Abstract
Ovine scrapie is a fatal neurodegenerative disorder that may be transmitted through exposure to infected uterine and placental tissues. Susceptibility to scrapie is primarily controlled by polymorphisms in the prion protein (PrP) gene. Scrapie in the U.S. Suffolk breed and in many breeds in Europe occurs in sheep homozygous for glutamine (171QQ), but rarely in sheep heterozygous for glutamine and arginine (171QR) or homozygous for arginine (171RR) at codon 171 of the PrP gene. This study demonstrated that accumulation of PrPSc in uterine-placental epithelial cells in the placentome was determined by fetal PrP genotype and the pregnancy status of scrapie-infected ewes. PrPSc was detected in 171QQ placentomes of infected ewes, but not in placentomes of infected ewes pregnant with 171QR conceptuses or in the non-pregnant uterus of infected ewes. The distribution of PrPSc plaques in placentomes was temporally associated with stage of gestation. There was a tendency toward increased size and number of placentomal PrPSc plaques from the endometrial stalk (maternal side) to chorionic plate (fetal side). These results indicate that accumulation of PrPSc is eliminated or reduced to undetectable levels in reproductive and placental tissues if infected ewes are not pregnant or conceive conceptuses with a resistant PrP genotype.
Ovine scrapie is a fatal neurodegenerative disorder presumably caused by the aggregated proteinase K (PK) resistant form of the sheep prion (PrPSc), which is derived from an endogenous, PK-sensitive cellular precursor (PrPC) through conformational alteration (1–6). This host protein is encoded by a single copy gene (2) and expressed by many cell and tissue types (7–10). In several species, individuals with certain coding mutations in the prion gene are predisposed to prion disease (3, 11, 12). Susceptibility to scrapie infection in Suffolk sheep and Suffolk crosses is determined primarily by codon 171 of the prion gene (11, 13). Sheep homozygous for glutamine at codon 171 (171QQ) are susceptible to scrapie, whereas sheep heterozygous for glutamine and arginine (171QR) or homozygous for arginine (171RR) are usually resistant to scrapie (11, 13).
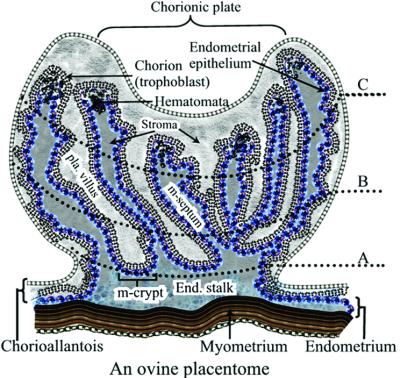
In sheep, PrPSc or scrapie infectivity is present primarily in tissues of the central and peripheral nervous system, lymphoreticular system, and placenta (9, 14–21). Recent studies of reproductive and placental tissues from scrapie-infected pregnant sheep demonstrated that PrPSc accumulates at the fetal–maternal interface (the placentome) and proposed that scrapie is not transmitted in utero, but may be transmitted from infected dams to their scrapie susceptible lambs during the perinatal period by exposure to infectious placental tissues (9). The ovine placentome is a natural chimera consisting of interdigitating uterine endometrial septae and placental chorionic villi (Fig. 1), in which the uterine cells carry the maternal genotype and the placental chorionic cells carry the fetal genotype. The unique structures of the placentome provide an applicable model for the study of the effect of prion genotype on scrapie transmission between tissues with intimate physical contact within a natural host for scrapie. The advantages of this model are that the placentomal PrPSc in the infected ewes is abundant for detection and PrP genotypes of the placentomal components can be controlled by breeding.
Figure 1.
A schematic illustration of a medial cross section of an ovine placentome. The placentome is a natural chimera, consisting of interdigitating uterine (placentomal or caruncular endometrium) and placental (placentomal or cotyledonary chorioallantois) tissues. The medial cross section of a placentome is divided into three zones: zones A, B, and C. Zone A contains maternal endometrial crypts and the distal ends of the cotyledonary villous tree and is located closest to the endometrial stalk and myometrium, zone B is the intermediate area, and zone C is the distal (relative to myometrium) portion of the placentome containing the distal ends of endometrial septae and the chorionic plate. Zone C is typically characterized by structures known as arcade system or hematomata. The placentomal endometrium consists of the uterine superficial epithelium and associated stroma, but no glands. Interplacentomal endometrium consists of loosely attached intercotyledonary chorioallantois and intercaruncular endometrium, which is rich in glands. Pla. villus, placental villus; m-septum, maternal endometrial septum; m-crypt, maternal crypt; end. stalk, endometrial stalk.
This study determined the effects of pregnancy stage, fetal PrP genotype, and pregnancy status on PrPSc accumulation at the fetal–maternal interface of scrapie-infected ewes. This study showed that accumulation of PrPSc in both uterine epithelial and placental trophoblast cells is determined by PrP genotype of the fetus and the pregnancy status of the infected ewe, and that the pattern of PrPSc plaque distribution in placentomes is associated with gestational stage. These results suggest that scrapie transmission through uterine-placental tissues may be reduced or eliminated if the infected ewes are not pregnant or conceive conceptuses with a resistant genotype.
Materials and Methods
Animals, Tissue Collection, and Processing.
All ewes (n = 61) in this study were either uninfected or naturally infected 171QQ Suffolk or Suffolk × Hampshire sheep. The scrapie status of the fetal, young, and adult ewes was determined before euthanasia, at necropsy, or at lambing by immunohistochemistry (22). Of 32 peripheral lymphoid tissue [live animal test of the third-eyelid lymphoid tissue (22)] PrPSc-positive ewes used in this study, 19 ewes exhibited clinical symptoms including pruritis, wool loss, weight loss, and/or ataxia. Eleven scrapie-infected ewes were mated to a 171QQ ram, 6 scrapie-infected ewes were mated to a 171RR ram, 25 uninfected ewes were mated to a 171QQ ram, and 15 scrapie-infected and 4 uninfected ewes were not mated. Infected ewes pregnant with 171QQ conceptuses were euthanized either at early (days 40–60; n = 8), mid- (days 80–100; n = 1), or term (Days 140–150; n = 2) pregnancy. All infected ewes pregnant with 171QR conceptuses were euthanized at term pregnancy, except for one that was euthanized at mid-pregnancy. All uninfected pregnant ewes were euthanized between days 40 and 140 of pregnancy. The uninfected ewes had no reported exposure to scrapie, and PrPSc was not detected in brain or tonsil by Western blotting or immunohistochemistry. The genotypes of fetuses/lambs and placentae were determined by sequencing of the prion gene.
Brainstem, tonsil, placentomes, interplacentomal endometrial-chorioallantois, ovary, oviduct, mammary gland, and bladder collected at euthanasia were fixed in 10% buffered formalin and processed for immunohistochemistry as described previously (22). Maternal brainstem, tonsil, dissected uterine caruncular (caruncles) and interplacentomal (intercaruncular) endometria, placental cotyledonary (cotyledons) and interplacentomal (intercotyledonary) chorioallantois, ovary, oviduct, mammary gland, and bladder of pregnant ewes, and fetal brain, kidney and bladder at different stages of pregnancy were collected and snap-frozen in liquid nitrogen and stored at −80°C until use. For each fetal unit, 2–3 placentomes and dissected caruncles and cotyledons from 3–6 placentomes were collected from each of the three areas occupied by the head, mid-body and tail of a fetus. A schematic illustration of the ovine placentome is shown in Fig. 1. Animal Care and Use Protocols were approved by the Institutional Animal Care and Use Committee of Washington State University.
Tissue Homogenate Preparation and Western Blot Analysis.
Tissue homogenates were prepared with (for PrPSc detection) or without (for PrPC and/or PrPSc detection) the use of 10% sarkosyl and ultracentrifugation as described (9). Briefly, tissue was homogenized and incubated for 30 min at 37°C in 10 mM Tris⋅HCl (pH 7.5), 5 mM MgCl, and DNase (200 μg/g wet tissue), followed by 30 min incubation at room temperature in sarkosyl solution (10 mM Tris⋅HCl, pH 7.5/20% sarkosyl). The homogenate was centrifuged at 6,000 × g for 10 min, and the resultant supernatant was centrifuged at 348,000 × g for 50 min at room temperature. The pellet was dissolved in 10 mM Tris⋅HCl (pH7.5) and treated with or without PK (20 μg/ml) at 37°C for 30 min. The PK-treated mixture was centrifuged at 279,000 × g for 30 min. The pellet was resuspended in 10 mM Tris⋅HCl and boiled in sample buffer before Western blotting.
Western blot procedure was performed (9, 23). Briefly, tissue homogenates were analyzed by 12% SDS/PAGE (Invitrogen), followed by transfer to poly(vinylidene difluoride) (PVDF) membranes (Amersham Pharmacia) before detection by anti-prion mAb F99/97.6.1 (22) and goat anti-mouse IgG-horseradish peroxidase (HRPO) (Southern Biotechnology Associates), and developed with a chemiluminescence substrate (Amersham Pharmacia).
Immunohistochemistry.
Immunohistochemistry was performed as previously described (22). Briefly, tissues were fixed in 10% neutral buffered formalin for 2–7 days, treated with 95% formic acid for 1 h, rinsed with water, and fixed for an additional 24 h before embedding in paraffin. Tissue sections placed on glass slides were deparaffinized, hydrated, and autoclaved in Target Retrieval Solution (pH 6; DAKO) at 121°C for 30 min. Slides were stained by using an automated stainer (Ventana Medical Systems, Tucson, AZ). Anti-prion mAbs F99/97.6.1 and F89/160.1.5 (22, 23) were used as primary reagents, followed by the application of a biotinylated secondary antibody, streptavidin-alkaline phosphate complex and alkaline phosphatase substrate (Ventana Medical Systems). Controls included isotype-matched irrelevant IgG and blocking of the mAbs with synthetic peptides to which the mAbs were raised. All slides were counterstained with hematoxylin. Photomicrographs were taken with a microscope equipped with a digital camera.
The medial cross section of a placentome was divided into three zones in parallel to the myometrium (Fig. 1). Zone A is proximal to the myometrium and composed of endometrial-chorioallantoic interdigitation at the base of the placentome; zone C is composed of the distal portion of the interdigitating tissue, which contains the arcade systems or hematomata; and zone B is the intermediate area. All stained PrPSc plaques within each zone of the entire medial sections of placentomes of early, mid-, and term pregnant scrapie-infected ewes were enumerated by using a bright field microscope.
Statistical Analysis.
Percent total PrPSc plaques from different zones of placentomes of infected ewes with 171QQ conceptuses during early and term pregnancies were analyzed by one-way ANOVA with a Student-Newman Keuls multiple comparisons test. Probability values of P < 0.05 were used.
Results
Detection Of PrPSc in Placentomes Consisting of 171QQ Uterine Caruncular Endometrium and 171QQ Placental Cotyledonary Chorioallantois from Scrapie-Infected Ewes.
Scrapie-infected sheep were defined by PrPSc accumulation in brain and/or lymphoid tissue by using an immunohistochemistry assay (22). Uterine and placental tissues collected during various stages of pregnancy were assayed for PrPSc by immunohistochemistry. PrPSc was detected in placentomes consisting of 171QQ uterine (caruncular endometrium) and 171QQ placental (cotyledonary chorioallantois) tissues collected from 10 of 11 of the pregnant (day 40–term post mating) scrapie-infected ewes (Fig. 2 A and D). PrPSc was not detected in 171QQ placentomes of 1 of 11 pregnant scrapie infected ewes (Table 1). This placentomal PrPSc-negative sheep was bred before arriving at this U.S. Department of Agriculture/Agricultural Research Service station; therefore, the gestational stage at euthanasia was unknown. However, we determined that this sheep was at an early stage of pregnancy when euthanized (data not shown). PrPSc was not detected in maternal intercaruncular endometrium, intercotyledonary chorioallantois, amniotic membrane, myometrium, ovary, oviduct, bladder, or mammary gland, fetal bladder, kidney, and brain of these pregnant scrapie-infected ewes (data not shown), or 171QQ placentomes of uninfected ewes (n = 25; Table 1). Interestingly, no PrPSc was detected in placentomes of 171QQ uterine and 171QR placental tissues from scrapie-infected ewes (n = 6; Table 1).
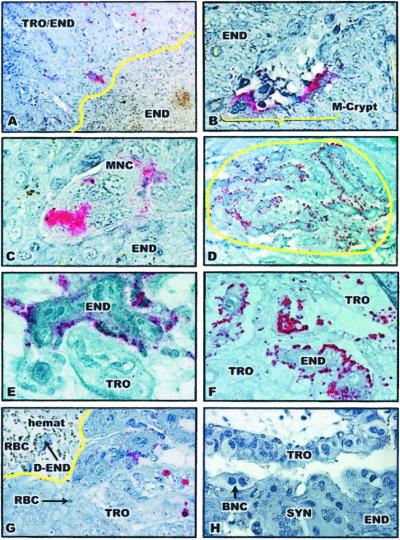
Figure 2.
Immunohistochemical analysis of PrPSc in placentomes of scrapie-infected ewes. The presence of pink staining is indicative of the presence of PrPSc. TRO/END, trophoblast-endometrium interdigitation; TRO, trophoblast; END, endometrium; m-crypt, maternal crypt; MNC, multinucleated cells; RBC, maternal erythrocytes; hemat, hematomata; D-END, degenerating endometrium; ALL, allantoic cavity; BNC, binucleate cells; SYN, endometrial syncytium. (A) PrPSc plaques in the 171QQ placentome of a scrapie-infected ewe at day 40 of pregnancy (×100). PrPSc was seen only in endometrial/placental interdigitation, not in endometrium alone. The yellow line represents the border between maternal endometrium and trophoblast/endometrium interdigitation at the fetal–maternal interface at the bottom of the maternal crypt. (B) PrPSc in cells in the maternal crypt (m-crypt) in the 171QQ placentome of an early pregnant scrapie-infected ewe (×200). (C) PrPSc in a multinucleated cell (MNC) located in the maternal crypt in the 171QQ placentome of an early pregnant scrapie-infected ewe (×400). (D) A PrPSc plaque in zone C of a 171QQ placentome of a term pregnant scrapie-infected ewe (×100). The yellow circle defines the PrPSc plaque. (E) PrPSc in maternal endometrial epithelial cells in the 171QQ placentome of a term pregnant scrapie-infected ewe (×400). (F) PrPSc in both endometrial epithelial cells (END) and trophoblast cells (TRO) in the 171QQ placentome of a term pregnant scrapie-infected ewe (×200). (G) PrPSc in placental trophoblast cells (TRO) located along the hematomata (hemat) (×400). Phagocytosed RBCs (RBC and arrow) were apparent in the placental trophoblast cells. The yellow line defines part of the hematomata containing maternal blood (RBC) and degenerating endometrium (D-END). (H) A placentomal cross section near the endometrial stalk stained with an isotype-matched mAb (×400).
Table 1.
Correlation between the presence of PrPSc in placentomes and fetal prion genotypes or pregnancy status of the scrapie infected ewes
| Scrapie status | Genotype
|
No. of dams | Clinical disease | Pregnancy status | PrPSc accumulation in tissues
|
|||
|---|---|---|---|---|---|---|---|---|
| Dam | Fetus | Bra + lym | Plact | Np-ute | ||||
| Uninfected | 25 | 0/25 | Px | 0/25 | 0/25 | — | ||
| — | 4 | 0/4 | NP | 0/4 | — | 0/4 | ||
| Infected | 11 | 6/11 | Px | 11/11 | 10/11 | — | ||
| QR | 6 | 2/6 | Px | 6/6 | 0/6 | — | ||
| — | 15 | 11/15 | NP | 15/15 | — | 0/15 | ||
Uninfected, Animals that had no reported exposure to scrapie and disease status confirmed by Western blotting and immunohistochemistry of postmortem brain and lymphoid tissues; Infected, animals that either exhibited clinical signs of scrapie or had detectable PrPSc in the lymphoid tissue (based on a live animal third eyelid test) prior to experiment. Genotype, Allelic coding amino acid variation at codon 171 of the prion gene; QQ, homozygous for glutamine; QR, heterozygous for glutamine and arginine; RR, homozygous for arginine. Clinical disease, The number of sheep with clinical scrapie vs. total number of infected sheep used (preclinical sheep had PrPSc in brain and/or lymphoid tissues but no clinical signs, which include rubbing and wool loss, progressive weight loss, and ataxia). Pregnancy status, Px, pregnant; NP, non-pregnant. Bra + lym, PrPSc detected in postmortem brain and lymphoid tissue; plact, placentome; np-ute, non-pregnant uterus.
All ewes studied were between 2 and 5 years old and had PrPSc in the brain and lymphoid tissue. Eight of these 18 ewes exhibited clinical signs of scrapie infection at euthanasia. However, no correlation was found between the presence of PrPSc in placentomes and age, stage of pregnancy, or clinical status of the dams (Table 1).
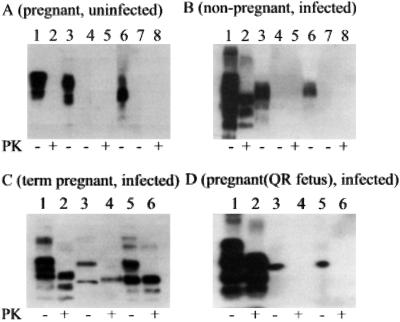
We confirmed the presence of PrPSc by Western blotting in 171QQ placentomal endometrium and chorioallantois collected at term pregnancy (Fig. 3C, lanes 4 and 6). PrPSc was not detected in uterine or placental tissues of uninfected ewes (Fig. 3A, lanes 5 and 8) or in scrapie-infected ewes pregnant with 171QR conceptuses (Fig. 3D, lanes 4 and 6). PrPC in brain, uterine, and placental tissues of uninfected sheep (Fig. 3A, lanes 1, 3, and 6) and PrPSc in brain of scrapie-infected sheep (Fig. 3 B–D, lane 2) were readily detectable by Western blotting using direct tissue homogenates without further enrichment (9). However, PrPSc was not consistently detectable in the uterine and placental tissues unless prepared by ultracentrifugation in the presence of sarkosyl (9, 17). Furthermore, PrPSc was not detectable by Western blotting in uterine and placental tissues of early pregnant (day 40–60 post mating) scrapie-infected ewes (data not shown). Therefore, uterine and placental tissues collected from term pregnant ewes were prepared for Western blotting by ultracentrifugation in the presence of sarkosyl.
Figure 3.
Western blot of PrPSc in tissue homogenates of brainstem, uterine caruncular endometrium, and placental cotyledonary chorioallantois of pregnant and non-pregnant ewes with or without scrapie. Protein from 0.5 mg brainstem or 30 mg endometrium, chorioallantois, and other tissues was loaded in each lane. PK (20 μg/ml) treatment at 37°C for 30 min of each tissue homogenate is indicated at the bottom of each panel (+). (A) Western blot of tissue homogenates prepared from brainstem (lanes 1 and 2), uterine caruncular endometrium (lanes 3–5), and cotyledonary chorioallantois (lanes 6–8) of an uninfected ewe (171QQ) pregnant (day 145 post mating) with a 171QQ conceptus. Samples in lanes 4, 5, 7, and 8 were prepared with sarkosyl and ultracentrifugation. Tissue homogenates were with (lanes 2, 5, and 8) or without (lanes 1, 3, 4, 6, and 7) PK treatment. (B) Western blot of tissue homogenates of brainstem (lanes 1 and 2), endometrium (lanes 3–5), and myometrium (lanes 6–8) of a non-pregnant scrapie-infected ewe (171QQ). Samples in lanes 4 and 5 and in lanes 7 and 8 were prepared with sarkosyl and ultracentrifugation. Tissue homogenates were with (lanes 2, 5, and 8) or without (lanes 1, 3, 4, 6, and 7) PK treatment. (C) Western blot of tissue homogenates prepared from brainstem, caruncular endometrium, and cotyledonary chorioallantois of a scrapie-infected ewe (177QQ) pregnant with a 171QQ conceptus. Lanes 1 and 2, brainstem prepared without sarkosyl and ultracentrifugation; lanes 3 and 4, uterine caruncular endometrium prepared with sarkosyl and ultracentrifugation; and lanes 5 and 6, placental cotyledonary chorioallantois prepared with sarkosyl and ultracentrifugation. Tissue homogenates were with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) PK treatment. (D) Western blot of tissue homogenates prepared from brainstem, caruncular endometrium, and cotyledonary chorioallantois of a scrapie-infected ewe (177QQ) pregnant with a 171QR conceptus. Lanes 1 and 2 (brainstem), prepared without sarkosyl and ultracentrifugation; lanes 3 and 4 (uterine caruncular endometrium), prepared with sarkosyl and ultracentrifugation; and lanes 5 and 6, placental cotyledonary chorioallantois prepared with sarkosyl and ultracentrifugation. Tissue homogenates were with (lanes 2, 4, and 6) or without (lanes 1, 3, and 5) PK treatment.
PrPSc Was Not Detected in Reproductive Tissues of Non-Pregnant, 171QQ, Scrapie-Infected Ewes.
PrPSc accumulation in the reproductive tissues in 15 scrapie-infected and 4 uninfected non-pregnant 171QQ ewes was determined (Table 1). All scrapie-infected ewes had PrPSc accumulation in brain and lymphoid tissue and some (11 of 15) exhibited clinical signs of scrapie (Table 1). No PrPSc was detected in brain or lymphoid tissues of uninfected non-pregnant ewes (data not shown). Homogenates of tissues from infected ewes were prepared with or without ultracentrifugation in the presence of sarkosyl. PrPSc was not detected by either Western blotting (Fig. 3B, lanes 5 and 8) or immunohistochemistry (data not shown) in brain, maternal caruncular endometrium and myometrium, intercaruncular endometrium, ovary, oviduct, bladder, or mammary gland of non-pregnant scrapie-infected ewes, nor in any of the reproductive tissues of non-pregnant uninfected ewes (data not shown).
Distribution of PrPSc Plaques in 171QQ Placentomes of Scrapie-Infected Ewes Is Associated with Stage of Pregnancy.
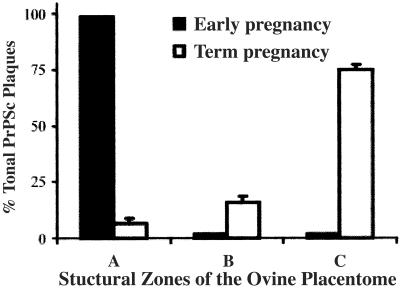
Ten scrapie-infected ewes tested positive for PrPSc in placentomes collected between day 40 post mating and term (Table 1). PrPSc was localized as plaques in placentomes of these ewes by immunohistochemistry (Fig. 2 A and D), and the size and location of PrPSc plaques in these placentomes varied depending on gestational stage (Table 2). PrPSc plaques were seen in placentomal areas where both placental villi and endometrial septae were present, but not in placentomal regions with endometrial tissue alone (Fig. 2A). PrPSc plaques in early pregnant placentomes (days 40–60 post mating; Fig. 2A) were small relative to the plaques in term placentomes (Fig. 2D) and were located in the area of maternal endometrial crypts of zone A (Fig. 4; Table 2). PrPSc staining in term pregnant placentomes was a mixture of small and large plaques located primarily in zone C near the chorionic plate of the placentome (Table 2; Fig. 4). The PrPSc plaques in zone C (75% of total plaques) were more abundant (P < 0.001) than those in zone B (16% of total plaques) and/or zone A (7% of total plaques) of term pregnant placentomes of scrapie-infected ewes (Fig. 4). Zone B of the placentome had primarily small PrPSc plaques (data not shown).
Table 2.
Correlation between stage of pregnancy and spatial distribution of PrPSc in placentomes of pregnant scrapie-infected ewes
| PrPSc | Stage of pregnancy
|
||
|---|---|---|---|
| Early (n = 7) | Mid (n = 1) | Term (n = 2) | |
| Pattern of PrPSc distribution | Numerous small plaques in multinucleate cells | Mostly small plaques and few large plaques | Primarily large plaques spanning several villi |
| Location of PrPSc plaque | Maternal crypts of zone A | Primarily maternal crypt of zone A and some in zone B | Primarily zone C |
PrPSc was defined by immunohistochemistry. Early pregnancy, days 40–60 post mating; mid-pregnancy, days 80–100 post mating; term pregnancy, days 140–150 post mating.
Figure 4.
Distribution of PrPSc plaques in zones A, B, and C of the placentome of ewes infected with scrapie during early (n = 7) and term (n = 2) stages of pregnancy. The total number of PrPSc plaques in medial sections of two separate placentomes of each scrapie-infected ewe was counted microscopically. The results are expressed as the percentage of total PrPSc plaques.
Cellular Localization of PrPSc in Uterine Caruncular Endometrial Epithelium and Placental Cotyledonary Chorioallantoic Trophoblast of 171QQ Placentomes.
Cellular localization of PrPSc in placentomes of scrapie-infected ewes is shown in Fig. 2. Both mAbs used in the present study bound with identical patterns; the results obtained by using mAb F99/97.6.1 are shown. As shown in Fig. 2 B and C for placentomes collected from early pregnant scrapie-infected ewes, PrPSc staining was primarily associated with uterine endometrial epithelium, trophoblast cells, and multinucleate cells in zone A of the maternal endometrial crypts. Very little mAb binding was found in zones B and C of placentomes from early pregnant scrapie-infected ewes. PrPSc was not detected in the stroma or blood vessels (data not shown). No particular uterine or placental structures were associated with this distribution pattern.
In term pregnant scrapie-infected ewes, PrPSc was clearly detected in single cell layers of the endometrial epithelium and placental trophoblast cells (Fig. 2 E, F, and G), as well as in the uterine syncytium (data not shown), a structure formed by fusion of endometrial epithelial cells during normal pregnancy (24). Although staining for PrPSc was present in both uterine epithelium and trophoblast cells throughout the placentome, there was a tendency toward increased accumulation of PrPSc in trophoblast cells in zone C rather than zones A and B in scrapie-infected ewes. In addition, abundant PrPSc was detected in chorionic trophoblast cells around the hematomata, which contains maternal blood as a result of degeneration of the distal ends of the maternal endometrial septae in this region (refs. 24 and 25; Fig. 2G).
PrPSc was not detected by immunohistochemistry in endometrial glands, which were abundant in the interplacentomal region but absent in the placentomal region, nor in intercaruncular endometrium, myometrium, or the interplacentomal endometrial-chorioallantois of scrapie-infected ewes (data not shown). PrPSc was not found in replicate tissue sections when isotype-matched irrelevant IgG mAbs or mAbs absorbed with peptide-antigen were used as the primary antibody (Fig. 2H).
Discussion
Susceptibility to scrapie in Suffolk and Suffolk cross-bred sheep is determined primarily by codon 171 of the PrP gene. Sheep with a 171QQ genotype are susceptible to scrapie, and those with a 171QR or 171RR genotype are resistant to scrapie (11, 13), with a few exceptions (26, 27). The present study investigated the effects of offspring genotype and pregnancy on PrPSc accumulation at the fetal–maternal interface of scrapie-infected sheep and possible transmission between tissues of different genotypes by using the ovine placentome as a natural chimeric model.
PrPSc was present at the fetal-maternal interface, endometrium, and the apposing placental chorioallantois in 171QQ placentomes of most (10/11) infected ewes with PrPSc in the brain and lymphoid tissue. PrPSc was not detected in the uterine tissues of non-pregnant infected ewes, nor in the uterine or placental tissues of infected ewes carrying 171QR fetuses. PrPSc was not detected in 171QQ placentomes from one scrapie-infected ewe. Based on placentomal morphology, this sheep was considered at an early stage of pregnancy when euthanized, suggesting that detectable levels of PrPSc had not yet accumulated in placentomes. The results of this study suggest that the accumulation of PrPSc in the uterus is pregnancy dependent and that PrPSc accumulation in the placentome requires the presence of a 171QQ conceptus. These observations support earlier reports that transmission is related to lambing (9, 14, 17) and suggest that transmission may be reduced or eliminated if infected ewes are not pregnant or conceive offspring with a scrapie-resistant genotype. The mechanisms by which pregnancy affects PrPSc deposition in the uterus of genetically susceptible sheep are not known. We showed previously that PrPC expression is up-regulated in pregnant caruncular endometrium where intimate contact of placental and uterine tissues occur, suggesting that molecules expressed by the cotyledonary trophoblast cells, uterine–placental cell–cell contact in the placentome, and/or the presence of pregnancy-associated hormones are important for PrPC expression and PrPSc accumulation in the caruncular endometrium during pregnancy (9). It is intriguing that PrPSc was absent from the uterine tissue of infected ewes pregnant with 171QR conceptuses. This “blockage” of in vivo PrPSc formation in the presence of placental 171QR and placental/maternal 171QQ PrP may be similar to the interference of PrPSc formation by heterologous PrP molecules shown in vitro (28, 29). One may speculate that placental 171QR PrP expressed by trophoblast cells may interact with uterine PrPSc through cell–cell contact and inhibit the conversion process. In vitro and in vivo studies to examine the role of cell-free and cell-associated trophoblast-derived factors in PrPSc propagation in uterine cells and the systemic effects of pregnancy in the absence of cell-to-cell contact with trophoblast will provide additional information on this critical transmission step. The efficient conversion of PrP in the placentome (detectable during 40–145 days post mating) compared with the lymphoid tissues (10–14 months after exposure during the perinatal period) or the brain (2 to 3 years after exposure) (15) provides a useful model system for examining PrP conversion under natural conditions.
This study demonstrated that PrPSc was localized to the ovine uterine and placental epithelial cells. Because no antibodies to specific markers of these epithelial cells were available, the maternal or fetal/placental origin of these cells at the interface during early pregnancy was estimated by location and morphology under light microscopy. PrPSc was detected in the uterine cells, mononuclear trophoblast cells, and the multinucleated cells formed by fusion between uterine epithelial and placental trophoblast cells. However, PrPSc appeared more abundant in the endometrial epithelial cells than in trophoblast cells in zones A and B of the placentome during early and mid pregnancy. The binucleate cells that originate from the trophoblast cells and invade massively into the endometrium were generally not associated with PrPSc staining (30). PrPSc was clearly present in both uterine and trophoblast cells in zone C of the prepartum placentome particularly. Abundant PrPSc was present in trophoblast cells in zone C near the allantoic cavity. The translocation of the PrPSc from primarily uterine epithelial cells in zone A during early pregnancy to the placental trophoblast cells in zone C at term pregnancy may represent a critical step in the transmission of the scrapie agent from the maternal tissue to placental tissue.
Interestingly, placentomal PrPSc was distributed in discrete plaques. The ovine placentome appears to be a homogenous interdigitation between the endometrial septae and chorioallantoic villi, and the PrPSc plaques in placentomes were not associated with any identifiable landmark structures. However, the spatial distribution pattern of the PrPSc plaques was highly associated with stage of pregnancy. The transition from smaller PrPSc plaques in zone A near the endometrial stalk during early pregnancy to relatively larger PrPSc plaques in zone C near the chorionic plate at term pregnancy may represent a temporal process of PrPSc accumulation and maternal-placental PrPSc transmission. The mechanism of such a transient change in plaque size and location are currently unknown. The fact that placentomal PrPSc plaques of early pregnancy were formed primarily in the maternal endometrial crypts containing the top of the chorionic villi, but not in the endometrium without the chorionic villous tree, supports the concept that the presence of placental tissue is critical for PrPSc accumulation. At term pregnancy, PrPSc plaques were located in the vicinity of the structure known as hematomata in zone C of the placentome. The formation of ovine placentomes is initiated at the time of attachment of uterine caruncles to placental cotyledons and is gradually completed by mid-pregnancy through uterine-placental interdigitation when the distal ends of the endometrial septae and chorionic villi reach the chorionic plate and endometrial stalk, respectively (Fig. 1). During the second half of pregnancy in sheep, the distal ends of the endometrial septae degenerate through an unknown mechanism, and maternal blood leaks through the broken blood vessels into the space surrounded by the intact trophoblast cells of the chorionic villi, leading to the formation of hematomata or the arcade systems in the placentome (24). By term pregnancy, particularly immediately before lambing, massive bleeding is seen in all placentomes, appearing as a dark brown color in the cotyledonary chorioallantois. Thus, hematomata contains maternal blood and degenerated and degenerating endometrial tissue. The trophoblast cells of the hematomata are highly phagocytic, and are thought to play an important role in nonspecific uptake of maternal endometrial secretions, cellular debris, and maternal blood, especially red blood cells, as a source for nutrients of the fetus (24, 25, 31). Indeed, the phagocytosis of maternal red blood cells by placental trophoblast cells was seen in both normal and scrapie-infected animals. Because phagocytosis at the fetal–maternal interface is a nonspecific process, this may be one way for the placental trophoblast cells to acquire PrPSc from the degenerating endometrial cells in the hematomata.
This study demonstrated that offspring genotype and pregnancy are determining factors for PrPSc accumulation at the fetal–maternal interface of scrapie-infected ewes. The characteristic spatial distribution of PrPSc at the interface during different stages of pregnancy may indicate a temporal course of maternal–placental transmission of scrapie. This study presented evidence that scrapie may not be transmissible between infected genetically susceptible uterine tissue and placental tissue with a resistant genotype using the natural chimeric model, the ovine placentome. The results of this study provide useful information for further understanding of scrapie transmission in species with extraneural accumulation of PrPSc. The nature of the PrPSc converting process in the pregnant uterus, the possible inhibitory effect of placental 171QR PrP molecules on PrPSc formation in the uterus, the identity of the pregnancy-related cells or molecules that allow the rapid accumulation of PrPSc in the placentome, and the source of PrPSc in phagocytic trophoblast cells remain to be determined. Additional in vivo studies with sheep at different stages of gestation resulting from genetically controlled mating as well as in vitro studies with uterine epithelial and trophoblast cell lines will provide a better understanding of cellular and molecular basis for transmission of scrapie, a prototype transmissible spongiform encephalopathy.
Acknowledgments
We thank Dr. Charles Leathers for critical review of the manuscript; Anne Anderson, Will Harwood, and Amy Lyda for excellent technical assistance; and Dwayne Chandler, Emma Karel, and Peter Steiner for assistance with animal care and necropsy. This research was supported by grants from the U.S. Department of Agriculture/Agricultural Research Service (CWU 5348-32000-015-00D).
Abbreviations
- PrP
prion protein
- PrPC
cellular PrP
- PrPSc
scrapie PrP
- 171QQ
-RR, or -QR, homozygous for glutamine or arginine, or heterozygous for glutamine and arginine at codon 171 of the PrP gene
- PK
proteinase K
References
- 1.Prusiner S B. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Oesch B, Westaway D, Walchli M, McKinley M P, Kent S B, Aebersold R, Barry R A, Tempst P, Teplow D B, Hood L E. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 3.Basler K, Oesch B, Scott M, Westaway D, Walchli M, Groth D F, McKinley M P, Prusiner S B, Weissmann C. Cell. 1986;46:417–428. doi: 10.1016/0092-8674(86)90662-8. [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi A, Weissmann C. Haemophilia. 1998;4:619–627. doi: 10.1046/j.1365-2516.1998.440619.x. [DOI] [PubMed] [Google Scholar]
- 5.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caughey B. Trends Biochem Sci. 2001;26:235–242. doi: 10.1016/s0968-0004(01)01792-3. [DOI] [PubMed] [Google Scholar]
- 7.Bendheim P E, Brown H R, Rudelli R D, Scala L J, Goller N L, Wen G Y, Kascsak R J, Cashman N R, Bolton D C. Neurology. 1992;42:149–156. doi: 10.1212/wnl.42.1.149. [DOI] [PubMed] [Google Scholar]
- 8.Horiuchi M, Yamazaki N, Ikeda T, Ishiguro N, Shinagawa M. J Gen Virol. 1995;76:2583–2587. doi: 10.1099/0022-1317-76-10-2583. [DOI] [PubMed] [Google Scholar]
- 9.Tuo W, Zhuang D, Knowles D P, Cheevers W P, Sy M S, O'Rourke K I. J Biol Chem. 2001;276:18229–18234. doi: 10.1074/jbc.M008887200. [DOI] [PubMed] [Google Scholar]
- 10.Liu T, Li R, Wong B S, Liu D, Pan T, Petersen R B, Gambetti P, Sy M S. J Immunol. 2001;166:3733–3742. doi: 10.4049/jimmunol.166.6.3733. [DOI] [PubMed] [Google Scholar]
- 11.Westaway D, Zuliani V, Cooper C M, Da Costa M, Neuman S, Jenny A L, Detwiler L, Prusiner S B. Genes Dev. 1994;8:959–969. doi: 10.1101/gad.8.8.959. [DOI] [PubMed] [Google Scholar]
- 12.Prusiner S B, Scott M R. Annu Rev Genet. 1997;31:139–175. doi: 10.1146/annurev.genet.31.1.139. [DOI] [PubMed] [Google Scholar]
- 13.O'Rourke K I, Holyoak G R, Clark W W, Mickelson J R, Wang S, Melco R P, Besser T E, Foote W C. J Gen Virol. 1997;78:975–978. doi: 10.1099/0022-1317-78-4-975. [DOI] [PubMed] [Google Scholar]
- 14.Pattison I H, Hoare M N, Jebbett J N, Watson W A. Vet Rec. 1972;90:465–468. doi: 10.1136/vr.90.17.465. [DOI] [PubMed] [Google Scholar]
- 15.Hadlow W J, Kennedy R C, Race R E. J Infect Dis. 1982;146:657–664. doi: 10.1093/infdis/146.5.657. [DOI] [PubMed] [Google Scholar]
- 16.Van Keulen L J, Schreuder B E, Meloen R H, Poelen-van den Berg M, Mooij-Harkes G, Vromans M E, Langeveld J P. Vet Pathol. 1995;32:299–308. doi: 10.1177/030098589503200312. [DOI] [PubMed] [Google Scholar]
- 17.Race R, Jenny A, Sutton D. J Infect Dis. 1998;178:949–953. doi: 10.1086/515669. [DOI] [PubMed] [Google Scholar]
- 18.Van Keulen L J, Schreuder B E, Vromans M E, Langeveld J P, Smits M A. J Comp Pathol. 1999;121:55–63. doi: 10.1053/jcpa.1998.0300. [DOI] [PubMed] [Google Scholar]
- 19.Beekes M, McBride P A. Neurosci Lett. 2000;278:181–184. doi: 10.1016/s0304-3940(99)00934-9. [DOI] [PubMed] [Google Scholar]
- 20.Heggebo R, Press C M, Gunnes G, Inge L K, Tranulis M A, Ulvund M, Groschup M H, Landsverk T. J Gen Virol. 2000;81:2327–2337. doi: 10.1099/0022-1317-81-9-2327. [DOI] [PubMed] [Google Scholar]
- 21.Andreoletti O, Berthon P, Marc D, Sarradin P, Grosclaude J, van Keulen L, Schelcher F, Elsen J M, Lantier F. J Gen Virol. 2000;81:3115–3126. doi: 10.1099/0022-1317-81-12-3115. [DOI] [PubMed] [Google Scholar]
- 22.O'Rourke K I, Baszler T V, Besser T E, Miller J M, Cutlip R C, Wells G A, Ryder S J, Parish S M, Hamir A N, Cockett N E, et al. J Clin Microbiol. 2000;38:3254–3259. doi: 10.1128/jcm.38.9.3254-3259.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O'Rourke K I, Baszler T V, Miller J M, Spraker T R, Sadler-Riggleman I, Knowles D P. J Clin Microbiol. 1998;36:1750–1755. doi: 10.1128/jcm.36.6.1750-1755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wimsatt W A. Am J Anat. 1950;87:391–457. doi: 10.1002/aja.1000870304. [DOI] [PubMed] [Google Scholar]
- 25.Mossman H W. Vertebrate Fetal Membranes. New Brunswick, NJ: Rutgers Univ. Press; 1987. [Google Scholar]
- 26.Ikeda T, Horiuchi M, Ishiguro N, Muramatsu Y, Kai-Uwe G D, Shinagawa M. J Gen Virol. 1995;76:2577–2581. doi: 10.1099/0022-1317-76-10-2577. [DOI] [PubMed] [Google Scholar]
- 27.Tranulis M A, Osland A, Bratberg B, Ulvund M J. J Gen Virol. 1999;80:1073–1077. doi: 10.1099/0022-1317-80-4-1073. [DOI] [PubMed] [Google Scholar]
- 28.Priola S A, Caughey B, Race R E, Chesebro B. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horiuchi M, Priola S A, Chabry J, Caughey B. Proc Natl Acad Sci USA. 2000;97:5836–5841. doi: 10.1073/pnas.110523897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wooding F B, Chambers S G, Perry J S, George M, Heap R B. Anat Embryol. 1980;158:361–370. doi: 10.1007/BF00301823. [DOI] [PubMed] [Google Scholar]
- 31.Dantzer V. In: Encyclopedia of Reproduction. Knobil E, Neil J, editors. San Diego: Academic; 1998. pp. 18–28. [Google Scholar]