan-Un K C, McMorrow T, et al. Hum Mol Genet. 2001;10:371–382. doi: 10.1093/hmg/10.4.371. [DOI] [PubMed] [Google Scholar]
- 14.Stojanovic N, Florea L, Riemer C, Gumucio D, Slightom J, Goodman M, Miller W, Hardison R. Nucleic Acids Res. 1999;27:3899–3910. doi: 10.1093/nar/27.19.3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy S, Hannenhalli S, Workman C. Bioinformatics (Oxford) 2001;17:871–877. doi: 10.1093/bioinformatics/17.10.871. [DOI] [PubMed] [Google Scholar]
- 16.Dubchak I, Brudno M, Loots G G, Pachter L, Mayor C, Rubin E M, Frazer K A. Genome Res. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayor C, Brudno M, Schwartz J R, Poliakov A, Rubin E M, Frazer K A, Pachter L S, Dubchak I. Bioinformatics (Oxford) 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz S, Zhang Z, Frazer K A, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. Genome Res. 2000;10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carter R J, Dubchak I, Holbrook S R. Nucleic Acids Res. 2001;29:3928–3938. doi: 10.1093/nar/29.19.3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vicente-Carbajosa J, Moose S P, Parsons R L, Schmidt R J. Proc Natl Acad Sci USA. 1997;94:7685–7690. doi: 10.1073/pnas.94.14.7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Forde B, Heyworth A, Pywell J, Kreis M. Nucleic Acids Res. 1985;13:7327–7339. doi: 10.1093/nar/13.20.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altschul S F, Madden T L, Schaeffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusova T A, Madden T L. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 24.Becraft P, Freeling M. Plant Cell. 1991;3:801–808. doi: 10.1105/tpc.3.8.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sylvester A W, Cande W Z, Freeling M. Development (Cambridge, UK) 1990;110:985–1000. doi: 10.1242/dev.110.3.985. [DOI] [PubMed] [Google Scholar]
- 26.Becraft P W, Bongard-Pierce D K, Sylvester A W, Poethig R S, Freeling M. Dev Biol. 1990;141:220–232. doi: 10.1016/0012-1606(90)90117-2. [DOI] [PubMed] [Google Scholar]
- 27.Moreno M A, Harper L C, Krueger R W, Dellaporta S L, Freeling M. Genes Dev. 1997;11:616–628. doi: 10.1101/gad.11.5.616. [DOI] [PubMed] [Google Scholar]
- 28.Clayton W D, Renvoize S A. Genera Graminum: Grasses of the World. London: Her Majesty's Stationery Office; 1986. [Google Scholar]
- 29.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ansari-Lari M A, Oeltjen J C, Schwartz S, Zhang Z, Muzny D M, Lu J, Gorrell J H, Chinault A C, Belmont J W, Miller W, Gibbs R A. Genome Res. 1998;8:29–40. [PubMed] [Google Scholar]
- 31.Gaut B S, Morton B R, McCaig B C, Clegg M T. Proc Natl Acad Sci USA. 1996;93:10274–10279. doi: 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freeman S, Herron J C. Evolutionary Analysis. Upper Saddle River, NJ: Prentice–Hall; 2001. [Google Scholar]
- 33.Li W-H, Graur D. Fundamentals of Molecular Evolution. Sunderland, MA: Sinauer; 1991. [Google Scholar]
- 34.Hardison R C, Oeltjen J, Miller W. Genome Res. 1997;7:959–966. doi: 10.1101/gr.7.10.959. [DOI] [PubMed] [Google Scholar]
- 35.Hogenesch J B, Ching K A, Batalov S, Su A I, Walker J R, Zhou Y, Kay S A, Schultz P G, Cooke M P. Cell. 2001;106:413–415. doi: 10.1016/s0092-8674(01)00467-6. [DOI] [PubMed] [Google Scholar]
- 36.Messing J, Llaca V. Proc Natl Acad Sci USA. 1998;95:2017–2020. doi: 10.1073/pnas.95.5.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.