Abstract
The self-induced formation of the disease-associated, protease-resistant prion protein (PrP-res) from the normal protease-sensitive isoform (PrP-sen) appears to be a key event in the pathogenesis of transmissible spongiform encephalopathies. The amino acid sequence specificity of PrP-res formation correlates with, and may account for, the species specificity in transmission of transmissible spongiform encephalopathy agents in vivo. To analyze the mechanism controlling the sequence specificity of PrP-res formation, we compared the binding of PrP-sen to PrP-res with its subsequent acquisition of protease resistance by using cell-free systems consisting of heterologous versus homologous mouse and hamster PrP isoforms. Our studies showed that heterologous PrP-sen can bind to PrP-res with little conversion to the protease-resistant state and, in doing so, can interfere with the conversion of homologous PrP-sen. The interference occurred with molar ratios of homologous to heterologous PrP-sen molecules as low as 1:1. The interference was due primarily to the inhibition of conversion, but not the binding, of the homologous PrP-sen to PrP-res. The results provide evidence that the sequence specificity of PrP-res formation in this model is determined more by the conversion to protease resistance than by the initial binding step. These findings also imply that after the initial binding, further intermolecular interactions between PrP-sen and PrP-res are required to complete the process of conversion to the protease-resistant state.
Transmissible spongiform encephalopathies (TSEs) are fatal neurodegenerative diseases which include scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, and Creutzfeldt–Jakob disease (CJD) in humans. TSEs have a long asymptomatic incubation period followed by a relatively short clinical phase. They are characterized by the accumulation of a disease-specific, protease-resistant isoform of prion protein (PrP), designated PrP-res or PrPSc, in the central nervous system. PrP-res is generated posttranslationally from the normal, protease-sensitive isoform of PrP (PrP-sen or PrPC) (1, 2). Despite the fact that PrP-res is derived from a host-encoded protein, several lines of evidence suggest that PrP-res is a major component of the infectious agent or the agent itself (reviewed in refs. 3–5). Consistent with the latter possibility are demonstrations in cell-free systems that PrP-res can induce the conversion of PrP-sen to a form that is protease-resistant like PrP-res (6). This reaction is highly specific in ways that mimic the species- and strain-specific aspects of TSE diseases in vivo (6–9). This apparently autocatalytic converting activity of PrP-res correlates with scrapie infectivity (10). However, no noninfectious form of PrP has been converted demonstrably to an infectious agent in vitro, so the full identity of the TSE infectious agent, or prion, remains uncertain.
TSE agents can sometimes be transmitted from one host species into another. For instance, bovine spongiform encephalopathy apparently has passed from cattle into humans via the food supply to cause new variant Creutzfeldt–Jakob disease (11, 12). Often, however, there are significant “species barriers,” i.e., inefficiencies in interspecies transmissions that are manifested by a lack of clinical disease or prolonged incubation periods in the recipient species (13–16). Transgenic mouse studies suggested that species barriers result from differences in PrP amino acid sequence between the donors and recipients of infection (17). Furthermore, the conversion of PrP-sen to the protease-resistant form in in vitro systems is strongly influenced by amino acid sequence differences between the PrP-sen and PrP-res molecules (7, 9, 18–20).
In cell-free reactions in which PrP-res induces the conversion of PrP-sen to the protease-resistant form, two steps have been segregated kinetically; first, the binding of PrP-sen to PrP-res, and second, its acquisition of protease resistance (21, 22). This observation raises the question of which step requires amino acid sequence compatibility between PrP-sen and PrP-res. To address this question, we have quantitatively compared the initial binding and overall cell-free conversion reactions between mouse and hamster PrP isoforms. Binding of heterologous PrP-sen to PrP-res was observed with little conversion to the protease-resistant state. Furthermore, such binding of heterologous PrP-sen interfered with the PrP-res-induced conversion of homologous PrP-sen. These results provide insight not only into the molecular control of species specificity but also the basic mechanism of PrP-res formation itself.
Materials and Methods
Cells and Purification of PrP-sen.
The PrP-sen molecules used throughout this study were modified to lack the glycosyl-phosphatidylinositol anchor and were expressed in PA317 and ψ2 mouse fibroblasts (6, 23). The hamster PrP-sen was of the Syrian hamster PrP sequence. The mouse PrP-sen was further mutated from leucine and valine at residues 108 and 111, respectively, to methionines to make it reactive with the monoclonal antibody 3F4 (24); thus, we designate this molecule as mouse(3F4) PrP-sen in what follows. Metabolic labeling of the cells with [35S]methionine and purification of 35S-labeled PrP-sen by immunopurification with 3F4 antibody were performed as described previously (25) except for the elution of 35S-labeled PrP-sen from staphylococcal protein A-Sepharose immunocomplexes; PrP-sen was eluted with 0.1 M acetic acid (pH 2.8) and kept at 4°C until use. Unlabeled PrP-sen was also purified from cells with the same procedure. The amounts (by weight) of unlabeled and 35S-labeled PrP-sen used in various experiments were estimated by immunoblotting.
Purification of PrP-res.
PrP-res was purified without proteinase K (PK) treatment from the brains of hamsters infected with 263K strain (26) or brains of mice infected with mouse-adapted sheep scrapie (Obihiro strain) (27) as described previously (28).
Cell-Free Conversion Reactions.
Cell-free conversion reactions without the use of guanidine hydrochloride (Gdn⋅HCl) were performed as described elsewhere (22). Briefly, purified PrP-res was diluted with deionized water and sonicated for 15 sec, and then 100 ng of PrP-res was mixed with 35S-PrP-sen (20,000 cpm; ≈2 ng) in 20 μl of reaction mixture containing 200 mM KCl, 1.25% Sarkosyl, 5 mM MgCl2, and 50 mM citrate buffer (pH 6.0). The molar ratio of 35S-PrP-sen to PrP-res was estimated to be ≈1:50 by immunoblotting. In the experiments in Figs. 3 and 4 and Table 1, 100 mM citrate buffer was used to maintain the pH of the reaction at 6.0. The reaction mixtures were incubated at 37°C for 3 days. Nine-tenths of the reaction mixtures was treated with 20 μg/ml PK (50 mM Tris⋅HCl, pH 8.0/150 mM NaCl in 100 μl) at 37°C for 45 min. The remaining one-tenth of the reaction mixture was analyzed without PK treatment. Digestion by PK was stopped by adding Pefabloc (Boehringer Mannheim) to 2 mM, 20 μg/ml thyroglobulin (carrier), and then 4 vol of methanol. Methanol-precipitated pellets were subjected to SDS/PAGE using precast acrylamide gels (Novex), and radioactive proteins were visualized and quantified with a PhosphorImager (Molecular Dynamics).
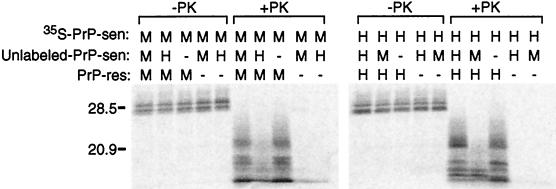
Figure 3.
Interference of protease-resistant 35S-PrP formation by unlabeled heterologous PrP-sen. The species of the PrP components (35S-PrP-sen, unlabeled PrP-sen, and PrP-res) in the reactions are indicated by H (hamster), M [mouse or mouse(3F4)], and − (solvent control). −PK and +PK indicate PK-untreated and PK-treated samples, respectively. The molar ratio of 35S-PrP-sen to unlabeled PrP-sen was 1:16. The relative amount of 35S-PrP-sen versus unlabeled PrP-sen was determined by quantitative immunoblot analysis.
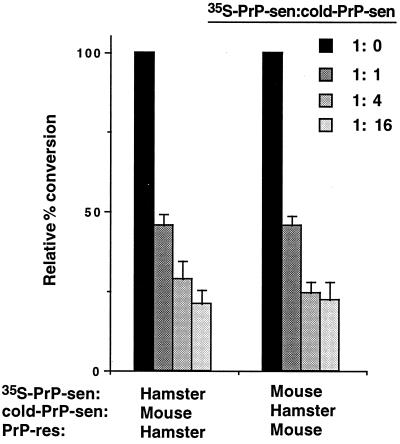
Figure 4.
Dose-dependent interference of protease-resistant 35S-PrP formation by unlabeled (“cold”) heterologous PrP-sen. The left bars show the cell-free conversion reactions with hamster 35S-PrP-sen and PrP-res, whereas the right bars show those with mouse(3F4) 35S-PrP and PrP-res. The conversion reactions were performed in the presence of increasing amounts of unlabeled heterologous PrP-sen as indicated. The percent conversion obtained from the control conversion reactions (without unlabeled PrP-sen, the ratio of 35S-PrP-sen to unlabeled PrP-sen ratio was 1:0) was assigned as 100% and the relative percent conversions as compared with control reactions are shown. Means and standard deviations of triplicate samples are indicated.
Table 1.
Binding of 35S-PrP-sen to PrP-res in the presence of unlabeled heterologous or homologous PrP-sen
| PrP in reaction
|
35S-PrP bound, %* | ||
|---|---|---|---|
| 35S-PrP-sen | Unlabeled PrP-sen† | PrP-res | |
| Mouse(3F4) | Mouse(3F4) | Mouse | 66.8 ± 4.6 |
| Mouse(3F4) | Hamster | Mouse | 61.2 ± 1.0 |
| Mouse(3F4) | — | Mouse | 70.7 ± 0.9‡ |
| Hamster | Hamster | Hamster | 71.0 ± 4.1 |
| Hamster | Mouse(3F4) | Hamster | 64.8 ± 4.4 |
| Hamster | — | Hamster | 76.7 ± 1.6‡ |
Percentage of 35S-PrP in the pelleted fraction relative to total 35S-PrP in the reaction mixture (sum of unbound and pelleted fractions). Background binding (without PrP-res) was 8.8% ± 1.5%.
Ratio of 35S-PrP-sen to unlabeled PrP-sen is 1:16.
These means are marginally statistically significant (P < 0.05) from those of the corresponding reactions with the excess unlabeled heterologous PrP-sen. The other differences were not significant. These values are also slightly higher than the corresponding binding percentages shown in Fig. 2. However, the conditions of the binding reaction were also somewhat different (twice the sodium citrate concentration stated in Materials and Methods) because of the technical requirements of having these reactions control for the experiments in which 16-fold more total PrP-sen was added compared with the reactions in Fig. 2.
Binding Analysis.
One hundred nanograms of PrP-res and purified 35S-PrP-sen (20,000 cpm and ≈2 ng, unless specified otherwise) were mixed in 20 μl of the same buffer as described for the conversion reactions and incubated at 37°C for 3 days. After incubation, reaction mixtures were centrifuged at 12,000 × g for 15 min. The supernatant was saved as the unbound fraction. The resulting pellet was washed once with 180 μl of the same buffer and centrifuged again. The final pellet was dissolved with 20 μl of 1% SDS. The radioactivities in unbound and pelleted (bound) fractions were quantified in a scintillation counter, and 35S-PrP in both fractions was analyzed by SDS/PAGE and PhosphorImager analysis.
Immunoblotting.
Transfer of the proteins from acrylamide gels to Immobilon-P membranes (Millipore) was performed as described (29). PrP on the membrane was visualized by using an anti-PrP synthetic peptide (amino acids 89–103) serum and ECF Western blotting reagents (Amersham), and quantified with a PhosphorImager instrument (Molecular Dynamics).
Results
Species Specificity of Gdn⋅HCl-Free Conversion Reactions.
In previous cell-free reactions in the presence of Gdn⋅HCl, heterologous conversion reactions between mouse and hamster PrP isoforms were inefficient, mimicking the poor interspecies transmissions of scrapie between mice and hamsters (7, 23). Since we recently found Gdn⋅HCl-free conditions for cell-free conversion reactions (22), we first examined whether species specificity could be observed under these more physiological conditions (Fig. 1). In these experiments, we used mouse(3F4) PrP-sen because it has exhibited conversion specificity similar to that of wild-type mouse PrP-sen (7, 23) and can be isolated under conditions identical to those used for hamster PrP-sen, i.e., with the same 3F4 antibody.
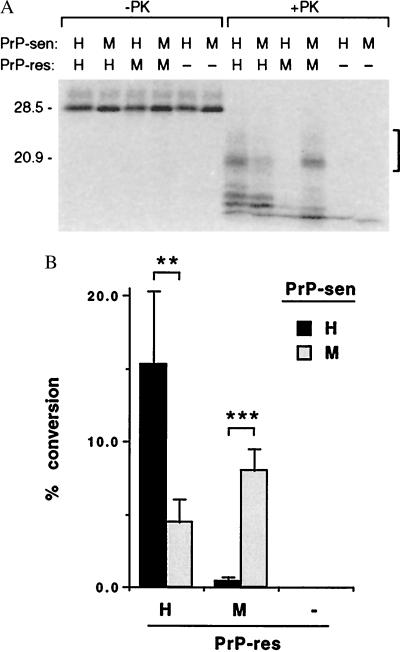
Figure 1.
Formation of protease-resistant 35S-PrP by 35S-PrP-sen with homologous and heterologous PrP-res under Gdn⋅HCl-free conditions. (A) The species of PrP-sen and PrP-res are indicated above the figure as H (hamster), M [mouse PrP-res or mouse(3F4)-PrP-sen]. Controls lacking PrP-res are indicated by −. −PK and +PK indicate PK-untreated and PK-treated samples, respectively. The major band of the 35S-PrP-sen (lacking the glycosyl-phosphatidylinosital anchor) was unglycosylated and comigrated with the 28.5-kDa marker. The protease-resistant 35S-PrP bands quantified by PhosphorImager analyses are indicated by the bracket on the right side. The bands lying below the bracket are partial conversion products, the formation of which appears to be less species dependent in this and previous studies (7). Molecular mass markers are on the left (in kDa). (B) Quantitation of the formation of protease-resistant 35S-PrP. The means and standard deviations of five independent experiments are indicated. Statistical significance of the differences between the means is designated by ** and *** for P < 0.01 and P < 0.001, respectively, in an unpaired t test. −, Without PrP-res.
As observed previously (6, 22), 35S-labeled hamster PrP-sen was converted to the expected partially protease-resistant forms (≈7 kDa smaller after PK digestion than the original 35S-PrP-sen) in the presence of homologous hamster PrP-res but not in its absence (Fig. 1A). Compared with this positive control reaction, the conversion of heterologous mouse(3F4) 35S-PrP-sen into the protease-resistant form by hamster PrP-res was ≈70% less efficient (P < 0.01, Fig. 1). Furthermore, the conversion of hamster 35S-PrP-sen by mouse PrP-res was ≈95% less efficient than the more homologous¶ reaction between mouse(3F4) 35S-PrP-sen/mouse PrP-res (P < 0.001, Figs. 1). Thus, the relative inefficiencies of the heterologous conversion reactions under the near-physiological conditions correlated well not only with the previously reported reactions with Gdn⋅HCl but also with the poor transmissibilities of scrapie between these two species in vivo.
Binding of PrP-sen to Heterologous PrP-res.
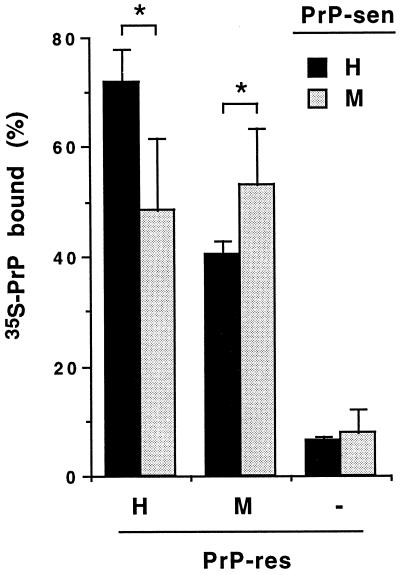
To determine whether the inefficient conversion of PrP-sen with heterologous PrP-res was due to poor initial binding of PrP-sen to PrP-res, the binding of 35S-PrP-sen to PrP-res was analyzed. As described previously (22), PrP-res was incubated with the 35S-PrP-sen and then was pelleted, along with any bound 35S-PrP-sen, by centrifugation. More than 95% of the PrP-res is pelleted by this procedure (data not shown), whereas in the absence of PrP-res, <10% of the 35S-PrP-sen appeared in the pellet (Fig. 2). Fig. 2 shows that after 3-day incubations, the binding of 35S-PrP-sen to PrP-res in heterologous combinations was 70–74% as efficient as with the homologous combinations (P < 0.05). These results suggest that the much less efficient conversions observed with heterologous mixtures of PrP-sen and PrP-res (Fig. 1) can be attributed only partly to a lack of binding between the two isoforms. Thus, it is likely that the sequence specificity of the conversion reaction is determined to a large extent by events in the subsequent step in which PrP-sen acquires protease resistance.
Figure 2.
Binding of 35S-PrP-sen to homologous and heterologous PrP-res. Radioactivities of unbound and pelleted (bound) fractions were determined as described in the text. The percentages of the radioactivity in the pelleted fraction relative to total radioactivity (sum of unbound and pelleted fractions) are expressed as 35S-PrP bound to PrP-res in the graph. Without PrP-res (−), <10% of total radioactivity was detected in the pelleted fraction. Means ±SD of four independent experiments are indicated. Marginal statistical significance (P < 0.05) of the differences between the means is designated by *.
Interference of PrP-res Formation by Heterologous PrP-sen.
Because the heterologous PrP-sen molecules that were poorly converted to the protease-resistant form were still able to bind to PrP-res, we analyzed whether they could also interfere with the binding and/or conversion of homologous PrP-sen molecules. In these reactions, the homologous 35S-PrP-sen and PrP-res (unlabeled) species were mixed with increasing amounts of unlabeled PrP-sen molecules (homologous or heterologous) and the efficiencies of conversion were determined (Fig. 3). No reduction in the formation of protease-resistant 35S-PrP was observed with ≤16-fold excess of homologous PrP-sen, indicating that the conversion reaction was not saturated with substrate. By contrast, when the unlabeled heterologous PrP-sen was added, formation of the protease-resistant 35S-PrP from the homologous 35S-PrP-sen was greatly reduced (Fig. 3), even at a 1:1 ratio of unlabeled (heterologous) PrP-sen to 35S-labeled (homologous) PrP-sen (Fig. 4). This indicated that heterologous PrP-sen molecules could interfere with conversion reactions between homologous PrP-sen and PrP-res molecules without themselves being converted. The fact that interference was observed even at 1:1 ratios of homologous to heterologous PrP-sen provided evidence that mouse(3F4) and hamster PrP-sen have sufficiently similar affinities to compete with one another for the conversion-inducing sites on either mouse or hamster PrP-res.
Binding of PrP-sen to PrP-res Is Not Blocked by Heterologous PrP-sen.
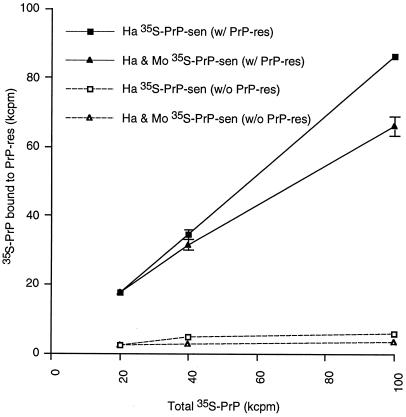
Because unlabeled heterologous PrP-sen interfered with the formation of homologous protease-resistant 35S-PrP, we examined whether unlabeled heterologous PrP-sen could also interfere with the binding of 35S-labeled PrP-sen to PrP-res. As expected from the previous conversion experiment in Fig. 3, the addition of a 16-fold excess of unlabeled homologous PrP-sen did not reduce the binding of the homologous 35S-PrP-sen (Table 1). Interestingly, however, the same was true with the addition of unlabeled heterologous PrP-sen. Thus, despite the interference of protease-resistant 35S-PrP formation by heterologous unlabeled PrP-sen (>75% inhibition, Figs. 3 and 4), the binding of the homologous 35S-PrP-sen was not blocked by more than ≈15% (Table 1). To confirm that the excess heterologous PrP-sen also binds to PrP-res in the presence of homologous PrP-sen, increasing amounts of mouse(3F4) 35S-PrP-sen were added to reaction mixtures containing constant amounts of 35S-labeled hamster PrP-sen and unlabeled hamster PrP-res. The total amount of 35S-PrP pelleted with hamster PrP-res increased with the addition of mouse(3F4) 35S-PrP-sen (Fig. 5), indicating that mouse(3F4) PrP-sen binds to hamster PrP-res even in the presence of hamster PrP-sen. Thus, both homologous and heterologous PrP-sen can bind simultaneously to PrP-res. The ability of the heterologous PrP-sen to interfere with the conversion of homologous PrP-sen therefore appears to be due primarily to inhibition of acquisition of protease resistance, rather than the binding, by the homologous PrP-sen.
Figure 5.
Dose-dependent binding of 35S-PrP-sen to PrP-res. A total of 20, 40, and 100 kcpm of hamster (Ha) 35S-PrP-sen or the mixture of hamster and mouse(3F4) (Mo) 35S-PrP-sen (see below) were incubated with (solid lines) or without (dashed lines) hamster PrP-res. The binding of 35S-PrP-sen to PrP-res was analyzed by sedimentation analysis as described in the text. The 100-kcpm sample consisted of 20 kcpm of hamster and 80 kcpm of mouse(3F4) 35S-PrP-sen. The 40-kcpm sample consisted of 20 kcpm of hamster and 20 kcpm of mouse(3F4) 35S-PrP-sen. The 20-kcpm sample consisted only of hamster 35S-PrP-sen. The horizontal axis indicates the 35S-PrP-sen added to the reactions (kcpm), whereas the vertical axis indicates 35S-PrP detected in the pelleted fractions. The recoveries of input 35S-PrP varied from 70% to 85%. Each point represents a mean ±SD of triplicates.
Discussion
The appearance of new variant Creutzfeldt–Jakob disease as a likely consequence of bovine spongiform encephalopathy transmission from cattle to humans has heightened the importance of understanding the mechanisms controlling the interspecies transmissibility of TSE agents. Clearly, PrP amino acid sequence differences between donor and recipient species can play an important modulatory role in vivo (17). Similarly, strong sequence compatibility requirements can be seen for PrP-res formation in scrapie-infected tissue culture cells and cell-free reactions (7, 9, 18–20). Studies in cell-free systems also have shown that the conversion of PrP-sen to the protease-resistant state results from direct interactions between PrP-sen and PrP-res aggregates. The binding between PrP-sen and PrP-res is highly selective with respect to other proteins (22); however, as we have shown here, binding between heterologous PrP isoforms can occur readily. Interestingly, the subsequent step in which bound PrP-sen acquires protease resistance depends more on PrP sequence homology and may not occur between heterologous PrP species. Thus, with mouse and hamster isoforms of PrP at least, it is the acquisition of protease resistance by PrP-sen rather than the initial binding of PrP-sen to PrP-res that primarily determines the sequence specificity of PrP-res formation.
Differences Versus Earlier Mouse–Hamster Conversions.
There are some differences in the species specificities of the conversion reactions observed in this study and previous studies. Conversions of wild-type mouse and mouse(3F4) PrP-sen [with glycosyl-phosphatidylinositol (GPI)] with PrP-res of the hamster scrapie 263K strain were not observed in the previous studies (7), and this is consistent with the lack of transmission of 263K hamster scrapie strain to mice (30). In contrast, 263K PrP-res did convert mouse(3F4) PrP-sen in this study, but the conversion efficiency of this combination remained much lower than that of the homologous combination. The differences in the experimental conditions, e.g., the presence of absence of Gdn⋅HCl or the use of GPI(−) PrP-sen instead of wild-type Pr-sen may have affected the results. However, the relative conversion efficiency in cell-free conversion reactions is still broadly consistent with the relative transmissibilities of TSE agents, suggesting that the cell-free conversion reaction used here is a useful tool for analyzing the mechanism of species specificity of PrP-res formation.
Sites of Interaction Between PrP-sen and PrP-res.
The segregation of the binding of PrP-sen to PrP-res from the acquisition of protease resistance by PrP-sen in the cell-free reactions provides insight into the details of the conversion process. One region containing three amino acid differences between mice and hamsters (mouse/hamster residues 138/139, 154/155, and 169/170) is known to strongly influence the species-specific formation of PrP-res (7, 18, 20, 31). However, our binding analyses have indicated that differences at these and other amino acid residues only slightly reduce the initial binding between mouse and hamster PrP isoforms. In a previous study, we found evidence that the initial binding of PrP-sen to PrP-res occurs through one or more surfaces adjacent to the C terminus in the three-dimensional structure of PrP-sen (22). One such surface contains residues 119–138 and others contain residues ≈165–174 and 206–223 of PrP-sen. Taken together, these findings suggest the following sequence of events in the conversion process: Binding occurs initially through one or more of these surfaces independent of possible mismatches at residue 138/139 or other residues. The subsequent conversion to protease-resistant PrP, however, requires further, more specific, intermolecular interactions and/or conformational changes that are strongly influenced by residues 138/139, 154/155, and/or 169/170. Indeed, this was suggested previously to be the case for residue 138/139 because, in PrP-sen, it is hydrophobic, internally disposed, and unlikely to take part in initial intermolecular interactions with PrP-res (32).
Stoichiometry of Conversion Sites per Unit of PrP-res.
We have also shown here that the formation of protease-resistant 35S-PrP in homologous reactions can be partially blocked by heterologous PrP-sen even when the latter is at ratios of 1:1 to the homologous 35S-PrP-sen and ≈1:50 to the PrP-res seed. This result suggests the following: (i) the heterologous, nonconvertible PrP-sen is able to compete effectively with the homologous PrP-sen for the conversion-inducing binding site; (ii) the ability of PrP-res to induce conversion of homologous PrP-sen is terminated once it is bound by conversion-incompetent heterologous PrP-sen (Fig. 6); and (iii) there are far fewer conversion-inducing sites than PrP-res molecules in the average PrP-res polymer under these conditions. The ≈50% inhibition of conversion that was observed with a 1:50 ratio of heterologous PrP-sen to PrP-res suggests that conversion-inducing binding sites for PrP-sen on PrP-res were roughly 50% occupied by the interfering PrP-sen. Thus, the number of conversion-inducing sites on the PrP-res polymers can be estimated to have been a maximum of ≈1 per 25 PrP-res molecules.
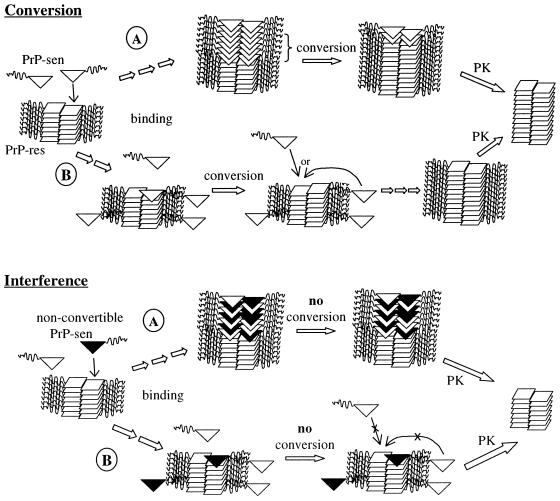
Figure 6.
Nucleated polymerization-based models for binding, conversion (Upper), and interference (Lower) phenomena. Single-site (A) and two-binding-site (B) pathways are shown. In the single-site model (Upper, A), rapid binding of PrP-sen (open triangles) is followed by a slower wave of conformational conversion of bound PrP-sen molecules to PrP-res (squares). The inclusion of nonconvertible PrP-sen (black triangles) among convertible PrP-sen molecules prevents propagation of the conversion through the bound PrP-sen molecules (Lower, A). In the two-site models (B), binding of PrP-sen can occur either at the conversion-inducing site (shown at the end of the PrP-res polymer) or at a nonconverting site (e.g., on the sides of the polymer). Interference with conversion (Lower, B) could occur by blockade of the conversion site by nonconvertible PrP-sen without blocking binding of either type of PrP-sen to the nonconverting sites. PK designates a proteinase K digestion step wherein PrP-sen is completely digested and the N-terminal octapeptide repeat domain (residues 23-≈90, the wavy lines in the PrP-sen and PrP-res structures) are removed from PrP-res.
Mechanistic Models for Interference.
There are at least two possible polymerization-based models (Fig. 6) for the observation that heterologous PrP-sen can interfere with the conversion of homologous PrP-sen (Figs. 3 and 4) without blocking the latter's binding to PrP-res (Table 1). One model assumes that there is only one type of site on the PrP-res polymer for binding and conversion. The heterologous PrP may be able to bind to that site and form part of a new binding domain for the next (homologous or heterologous) PrP-sen molecule. However, if the heterologous PrP-sen molecule cannot undergo the conformational change to PrP-res, it, in turn, is unable to propagate the conformational change to subsequently added PrP-sen molecules. In this way, PrP-res-induced copolymerization (i.e., binding) of both homologous and heterologous PrP-sen molecules might occur continuously without additional PrP-res formation. Alternatively, there may be two types of binding sites for PrP-sen on PrP-res, one that induces conversion to PrP-res and another that does not. Interference of conversion would then be due solely to the blockade of conversion-inducing sites without hindering continued binding to the other type of site. The interference in this scenario might resemble the action of capping proteins in actin polymerization (33).
The interference with formation of protease-resistant PrP observed in this study provides some mechanistic insights into previous studies with transgenic (Tg) mice and scrapie-infected cells. Mice transgenic for the hamster PrP gene are susceptible to hamster scrapie (17). However, the incubation period in hamster-PrP Tg mice that still express endogenous mouse PrP is much longer than the incubation periods in hamster-PrP Tg mice in which the mouse PrP has been ablated (17, 34, 35). Furthermore, expression of the hamster PrP gene in scrapie-infected mouse neuroblastoma cells abolished the authentic PrP-res formation (19). These studies imply that interactions between heterologous PrP-sen and PrP-res interfere with PrP-res formation and/or the propagation of scrapie agent. Since we have shown here that homologous PrP binding interactions do not appear to be greatly favored over heterologous interactions, it is likely that PrP-res formation is interrupted once conversion-incompetent heterologous PrP-sen binds to PrP-res. In vivo, this could lead to a delay in both PrP-res formation and onset of clinical disease.
General Implications for Interspecies TSE Transmissions.
In more general terms, the following scenarios for TSE transmissions between species in vivo are suggested. These scenarios assume that PrP-res formation is required for TSE agent replication. If the incoming agent-associated PrP-res can both bind and efficiently convert the host's PrP-sen to PrP-res, then no PrP-based barrier to transmission would be expected. If, at the opposite extreme, there were interspecies combinations of PrP-res and PrP-sen where no binding or conversion occurs, there would be complete resistance of the host to infection. An intermediate situation is suggested by our observations with the mouse and hamster PrP isoforms in which the incoming PrP-res can bind to the host's PrP-sen without converting it efficiently to PrP-res. A significant, but not necessarily complete, barrier to transmission would be expected. Such a phenomenon could be involved in recent studies showing that hamster scrapie infectivity can persist in wild-type, but not PrP-knockout mice, for their natural lifespan but without causing clinical illness (16). If binding between the PrP-res and PrP-sen occurs, there at least may be the opportunity for the subsequent induction of protease resistance in the host PrP-sen, even if this process were inefficient or stochastic. Once the new PrP-res is generated from the PrP-sen in the new host, there would be no sequence-based species barrier to the subsequent formation of PrP-res. PrP-res might then accumulate more efficiently and, in some cases, cause clinical disease within the lifespan of the host. Such a scenario might exist for transmissions of bovine spongiform encephalopathy into humans, where the efficiency of PrPBSE-induced conversions of human PrP-sen is very low but measurable (9).
Therapeutic Implications.
Two possible targets for anti-TSE compounds are suggested by the present study. Compounds that block either the binding of PrP-sen to PrP-res or the subsequent conversion of PrP-sen to PrP-res will potentially be anti-TSE compounds. Therefore, cell-free conversion reactions and simple binding assays that can be performed in multiwell plates may be good in vitro screening methods for anti-TSE compounds.
Abbreviations
- TSE
transmissible spongiform encephalopathy
- PrP
prion protein
- PrP-res and PrP-sen
protease-resistant and protease-sensitive isoforms of PrP
- PK
proteinase K
- Gdn⋅HCl
guanidine hydrochloride
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.110523897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.110523897
For simplicity, we will refer to reactions between the mouse(3F4) 35S-PrP-sen and mouse PrP-res as “homologous” despite the differences at residues 108 and 111. The term “heterologous” will be reserved for reactions between mouse [or mouse(3F4)] and hamster PrP molecules, where there are 12 [or 10] residue mismatches, including those that are known to greatly influence the specificity of conversion reactions (7, 18–20, 23).
References
- 1.Borchelt D R, Scott M, Taraboulos A, Stahl N, Prusiner S B. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caughey B, Raymond G J. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 3.Caughey B, Chesebro B. Trends Cell Biol. 1997;7:56–62. doi: 10.1016/S0962-8924(96)10054-4. [DOI] [PubMed] [Google Scholar]
- 4.Weissmann C. J Biol Chem. 1999;274:3–6. doi: 10.1074/jbc.274.1.3. [DOI] [PubMed] [Google Scholar]
- 5.Prusiner S B. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kocisko D A, Come J H, Priola S A, Chesebro B, Raymond G J, Lansbury P T, Caughey B. Nature (London) 1994;370:471–474. doi: 10.1038/370471a0. [DOI] [PubMed] [Google Scholar]
- 7.Kocisko D A, Priola S A, Raymond G J, Chesebro B, Lansbury P T, Jr, Caughey B. Proc Natl Acad Sci USA. 1995;92:3923–3927. doi: 10.1073/pnas.92.9.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bessen R A, Kocisko D A, Raymond G J, Nandan S, Lansbury P T, Jr, Caughey B. Nature (London) 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 9.Raymond G J, Hope J, Kocisko D A, Priola S A, Raymond L D, Bossers A, Ironside J, Will R G, Chen S G, Petersen R B, et al. Nature (London) 1997;388:285–288. doi: 10.1038/40876. [DOI] [PubMed] [Google Scholar]
- 10.Caughey B, Raymond G J, Kocisko D A, Lansbury P T., Jr J Virol. 1997;71:4107–4110. doi: 10.1128/jvi.71.5.4107-4110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruce M E, Will R G, Ironside J W, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, et al. Nature (London) 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 12.Hill A F, Desbruslais M, Joiner S, Sidle K C L, Gowland I, Collinge J, Doey L J, Lantos P. Nature (London) 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 13.Pattison I H, Jones K M. Res Vet Sci. 1968;9:408–410. [PubMed] [Google Scholar]
- 14.Dickinson A G. In: Slow Viruses in Animals and Man. Kimberlin R H, editor. Amsterdam: North Holland; 1976. pp. 209–241. [Google Scholar]
- 15.Kimberlin R H, Walker C A, Millson G C. Lancet. 1975;ii:1309–1310. doi: 10.1016/s0140-6736(75)90649-2. [DOI] [PubMed] [Google Scholar]
- 16.Race R, Chesebro B. Nature (London) 1998;392:770. doi: 10.1038/33834. (Lett.). [DOI] [PubMed] [Google Scholar]
- 17.Prusiner S B, Scott M, Foster D, Pan K M, Groth D, Mirenda C, Torchia M, Yang S L, Serban D, Carlson G A, et al. Cell. 1990;63:673–686. doi: 10.1016/0092-8674(90)90134-z. [DOI] [PubMed] [Google Scholar]
- 18.Scott M R, Kohler R, Foster D, Prusiner S B. Protein Sci. 1992;1:986–997. doi: 10.1002/pro.5560010804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priola S A, Caughey B, Race R E, Chesebro B. J Virol. 1994;68:4873–4878. doi: 10.1128/jvi.68.8.4873-4878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Priola S A, Chesebro B. J Virol. 1995;69:7754–7758. doi: 10.1128/jvi.69.12.7754-7758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DebBurman S K, Raymond G J, Caughey B, Lindquist S. Proc Natl Acad Sci USA. 1997;94:13938–13943. doi: 10.1073/pnas.94.25.13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horiuchi M, Chabry J, Caughey B. EMBO J. 1999;18:3193–3203. doi: 10.1093/emboj/18.12.3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chabry J, Priola S A, Wehrly K, Nishio J, Hope J, Chesebro B. J Virol. 1999;73:6245–6250. doi: 10.1128/jvi.73.8.6245-6250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R I, Wisniewski H M, Diringer H. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caughey B, Kocisko D A, Raymond G J, Lansbury P T. Chem Biol. 1995;2:807–817. doi: 10.1016/1074-5521(95)90087-x. [DOI] [PubMed] [Google Scholar]
- 26.Kimberlin R H, Walker C A. J Gen Virol. 1978;39:487–496. doi: 10.1099/0022-1317-39-3-487. [DOI] [PubMed] [Google Scholar]
- 27.Shinagawa M, Takahashi K, Sasaki S, Doi S, Sato G. Microbiol Immunol. 1985;29:543–551. doi: 10.1111/j.1348-0421.1985.tb00856.x. [DOI] [PubMed] [Google Scholar]
- 28.Caughey B W, Dong A, Bhat K S, Ernst D, Hayes S F, Caughey W S. Biochemistry. 1991;30:7672–7680. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 29.Caughey B, Raymond G J, Ernst D, Race R E. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimberlin R H, Walker C A, Fraser H. J Gen Virol. 1989;70:2017–2025. doi: 10.1099/0022-1317-70-8-2017. [DOI] [PubMed] [Google Scholar]
- 31.Scott M, Groth D, Foster D, Torchia M, Yang S L, DeArmond S J, Prusiner S B. Cell. 1993;73:979–988. doi: 10.1016/0092-8674(93)90275-u. [DOI] [PubMed] [Google Scholar]
- 32.Billeter M, Riek R, Wider G, Hornemann S, Glockshuber R, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:7281–7285. doi: 10.1073/pnas.94.14.7281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cooper J A, Schafer D A. Curr Opin Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- 34.Bueler H, Aguzzi A, Sailer A, Greiner R-A, Autenried P, Aguet M, Weissmann C. Cell. 1993;73:1339–1347. doi: 10.1016/0092-8674(93)90360-3. [DOI] [PubMed] [Google Scholar]
- 35.Race R, Oldstone M B A, Chesebro B. J Virol. 2000;74:828–833. doi: 10.1128/jvi.74.2.828-833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]