Abstract
The 140-kb a1-sh2 interval of the maize genome contains at least four genes (a1, yz1, x1, and sh2). Partial sequence analysis of two haplotypes has revealed many single nucleotide polymorphisms and InDel polymorphisms, including several large structural polymorphisms. The physical positions of 101 meiotic recombination breakpoints are not distributed uniformly across the interval and are instead concentrated within three recombination hot spots. Two of these recombination hot spots are genic (a1 and yz1) and one is apparently nongenic. The x1 gene is not a recombination hot spot. Thus, these results suggest that not all hot spots are genes and indicate that not all genes are hot spots. Two of the 101 recombination events arose by means of either noncrossover events involving conversion tract lengths of at least 17 kb or double-crossover events. Only one recombination breakpoint mapped to the ≈80-kb distal portion of the a1-sh2 interval that contains large amounts of repetitive DNA including retrotransposons; in this region the ratio of genetic to physical distance is less than 0.5% of the genome's average. These results establish that the retrotransposon faction of the maize genome is relatively inert recombinationally.
Homologous meiotic recombination recombines physically linked genetic material by means of reciprocal crossovers (COs) and unilateral noncrossovers (NCOs). According to the canonical double-strand break (DSB) repair model (1, 2) a meiotic recombination event of either type is initiated by a DSB and depending on how the recombination intermediate, a double holiday junction, is resolved, a CO or NCO results. Recently, a modified DSB repair model has been proposed in which an early commitment is made to enter either the CO or NCO pathway (3).
Bacterial, fungal, plant, and mammalian genomes all exhibit recombination “hot spots” and “cold spots,” where recombination rates per kb are much higher or lower than the genome average (4–6). Even though the sizes of the genomes of diverse eukaryotic organisms are quite different, the lengths of their genetic maps are fairly constant. Based on this observation, and the assumption (now being confirmed by genome sequencing projects) that these genomes contain similar numbers of genes, it was hypothesized that recombination occurs primarily in genes (7). Several observations are consistent with this hypothesis: (i) all recombination hot spots identified to date in the approximate 2,500-Mb and 5,289-cM (centimorgan) (Georgia Davis, personal communication) maize genome are genes (6), even though the bulk of this genome consists of repetitive DNA such as retrotransposons (8); (ii) gene-rich chromosomal regions of wheat (9–11) and barley (12) are more recombinationally active than gene-poor regions; and (iii) in Arabidopsis (13) and tomato (14, 15) recombination is suppressed in chromosomal regions near the gene-poor centromeres (16, 17).
Two alternative hypotheses have been proposed to explain the correlation between recombination rates and gene density (6). One is that genes per se are recombinationally hyperactive such that chromosomal regions with high gene densities are recombinationally hyperactive. The alternative hypothesis is that genes per se do not exhibit recombinational hyperactivity, but genes tend to cluster in recombinationally hyperactive chromosomal regions. Analyses of recombination within single genes cannot distinguish between these hypotheses. Instead, it is necessary to characterize the distribution of recombination events across an interval that contains a mixture of genic and nongenic regions.
We tested these hypotheses by characterizing the distribution of recombination events across the 140-kb multigenic a1-sh2 interval of the maize genome (18). We address the questions of whether all genes in this interval are recombination hot spots and whether all hot spots in this interval are genes. The a1-sh2 interval (GenBank accession nos. AF072704, AF347696, AF434192, and AF434193) is an ideal model for such studies because recombination events across this interval can be readily selected by phenotypic screens, and this interval contains a mixture of genic and nongenic regions.
Materials and Methods
Maize Genetic Stocks.
The A1-LC allele was derived from the inbred Line C and conditions a colored kernel phenotype. The a1∷rdt and a1-mum2 alleles contain transposons that disrupt the function of the a1 gene (19–21) and condition a colorless phenotype in the stocks used in this study. Kernels that carry functional Sh2 alleles are round whereas those homozygous for sh2 alleles are shrunken (22).
Restriction Fragment Length Polymorphism (RFLP) Markers.
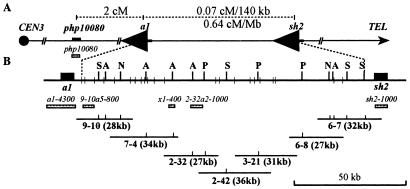
Seven cosmid subclones (in SuperCos1, Stratagene) of yeast artificial chromosome ASH-2 (18) constitute a contig spanning almost the entire 140-kb a1-sh2 interval (Fig. 1). The 9-10a5-800 and 2-32a2-1000 RFLP markers were isolated as plasmid subclones (in pBSK, Stratagene) from these cosmids. The single-copy 9-10a5 locus includes a 3′ portion of the yz1 gene and is detected with the 0.8-kb HindIII–SacIA insert released from pUCA5. The 1.0-kb fragment that detects the 2-32a2 locus was PCR-amplified from clone pUCA2 by using vector-derived primers. This probe detects several loci in the maize genome. One of them, a 5.5-kb BglII fragment, maps genetically to the 9-10a5/yz1-sh2 interval and is present in the a1-sh2 interval from the A1-LC Sh2 but not the a1∷rdt sh2 haplotype. The RFLP probes that detect the php10080, a1, and sh2 loci have been described (23–25). Computational analysis of the sequences of the rice and sorghum a1-sh2 intervals revealed a predicted gene (26, 27). Its maize homolog, x1, was identified and physically positioned in the a1-sh2 interval (see additional Methods, which are published as supporting information on the PNAS web site, www.pnas.org). The structure of the single-copy x1 gene was determined by sequencing clones that contain x1-cDNA and genomic DNA (unpublished work). A 400-bp x1 gene-specific probe was obtained by PCR amplification of a 1.4-kb cDNA clone (X-V3) by using primers that anneal to its last exon.
Figure 1.
Physical and genetic characterization of the a1-sh2 interval. (A) Genetic organization of the a1-sh2 interval on chromosome 3L. (B) A restriction map of the 140-kb a1-sh2 interval includes rare cutting restriction enzyme sites (labeled vertical bars; N = NotI; P = PacI; A = AscI; S = SfiI) and EcoRI sites (unlabeled short vertical bars). The a1 and sh2 genes are not drawn to scale. The sizes of the individual cosmids that comprise a contig of the a1-sh2 interval are shown (the proximal end of this contig is located within TD2, Fig. 2). Unlabeled short vertical bars on the cosmids represent the corresponding rare cutting restriction enzyme sites indicated in the restriction map. Shaded horizontal bars (not drawn to scale) represent RFLP markers.
Sequence of the a1-sh2 Interval.
Portions of the a1-sh2 interval from the two haplotypes (A1-LC Sh2 and a1∷rdt sh2) from which recombinants were isolated were sequenced. The a1 to yz1 interval from the a1∷rdt sh2 haplotype is 12,558 bp (GenBank accession no. AF072704). A 21,230-bp sequence that spans the a1 and yz1 interval was assembled from sequences of clones derived from the A1-LC Sh2 haplotype [GenBank accession nos. X05068 (24), AF363390, and AF363391] and two closely related haplotypes [a1-mum2 Sh2 (GenBank accession no. AF347696) and A1-LH82 Sh2 (GenBank accession no. AF434192)]. The structures of the A1-LC Sh2, a1-mum2 Sh2, and A1-LH82 Sh2 haplotypes are identical or nearly identical in the region between the a1 and yz1 loci (see additional Methods). Hence, the 21,230-bp sequence has been designated the A1-LC Sh2 haplotype.
The sequence of the x1 allele (GenBank accession no. AF434193) from the inbred LH82 was obtained by sequencing portions of cosmid Cos2-32. Partial sequences of the x1 alleles from the Line C and a1∷rdt sh2 stocks were obtained by sequencing PCR products amplified from the corresponding genomic DNAs (GenBank accession nos. AF434194 and AF434195).
Oligonucleotide Design.
PCR and sequencing primers were designed based on the sequences of the A1-LC Sh2 and a1∷rdt sh2 haplotypes. Allele-specific primers were designed according to InDel polymorphism (IDPs) between these haplotypes. Nonspecific universal primers also were designed that anneal to both haplotypes. In all instances primers designed based on the A1-LC Sh2 sequence behaved as expected when used to amplify genomic DNA from line C. Primer sequences are in the additional Methods.
Results
Sequence Analyses of the 140-kb a1-sh2 Interval.
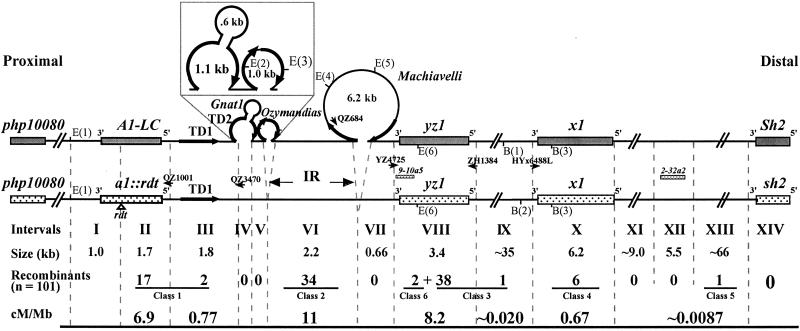
Analyses of large portions of the cosmid contig revealed the presence of two additional genes, yz1 and x1 (GenBank accession nos. AF434192 and AF434193, unpublished work). The A1-LC Sh2 and a1∷rdt sh2 haplotypes exhibit both small sequence heterologies and large structural polymorphisms (Fig. 2). The 1.1-kb TD1 and TD2 sequences comprise a tandem duplication that is present in the A1-LC Sh2 haplotype. TD2 contains a 0.6-kb novel MITE (miniature inverted repeat transposable element) (28) termed Gnat1. Gnat1-like sequences occur about 2,000 times in the maize genome (Y. Fu and P.S.S., unpublished observation). The a1∷rdt sh2 haplotype contains only TD1. Two retrotransposons, Ozymandias and Machiavelli, are present in the A1-LC Sh2 but not the a1∷rdt sh2 haplotype. Ozymandias is incomplete and contains only its two long terminal repeats and the primer-annealing site. Machiavelli is an apparently intact 6.2-kb Ty1/copia-like retrotransposon.
Figure 2.
The distribution of breakpoints associated with the 101 recombinants across the 140-kb a1-sh2 interval. The positions of six RFLP markers, php10080, a1, 9-10a5, x1, 2-32a2, and sh2 are indicated relative to the proximal and distal ends of chromosome 3. Intervals I–XIII are defined by the positions of RFLPs and IDPs. Interval IV consists of 1.7 kb in the A1-LC Sh2 haplotype and 0.4 kb in the a1∷rdt sh2 haplotype. Interval V is approximately 80 bp in both haplotypes. The numbers of recombinants that map to each individual interval and the resulting ratios of genetic to physical distance (cM/Mb) are shown. Figure is not drawn to scale. E = EcoRI. B = BglII.
Isolation of Recombination Events.
Meiotic recombination events that resolved within the 140-kb a1-sh2 interval were isolated from the test cross: A1-LC Sh2/a1∷rdt sh2 × a1∷rdt sh2/a1∷rdt sh2 based on their nonparental recombinant phenotypes. From a population of 249,000 progeny, 78 colored shrunken and 165 colorless round kernels were isolated, which presumably carried recombinant chromosomes, designated A1*sh2 and a1*Sh2. The genotypes of about half of these exceptional progeny were tested by backcrosses to the a1∷rdt sh2 stock. The fractions of the two classes of progeny that were verified to carry recombinant chromosomes were used to determine the numbers of actual recombinants among the progeny with nonparental phenotypes (Table 1). The genetic distance between a1 and sh2 is 0.070 ± 0.005 cM (Table 2, which is published as supporting information on the PNAS web site). Plants homozygous for the recombinant A1*sh2 and a1*Sh2 chromosomes were generated by self-pollinations. In total, 101 recombinants were recovered for analysis.
Table 1.
No. of recombinants isolated
| 1992
|
1993
|
|||
|---|---|---|---|---|
| Clsh | clrd | Clsh | clrd | |
| No. isolated | 30 | 108 | 48 | 57 |
| No. tested | 19 | 64 | 22 | 45 |
| No. confirmed* | 18 | 28 | 21 | 43 |
| Corrected no.† | 28 | 47 | 46 | 54 |
Clsh, colored shrunken kernels; clrd, colorless round kernels.
Nine of the confirmed recombinants were not analyzed because homozygotes were not available.
Corrected no. = no. isolated × (no. confirmed/no. tested). Corrected numbers were used to calculate the genetic distance between a1 and sh2.
Mapping Recombination Resolution Sites.
The resolution sites associated with each recombinant haplotype were mapped relative to molecular markers. Initially, the 101 recombinants were subjected to RFLP analysis with the markers php10080, a1-4300, 9-10a5/yz-800, x1-400, 2-32a2-1000, and sh2-1000. Such analyses revealed 10 different DNA hybridization patterns (Table 3, which is published as supporting information on the PNAS web site) that represent five classes of CO events (Fig. 2, classes 1–5) and one class of NCO or double crossover (DCO) events (class 6). The class 1 events resulted from reciprocal COs between the rdt transposon insertion site in a1 and E(2) in Ozymandias (Fig. 2); the class 2 events resulted from COs between E(2) and E(5) in Machivalli; COs between E(5) and B(2) near x1 resulted in the class 3 events; COs between B(2) and a polymorphic BglII site in the vicinity of 2-32a2 gave rise to the class 4 events; and COs that resolved between the polymorphic BglII site near 2-32a2 and a polymorphic EcoRI site in the vicinity of sh2 generated the class 5 event. The class 6 events arose by NCOs or DCOs in which part of the sequence in the a1∷rdt sh2 haplotype was replaced with that from the A1-LC Sh2 haplotype. The proximal breakpoints associated with these class 6 events mapped between php10080 and E(1) and the distal breakpoints in yz1.
Mapping Recombination Breakpoints to High Resolution.
The physical positions of breakpoints associated with recombinants in each of the hybridization classes were more precisely mapped by using IDPs and single nucleotide polymorphisms between the two parental haplotypes.
PCR amplifications using the a1∷rdt sh2-specific primers QZ1001 and QZ3470 (Fig. 2) coupled with primers that amplify both haplotypes mapped 17 of the 19 class 1 recombination breakpoints to the 1.7-kb interval II defined by the rdt transposon and the QZ1001 annealing site. The remaining two class 1 breakpoints mapped to the 1.8-kb interval III between the QZ1001 annealing site and TD2. Similarly, A1-LC Sh2-specific primers (QZ684, YZ4725, ZH1384, and HYx6488L) and an a1∷rdt sh2-specific primer (QZ3470) were used to map the recombination breakpoints in the other classes (Fig. 2). Thirty-four class 2 recombination breakpoints mapped to the 2.2-kb interval VI (designated the interloop region, IR) flanked by Ozymandias and Machiavelli retrotransposons. Thirty-eight of the 39 class 3 recombination breakpoints and the distal breakpoints of the two class 6 events resolved within the 3.4-kb interval VIII that includes yz1. The remaining class 3 breakpoint mapped to an approximately 35-kb interval between the ZH1384 and HYx6488L annealing sites (interval IX). All six class 4 breakpoints mapped in the 6.2-kb region containing the x1 gene (interval X). The single class 5 recombinant mapped to the ≈66-kb interval XIII between 2-32a2 and sh2.
Interval II.
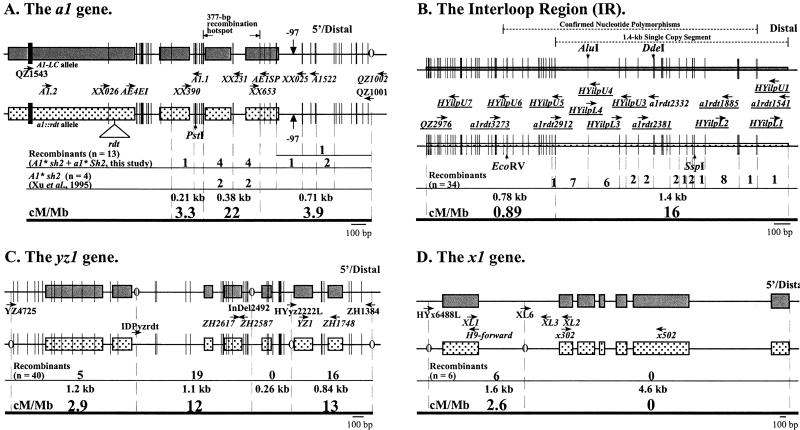
Seventeen recombination breakpoints mapped to the 1.7-kb interval II that is composed of the 5′ portion of the a1 gene. The ratio between interval II's genetic and physical distances is 6.9 cM/Mb, which is three times higher than the genome's average (2.1 cM/Mb). Therefore, as reported previously (18, 29), the a1 gene is a recombination hot spot. The 17 recombination breakpoints in interval II were further mapped relative to DNA sequence polymorphisms by cleaved amplified polymorphic sequences and sequence analyses. Interval II was PCR-amplified from plants homozygous for individual recombinant haplotypes (Fig. 3A). The resulting PCR products were subjected to PstI digestion as described by Xu et al. (23) and sequenced. These analyses established that 16 recombinant haplotypes had breakpoints distal to the diagnostic PstI site; only one breakpoint mapped proximal to this PstI site (Fig. 3A). Analysis of the PCR product sequences established the physical position of each recombination breakpoint relative to DNA polymorphisms that exist between the two parental haplotypes. As shown in Fig. 3A, 12 of the 17 recombination breakpoints mapped to the previously identified 377-bp hot spot within the a1 gene (23). Only four of the 17 breakpoints mapped between the hot spot and the QZ1001 annealing site. The ratio of genetic to physical distances in this 377-bp hot spot is 22 cM/Mb. Hence, this hot spot experiences approximately 10-fold more recombination per unit physical length than the genome's average.
Figure 3.
High-resolution mapping of recombination breakpoints within (A) the a1 gene (interval II), (B) the IR (interval VI), (C) the yz1 gene (interval VIII), and (D) the x1 gene (interval X). The A1-LC Sh2 haplotype (gray boxes) is positioned above the a1∷rdt sh2 haplotype (dotted boxes). Vertical bars and parentheses represent DNA sequence polymorphisms between the two haplotypes; IDPs indicated by parentheses were used to design allele-specific primers. The widths of vertical bars are proportional to the numbers of bases in a polymorphism. Polymorphisms in the region flanked by primers a1rdt3273 and a1rdt1541 (B) were confirmed to exist in the A1-LC Sh2 haplotype by sequencing the 34 recombinant alleles that mapped to the IR. The region from the A1-LC Sh2 and a1∷rdt sh2 haplotypes that are flanked by primers XL1 and x502 (D) were sequenced. Primers used in PCR and sequencing are indicated by horizontal arrows. Universal primers are indicated in italics. Primers used for RT-PCR are underlined. Allele-specific primers are positioned close to the haplotypes they amplify. The numbers of recombination breakpoints that resolved within each interval and the resulting ratios of genetic to physical distance (cM/Mb) are shown. The triangle represents the rdt transposon insertion in the a1∷rdt allele. Restriction enzyme sites used in cleaved amplified polymorphic sequence analyses are indicated.
Interval VI.
Thirty-four recombinant haplotypes contain breakpoints in the 2.2-kb interval VI (the IR). The breakpoints associated with these recombinant haplotypes were mapped relatively to four polymorphic restriction enzyme recognition sites (EcoRV, AluI, DdeI, and SspI) (Fig. 3B) between the parental haplotypes by cleaved amplified polymorphic sequence analyses. In addition, portions of the PCR-amplified IR from these 34 recombinants were sequenced (Fig. 3B). All but one of the 34 recombination breakpoints mapped to the 1.4-kb distal portion of the IR that is flanked by the a1rdt2912 and a1rdt1541 annealing sites (Fig. 3B). This 1.4-kb segment is single-copy in the maize genome (data not shown) and exhibits fewer sequence polymorphisms between the two parental haplotypes than the remainder of the 0.8-kb segment of the IR that is repetitive in the maize genome (Fig. 3B and data not shown).
Several computational approaches were used to test whether the IR contains a gene. blast analyses (blastn, blastx, and tblastx) of the IR sequence against GenBank revealed that IR-related sequences are absent from the rice and sorghum a1-sh2 intervals; the 1.4-kb single-copy distal portion of the IR does not exhibit significant sequence similarity to any GenBank accessions. None of four gene prediction algorithms (fgenesh, genemark.hmm, genscan, and glimmerr) predict any genes in the single-copy portion of the IR. Reverse transcription–PCR experiments were performed to test whether the IR is transcribed. Primers were designed in regions of the IR that contain potential ORFs and are flanked by predicted splice sites (Fig. 3B). No expression was detected in seedling, shoot, adult leaf, tassel, husk, or ear tissue. Thus, there is no evidence that the IR is genic. The ratio of genetic to physical distances in the entire IR is 11 cM/Mb. This is approximately five times higher than the genome's average. The 1.4-kb single-copy portion of the IR has a value of 17 cM/Mb, approximately eight times higher than the genome's average. This result establishes that this portion of the apparently nongenic IR is a recombination hot spot.
Interval VIII.
The breakpoints associated with the 40 recombination events in interval VIII (the yz1 gene) flanked by primer sites YZ4725 and ZH1384 were mapped to higher resolution by using three additional pairs of primers (ZH1748/ZH2617, HYyz2222L/ZH1748, and IDPyzrdt/ZH2587) (Fig. 3C). HYyz2222L is a A1-LC Sh2 haplotype-specific primer whereas IDPyzrdt is specific to the a1∷rdt sh2 haplotype. PCR amplification with ZH1748 and ZH2617 detects an IDP (InDel 2492) that is located in intron 3. These PCR analyses revealed that 16 breakpoints map to the 0.84-kb region defined by the annealing sites of ZH1384 and HYyz2222L and containing the first two exons. Another 19 recombinant breakpoints resolved in the 1.1-kb region flanked by Indel 2492 and the IDPyzrdt annealing site and containing exon 4 and 5. The remaining five recombination breakpoints map to the 1.2-kb region that includes the last two exons. The ratio of genetic to physical distances in yz1 is 8.2 cM/Mb, a value approximately four times higher than the genome's average. Hence, like all other maize genes studied to date, the yz1 gene is a recombination hot spot.
Interval X.
Six breakpoints resolved within the 6.2-kb interval X that contains the x1 gene. The polymorphic primer HYx6488L coupled with the universal primer H9-forward (Fig. 3D) amplifies genomic DNA from line C but not from the a1∷rdt sh2 stock. Because the resulting PCR product cosegregates with the a1-sh2 interval, it was used as a marker to map the recombination resolution sites within interval X. PCR amplification using another A1-LC Sh2 haplotype-specific primer, XL6, coupled with the universal primer XL3, revealed that all six recombination breakpoints mapped to the 3′ end of x1. The ratio of genetic to physical distances in the x1 gene is 0.67 cM/Mb, a value that is much lower than all other maize genes characterized to date and the genome average (2.1 cM/Mb). Hence, unlike all other maize genes studied to date, the x1 gene is not a recombination hot spot.
Intervals XI–XIV.
Only one breakpoint occurred in the 80-kb x1-sh2 interval. Hence, this region is nearly recombinationally inert. Indeed, the ratio of genetic to physical distances within this region (≈0.0087 cM/Mb) is less than 0.5% of the genome's average.
Discussion
The Retrotransposon Fraction of the Maize Genome Is Recombinationally Inert.
The positions of recombination resolution sites are not randomly distributed across the 140-kb a1-sh2 interval. Only one of the 101 recombination events resolved within the ≈80-kb x1-sh2 interval. The ratio of genetic to physical distances in this half of the a1-sh2 interval (0.0087 cM/Mb) is less than 0.5% that the genome's average. Based on hybridization data, this subinterval contains large amounts of repetitive DNA (data not shown), some of which is derived from retrotransposons (GenBank accession nos. AF464766 to AF464773). Hence, this result is consistent with the view that the retrotransposon fraction of the maize genome is not recombinationally active.
In contrast, all but eight of 101 recombination breakpoints resolved within the 21-kb a1-yz1 interval. This interval of the A1-LC Sh2 and a1∷rdt sh2 haplotypes exhibits three large structural polymorphisms that arose by tandem duplication events and/or transposon/retrotransposon insertions. None of the 93 recombination breakpoints that mapped in the a1-yz1 interval resolved within these three structural polymorphisms. This finding indicates that at least when hemizygous, retrotransposons can be recombinationally inert. Given that a large fraction of the maize genome is composed of retrotransposons and the highly polymorphic nature of this genome, this finding at least partially explains why recombination events generally cluster within genes.
Identification of a Recombination Hot Spot That Is Probably Not a Gene and a Gene That Is Not a Hot Spot.
Within the 21-kb a1-yz1 interval recombination resolution sites clustered into three recombination hot spots. No recombination hot spots are located in the remaining 120 kb of the a1-sh2 interval (i.e., yz1-sh2). Thus, these results establish that within the 140-kb multigenic a1-sh2 interval recombination hot spots cluster in a region (a1-yz1) that is larger than a single gene. Two of the recombination hot spots are genic (a1 and yz1) and one is apparently nongenic (the IR). Although Timmermans et al. (30) isolated a single recombination event in an apparently nongenic region, it had not previously been established that nongenic regions of a plant genome can serve as recombination hot spots. These data suggest that the hypothesis that all plant recombination hot spots are genic is not correct.
The 6.2-kb x1 gene exhibits a ratio of genetic to physical distance (0.67 cM/Mb) that is lower than any other characterized maize gene. Even within the most recombinationally active 3′ portion of x1, this ratio is only 2.6 cM/Mb, which is approximately equal to the genome's average (2.1 cM/Mb). Hence, the x1 gene cannot be considered a recombination hot spot. Although inversions can inhibit recombination, our mapping and sequence data are not consistent with the presence of an inversion in x1. Hence, the hypothesis that all genes are recombination hot spots can be rejected.
Some of the unique genic (a1 and yz1) and apparently nongenic (IR) sequences in the a1-sh2 interval are recombination hot spots. In contrast, the unique genic sequence x1 is not a recombination hot spot. This finding suggests that uniqueness within the genome is not a sufficient condition for high recombinational activity. Instead, it is likely that the recombinational activity of a sequence depends in part on its chromatin structure that in turn can be influenced by its regional environment. For example, hot spots for double-strand break initiation in Saccharomyces cerevisiae generally have open chromatin structure (31, 32). The finding that the maize recombination hot spot bz1 resides in a 32-kb gene-rich region without retrotransposons (33) is consistent with this view. The unique sequences in the a1-yz1 interval that host recombination hot spots may contribute synergistically to promote a chromatin structure that is accessible to the recombination machinery. In contrast, the single-copy x1 locus appears to be isolated from other unique regions, which may reduce its ability to form an open chromatin structure, thereby reducing its accessibility to the recombination machinery. However, the recombinational activity in x1 is still more than 30 times higher than its flanking regions (0.67 cM/Mb vs. 0.02 and 0.0087 cM/Mb). Similarly, the a1, yz1, and IR hotspots also exhibit substantially more recombinational activity than their flanking regions. Hence, these data suggest that regional chromatin structure is not sufficient to create recombination hot or cold spots.
Distribution of Recombination Breakpoints Within a1, yz1, and the IR.
The distribution of recombination breakpoints within recombinationally active maize genes varies. Breakpoints are distributed fairly uniformly in the bz1 and wx1 loci (34, 35). In contrast, breakpoints cluster at the 5′ ends of the a1 (ref. 23 and this study) and b1 loci (36), and the 3′ end of the r1 locus (37). The two hot spots defined in this study (IR and yz1 locus) also exhibit different patterns of breakpoint distribution. Within the IR, almost all breakpoints clustered at the distal portion, whereas breakpoints were distributed relatively uniformly across the yz1 locus, although its 3′ end is somewhat less recombinationally active than the remainder of the gene.
The variation in the distribution of recombination breakpoints within these hot spots may be caused by cis-acting modifiers that regulate the resolution of recombination intermediates. For example, sequence heterologies have a major effect on recombination in Saccharomyces (38) and other fungi (39). Fewer recombination events resolve in those regions of the bz1 locus that exhibit high densities of sequence heterologies (34). Consistent with these observations, recombination events preferentially resolved in regions with the highest levels of sequence identity within the a1 locus and the IR (Fig. 3 A and B). However, within the 1.4-kb distal portion of the IR, the density of recombination resolution sites is not correlated with the density of DNA sequence heterologies. In addition, even though the x1 alleles from the two parental haplotypes do not contain any sequence polymorphisms in the region between exons 2 and 6, no recombination events were observed in that portion of the x1 gene (Fig. 3D). Hence, although a high level of sequence identity may contribute to the recombinational activity of a sequence, it is not sufficient to define a recombination hot spot.
Effects of Transposon Insertions on Recombination.
Rates of intragenic recombination are suppressed in the vicinity of Ds and Mu1 insertions (23, 34, 40). In addition, Ds insertions are thought to alter the distribution of recombination breakpoints in the otherwise uniformly recombinogenic bz1 locus to create allele-specific hot and cold spots (34). In contrast, a preliminary analysis did not provide any evidence that a Mu1 insertion in the a1 gene alters the distribution of recombination event (23).
In this previous study, the positions of 15 recombination events isolated from the a1-mum2/a1∷rdt heterozygote were physically mapped within the 1.2-kb interval of the a1 gene that is defined by the Mu1 and rdt transposon insertions. All but one of these recombination events resolved within a 377-bp recombination hot spot. Xu et al. (23) compared this distribution of recombination events to those isolated from a directly comparable heterozygote that does not contain the Mu1 insertion in the a1 gene (A1-LC/a1∷rdt). This comparison is appropriate because, other than the Mu1 insertion, the A1-LC and a1-mum2 alleles have identical sequences (GenBank accession nos. X05068, AF363390, AF363391, and AF347696). All four of the recombination events isolated from an A1-LC/a1∷rdt heterozygote resolved within the 377-bp hot spot. In the current study the positions of an additional 10 intragenic recombination events isolated from the A1-LC/a1∷rdt heterozygote and that physically mapped within the 1.2-kb region studied by Xu et al. (23) were determined. All but two of these recombination events resolved within the 377-bp hot spot that experienced 10-fold more recombination than the genome's average.
Dooner and Martinez-Ferez (34) have suggested that large hemizygous insertions can suppress recombination in nearby regions and thereby create recombination hot spots. Indeed, they have observed that within the bz1 locus the lowest ratio of genetic to physical distances occurred in the “interval defined by the insertion and the closest point mutation.” Consistent with this observation, none of the 15 recombination breakpoints isolated from a1-mum2/a1∷rdt by Xu et al. (23) resolved within the interval defined by the Mu1 insertion at −97 in a1-mum2 and the closest polymorphism. However, at most only one of 17 recombination breakpoints isolated from a A1-LC/a1∷rdt heterozygote in the current study resolved within this interval. Because the A1-LC allele does not contain a Mu1 insertion, the Mu1 insertion in a1-mum2 cannot be responsible for the lack of recombination resolution events in the interval defined by positions −97 and +16. In summary, the relative distributions of recombination resolution sites in the 377-bp hot spot defined by Xu et al. (23) and the interval defined by positions −97 and +16 are not affected by the presence or absence of the Mu1 insertion. Hence, these data provide strong support for the view that unlike the Ds insertions in bz1 the Mu1 insertion in a1-mum2 does not affect the distribution of recombination resolution sites and is therefore not responsible for the recombination hot spot reported by Xu et al. (23).
Implication from Isolation of Two NCO or DCO Events.
NCOs unilaterally transfer genetic data from one chromatid to another. Hence, they differ from COs in that the latter event, but not the former, results in the exchange of flanking markers. In plants it is not usually possible to distinguish between NCOs and DCOs. The rate of closely linked DCOs is, however, expected to be low because in the absence of interference the probability of a DCO is the product of the probabilities of two single CO events. The two class 6 recombination events observed in this study (Fig. 2) could have arise by either DCOs or NCOs. Because the genetic distance between php10080 and sh2 is 2 cM it would be expected to observe two COs between a1 and php10080 among 101 individuals. However, the 101 individuals analyzed in this study each carried a recombination breakpoint between a1 and sh2. Hence, only if the rate of interference in this chromosomal region is very low would the two class 6 recombination events be likely to have arisen by DCOs. On the other hand, if the class 6 events represent NCOs, then they involve very long conversion tracts.
Conversion tracts of NCOs in Drosophila are relatively short and continuous (41), with a mean length of 350 bp. In contrast, long and interrupted conversion tracts (up to 5.9 kb) have been observed in Neurospora (42). Two apparent conversion tracts in the maize a1 gene are in excess of 590 and 787 bp (23). Another two apparent conversion tracts at the maize bz1 locus are between 965 and 1,165 bp and between 1.1 and 1.5 kb (34). If the class 6 events in the current study are NCOs, they are associated with much longer conversion tracts. One end point of each putative conversion tract is between php10080 and a1 and the other is within yz1. Hence, if the class 6 events are NCOs, they involve conversion tracts of at least 17 kb.
Supplementary Material
Acknowledgments
We thank Drs. Curt Hannah for the sh2 clones, Steve Dellaporta for the W22 cDNA library, and Monica Frey and Alfons Gierl for the C131A cDNA library. Lei Zhang and Dr. Yiji Xia constructed the cosmid contig, Dr. Lisa M. Weaver sequenced clones from interval XIII, and Yuan Zhang constructed and sequenced pCS-YZ. This research was supported by competitive grants from the United States Department of Agriculture–National Research Initiative Program (Awards 9701407 and 9901579 to P.S.S. and B.J.N., and Award 0101869 to P.S.S.). This is Journal Paper No J-19604 of the Iowa Agriculture and Home Economics Experiment Station, Project Nos. 3125, 3334, 3485, and 6502, supported by Hatch Act and State of Iowa funds.
Abbreviations
- CO
crossover
- NCO
noncrossover
- DCO
double crossover
- cM
centimorgan
- IDP
InDel polymorphism
- IR
interloop region
- RFLP
restriction fragment length polymorphism
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF072704, AF347696, AF363390, AF363391, U46063, AF434192 to AF434195, and AF464766 to AF464773).
See commentary on page 5763.
References
- 1.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 2.Sun H, Treco D, Szostak J W. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 3.Allers T, Lichten M. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 4.Lichten M, Goldman A S H. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- 5.Puchta H, Hohn B. Trends Genet. 1996;1:340–348. [Google Scholar]
- 6.Schnable P S, Hsia A-P, Nikolau B J. Curr Opin Plant Biol. 1998;1:123–129. doi: 10.1016/s1369-5266(98)80013-7. [DOI] [PubMed] [Google Scholar]
- 7.Thuriaux P. Nature (London) 1977;268:460–462. doi: 10.1038/268460a0. [DOI] [PubMed] [Google Scholar]
- 8.SanMiguel P, Tikhonov A, Jin Y-K, Melake-Berhan A, Springer P S, Edwards K J, Avramova Z, Bennetzen J L. Science. 1996;274:765–768. doi: 10.1126/science.274.5288.765. [DOI] [PubMed] [Google Scholar]
- 9.Gill K S, Gill B S, Endo T R, Boyko E V. Genetics. 1996;143:1001–1012. doi: 10.1093/genetics/143.2.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gill K S, Gill B S, Endo T R, Taylor T. Genetics. 1996;144:1883–1891. doi: 10.1093/genetics/144.4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faris J D, Haen K M, Gill B S. Genetics. 2000;154:823–835. doi: 10.1093/genetics/154.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Künzel G, Korzun L, Meister A. Genetics. 2000;154:397–412. doi: 10.1093/genetics/154.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Copenhaver G P, Browne W E, Preuss D. Proc Natl Acad Sci USA. 1998;95:247–252. doi: 10.1073/pnas.95.1.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanksley S D, Ganal M W, Prince J P, de Vicente M C, Bonierbale M W, Broun P, Fulton T M, Giovannoni J J, Grandillo S, Martin G B, et al. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frary A, Presting G G, Tanksley S D. Mol Gen Genet. 1996;250:295–304. doi: 10.1007/BF02174387. [DOI] [PubMed] [Google Scholar]
- 16.Moore G. Annu Rev Plant Physiol Plant Mol Biol. 2000;51:195–222. doi: 10.1146/annurev.arplant.51.1.195. [DOI] [PubMed] [Google Scholar]
- 17.The Arabidopsis Genome Initiative. Nature (London) 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 18.Civardi L, Xia Y J, Edwards K J, Schnable P S, Nikolau B J. Proc Natl Acad Sci USA. 1994;91:8268–8272. doi: 10.1073/pnas.91.17.8268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Reilly C, Shepherd N C, Pereira A, Schwarz-Sommer Z, Bertam I, Robertson D S, Peterson P A, Saedler H. EMBO J. 1985;4:877–882. doi: 10.1002/j.1460-2075.1985.tb03713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shepherd N S, Sheridan W F, Matters M G, Deno G. In: Plant Transposable Elements. Nelson O, editor. New York: Plenum; 1988. pp. 137–148. [Google Scholar]
- 21.Brown J J, Mattes M G, O'Reilley C, Shepherd N S. Mol Gen Genet. 1989;215:239–244. doi: 10.1007/BF00339723. [DOI] [PubMed] [Google Scholar]
- 22.Tsai C Y, Nelson O E. Science. 1966;151:341–343. doi: 10.1126/science.151.3708.341. [DOI] [PubMed] [Google Scholar]
- 23.Xu X J, Hsia A-P, Zhang L, Nikolau B J, Schnable P S. Plant Cell. 1995;7:2151–2161. doi: 10.1105/tpc.7.12.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarz-Sommer Z, Shepherd N, Tacke E, Gierl A, Rhode W, Leclercq L, Mattes M, Berndtgen R, Peterson P, Saedler H. EMBO J. 1987;6:287–294. doi: 10.1002/j.1460-2075.1987.tb04752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhave M R, Lawrence S, Barton C, Hannah L C. Plant Cell. 1990;2:581–588. doi: 10.1105/tpc.2.6.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Bennetzen J L. Plant Mol Biol. 1996;32:999–1001. doi: 10.1007/BF00041383. [DOI] [PubMed] [Google Scholar]
- 27.Chen M, SanMiguel P, Bennetzen J L. Genetics. 1998;148:435–443. doi: 10.1093/genetics/148.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wessler S R, Bureau T E, White S E. Curr Opin Genet Dev. 1995;5:814–821. doi: 10.1016/0959-437x(95)80016-x. [DOI] [PubMed] [Google Scholar]
- 29.Brown J, Sundaresan V. Theor Appl Genet. 1991;81:185–188. doi: 10.1007/BF00215721. [DOI] [PubMed] [Google Scholar]
- 30.Timmermans M C P, Das O P, Messing J. Genetics. 1996;143:1771–1783. doi: 10.1093/genetics/143.4.1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohta K, Shibata T, Nicolas A. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu T C, Lichten M. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 33.Fu H, Park W, Yan X, Zheng Z, Shen B, Dooner H K. Proc Natl Acad Sci USA. 2001;98:8903–8908. doi: 10.1073/pnas.141221898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dooner H K, Martinez-Ferez I M. Plant Cell. 1997;9:1633–1646. doi: 10.1105/tpc.9.9.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okagaki R J, Weil C F. Genetics. 1997;147:815–821. doi: 10.1093/genetics/147.2.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson G I, Kubo K M, Shroyer T, Chandler V L. Genetics. 1995;140:1389–1406. doi: 10.1093/genetics/140.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eggleston W B, Alleman M, Kermicle J L. Genetics. 1995;141:347–360. doi: 10.1093/genetics/141.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borts R H, Haber J E. Genetics. 1989;123:69–80. doi: 10.1093/genetics/123.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colot V, Maloisel L, Rossignol J-L. Cell. 1996;86:855–864. doi: 10.1016/s0092-8674(00)80161-0. [DOI] [PubMed] [Google Scholar]
- 40.Dooner H K. Genetics. 1986;113:1021–1036. doi: 10.1093/genetics/113.4.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hilliker A J, Harauz G, Reaume A G, Gray M, Clark S H, Chovnick A. Genetics. 1994;137:1019–1026. doi: 10.1093/genetics/137.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeadon P J, Catcheside D E A. Genetics. 1998;148:113–122. doi: 10.1093/genetics/148.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.