Abstract
Femtosecond time-resolved spectroscopy on model peptides with built-in light switches combined with computer simulation of light-triggered motions offers an attractive integrated approach toward the understanding of peptide conformational dynamics. It was applied to monitor the light-induced relaxation dynamics occurring on subnanosecond time scales in a peptide that was backbone-cyclized with an azobenzene derivative as optical switch and spectroscopic probe. The femtosecond spectra permit the clear distinguishing and characterization of the subpicosecond photoisomerization of the chromophore, the subsequent dissipation of vibrational energy, and the subnanosecond conformational relaxation of the peptide. The photochemical cis/trans-isomerization of the chromophore and the resulting peptide relaxations have been simulated with molecular dynamics calculations. The calculated reaction kinetics, as monitored by the energy content of the peptide, were found to match the spectroscopic data. Thus we verify that all-atom molecular dynamics simulations can quantitatively describe the subnanosecond conformational dynamics of peptides, strengthening confidence in corresponding predictions for longer time scales.
In the folding process, proteins and peptides acquire their native structures through a conformational dynamics involving a hierarchy of temporal and spatial scales (1–3). According to molecular dynamics (MD) simulations (4–7), the elementary events of this dynamics proceed on the picosecond time scale and comprise concerted rotations around the bonds at the Cα atoms. Experimentally, however, these events have been elusive to observation, as classical stopped-flow or jump techniques have a time resolution of milli- or, at best, microseconds. Recently, the nanosecond time scale has become accessible by laser-induced pH- or temperature-jump experiments (8–10) and by triplet–triplet energy transfer (11), as well as by Laue x-ray diffraction (12). For the subpicosecond time scale, on which peptides thermally fluctuate within their conformational substates, two-dimensional infrared spectroscopy has provided insights (13).
An experimental check of MD descriptions by ultrafast spectroscopy requires a polypeptide construct that contains a fast optical trigger capable of inducing specific conformational transitions at a well-defined starting time and a spectroscopic probe sensitive to the structural changes in the peptide. Unfortunately, biologically important photoactive proteins, which exhibit light-induced functional processes accompanied by conformational changes (14–16), are too large and complex to allow detailed comparisons with simulations.
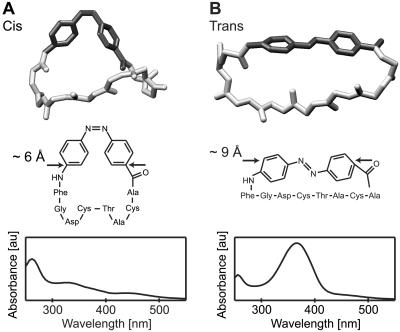
To bridge the gap between simulation-based theory and experiment, we have selected the photo-switchable peptide shown in Fig. 1, which is backbone-cyclized with a suitably functionalized (4-aminophenyl)azobenzoic acid (APB) (17, 18). The APB molecule has been chosen as optical trigger, because azobenzene dyes are known to photoisomerize at the central N—N bond within fractions of picoseconds at high quantum yields (19) and with large changes in geometry. The conformational space of the peptide moiety becomes restricted by the cyclization. The cis/trans-photoisomerization of APB induces significant conformational changes in that moiety, as shown by NMR analysis of the cis- and trans-isomers of the APB-peptide (17, 20). Because the two isomeric forms of the APB chromophore retain pronounced spectral differences in the peptide construct, a clear-cut spectroscopic distinction and a selective light-induced interconversion are possible (Fig. 1 Bottom). Furthermore, the APB-peptide is simple enough to allow an all-atom simulation of its conformational dynamics by an MD approach. For the cis- and trans-isomers, the simulations yield equilibrium structural ensembles, with conformational substates close to the NMR-derived structures (representative average MD structures are shown in Fig. 1 Top).
Figure 1.
(A) Structure of the cis-APB-peptide (Middle), corresponding absorption spectrum (Bottom), and snapshot from the MD simulations (Top), which represents the mean conformation of the molecule in the first picosecond. The peptide backbone (light gray) and the APB-group (dark gray) are shown. (B) Photoinduced cis/trans-isomerization leads to a considerable elongation of the APB moiety.
With these premises, the model system enables an integrated approach, which combines femtosecond time-resolved spectroscopy of the photoisomerization of the azobenzene and of the subsequent conformational relaxation of the peptide with MD simulations aiming at a quantitative description of these dynamical processes.
Materials and Methods
The synthesis of linear and cyclic APB-peptides was reported before (18). The sample was dissolved in DMSO to a final concentration of 0.8 mM (linear APB-peptide) and 2.3 mM (cyclic APB-peptide), respectively, and exchanged between individual laser shots by pumping the solution with sufficient flow rate through the fused silica cuvette (0.5-mm optical path length). The initial cis-isomeric state was prepared by continuous wave illumination with light between 320 and 380 nm (1-kW high pressure HgXe arc lamp with UG11 and WG320 filters; Schott, Mainz, Germany), supplied via a liquid light guide.
The visible pump–white light probe setup, including a multichannel detection system, has been described in detail (21). A 1-kHz regenerative chirped pulse-amplified Ti:sapphire system together with a noncollinear optical parametric amplifier (22) provided excitation at a central wavelength of 480 nm. In combination with a white light continuum (23) used for probing from 300 to 700 nm, a typical cross-correlation width (instrumental response function) of about 80 fs was achieved. The various steps for the data processing include: measurement of absorption changes for pure solvent and sample, dispersion correction, subtraction of signals because of solvent, normalization, global fitting of the final data set with constant rates, and calculation of response of pure initial states.
The MD simulations were based on a force field that extends the all-hydrogen peptide model CHARMM22 (24) by a multiple scale MD approach (MSMD) for the calculation of the electrostatic interactions and by a choice of additional parameters for various compounds. MSMD avoids the usual cutoff of the electrostatics. Instead, by using fast hierarchical multipole expansions (25, 26), the electrostatic interactions are calculated explicitly up to a distance given by the minimum image convention relevant for periodic boundary systems and from a reaction field model at larger distances. Thus, MSMD substantially extends the approach suggested before (27). Van der Waals interactions are explicitly calculated up to a distance of 10 Å. Parameters for the azobenzene and APB residue were derived from density functional theory (DFT) by using GAUSSIAN 98 (28) (B3LYP/6–31G+), those for the explicit description of the solvent DMSO from hybrid DFT/MD simulations (29). Periodic boundary conditions were imposed for solvent–solute systems contained in cubic boxes. Simulations were carried out at constant volume, temperature, and particle number with the solvent being coupled to a heat bath of 300 K at a coupling constant of 0.5 ps. Numerical integration was performed by a multiple time-step algorithm with a 1-fs integration time step. The lengths of all bonds involving hydrogens were held fixed by the shake algorithm (30).
Starting coordinates were taken from six NMR structures of cis-APB-peptide published in ref. 20 and from six arbitrary structures of cis-azobenzene. The solutes were placed in a 60-Å periodic DMSO box containing about 1,600 molecules at the experimental density of 1.1 g/cm3. The solvent energy was minimized for 200 fs and equilibrated for 30 ps while keeping the solute fixed, followed by a 50-ps equilibration of the whole system. The light-induced inversion reaction at one of the nitrogen atoms in the diazene group was modeled by a force field applying an in-plane torque and accounting for the 270-kJ/mol energy uptake by electronic excitation. After excitation, trajectories were generated from 1-ns runs, enabling the calculation of the total energies of the APB-peptide and of azobenzene, respectively.
Results and Discussion
To enable an understanding of the results of our spectroscopic pump-probe experiments, we first wish to sketch the sequence of events, which a priori are likely to happen considering the known photochemistry of azobenzene (19) and the properties (17, 20) of the cyclic APB peptides summarized in the Introduction. By irradiating a solution of cis-APB-peptides in the nπ*-band at 480 nm with a femtosecond light pulse, the azobenzene moiety is expected to isomerize in the first electronically excited state S1 at the N—N bond from cis to trans crossing into a vibrationally excited trans ground state S*0 through a conical intersection (step 1). Subsequently, the vibrational energy of S*0 will be dissipated, heating the peptide chain and the surrounding solvent (step 2). The geometrical change of APB resulting from its cis/trans-photoisomerization (elongation from 6 to 9 Å and planarization; see Fig. 1) will exert a strain on the peptide moiety. The strain will force this moiety to relax from its initial cis-configuration through a series of conformational transitions to the equilibrium trans-conformation (step 3). This relaxation can be viewed as a minimal model for certain stages of peptide folding.
Without the ring restriction, no conformational relaxation within the peptide moiety can be induced by photoisomerization of the APB dye, such that time-resolved spectroscopy will monitor the chromophore properties unperturbed by peptide relaxation. Therefore, a linear APB-peptide, in which the Gly-Phe-linkage (see Fig. 1) is missing, was also investigated. Fortunately, the static optical properties of the linear and cyclic APB-peptides are nearly identical. This identity allows monitoring of the peptide-folding process in the cyclic molecule (step 3) by comparing the time-resolved spectra of the linear and cyclic peptides. We note in addition that the nπ*-transition of azobenzene is unaffected by integration into the peptide. Therefore, the photoisomerization of the APB-peptides can also be compared with that of free azobenzene in solution by direct matching of spectral signatures at wavelengths larger than 480 nm.
In our setup of ultrafast pump-probe spectroscopy, the cis/trans-photoisomerization of APB is triggered by a 60-fs pulse, and the reactions are probed by a delayed white-light pulse of similar duration (20, 23). According to the static spectra depicted in Fig. 1, the formation of the trans-APB ground state (in step 1) should lead to a strong increase of the ππ*-absorption around 370 nm. Whenever absorption at wavelengths longer than 500 nm is observed, it has to be attributed to intermediates, because the relaxed ground states of both isomers do not absorb in this low-energy spectral range. Such intermediates may represent either the electronically excited state of APB (S1→Sn absorption; step 1), the hot ground state of trans-APB (S*0→S1 absorption; step 2), or the distorted cyclic trans-APB-peptide, whose internal strains relax by conformational transitions (S →S1 absorption; step 3).
→S1 absorption; step 3).
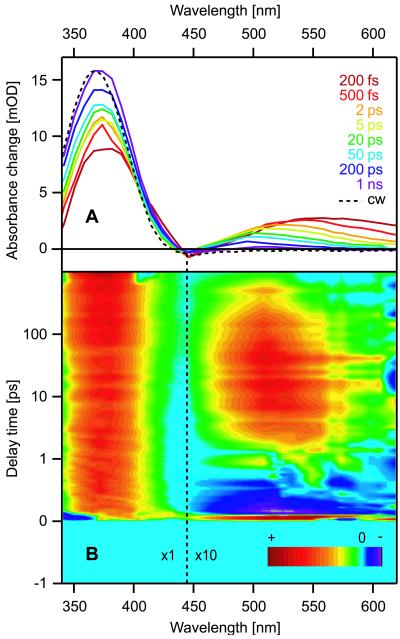
Fig. 3A reports the transient spectra of the cyclic APB-peptide obtained by photoexcitation of the cis-isomer; the spectra represent differences between time-delayed and initial absorption. With increasing time delays ranging from 200 fs to 1 ns, the transient spectra approach the static difference spectrum (dashed line) between the cyclic trans- and cis-APB-peptides. In particular, the strong ππ*-absorption of trans-APB near 370 nm builds up and undergoes a blue-shift with increasing time delay. After 1 ns, this process is not yet completed, which indicates ongoing slow relaxations toward the equilibrium conformation of the cyclic trans-APB-peptide. The incompleteness of relaxations is also apparent in the spectral region slightly above 500 nm, in which small signatures of intermediates still remain. To distinguish processes localized at the chromophore from those within the peptide moiety, this reaction dynamics will now be compared with that of the linear APB-peptide and that of free azobenzene in solution.
Figure 3.
(A) Transient spectra of cyclic APB-peptide showing the absorption change as a function of wavelength at certain delay times after the photoexcitation of the cis-isomer. The stationary difference spectrum (cis−trans) corresponds to the dashed line. (B) Difference in transient absorption changes between cyclic and linear APB-peptide as a function of wavelength and delay time. The color table and the scaling factor for the amplitudes (×10 for λpr > 445 nm) are indicated in the plot. The scale is linear for delay times between −1 and 1 ps and logarithmic for longer delay times.
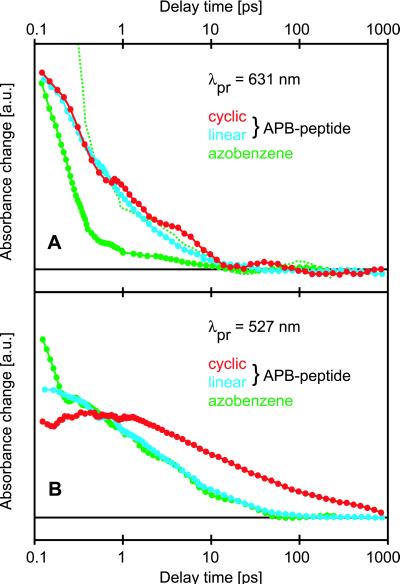
Fig. 2A exhibits transient absorption changes in the far-red spectral region at 631 nm probing the decay of the excited state and, thus, the duration of the photoisomerization (step 1). Here, the kinetics of the cyclic (red curve) and linear (blue curve) APB-peptides are almost identical. Both APB-kinetics can be modeled by a biexponential decay with time constants of about 300 fs and 3 ps (time constants derive from global fits to transients in the spectral range 340–640 nm). For free azobenzene (green curve), the photoisomerization is also biexponential (170 fs and 2 ps) (19), with a more pronounced fast component. Thus, the excited-state decay times of the chromophore are scarcely influenced by the attached peptide. Note that the 170-fs kinetics has been assigned to a ballistic motion funneling the molecule directly through a conical intersection from cis-S1 toward trans-S0, whereas the 2-ps channel has been ascribed (19) to a diffusive motion in S1 entailing a delayed internal conversion to S*0.
Figure 2.
Comparison of transient absorbance changes on cis/trans-isomerization between linear (blue), cyclic (red) APB-peptides, and azobenzene (green) at probing wavelengths in a spectral region devoid of any ground state absorption (A) and in the low-frequency side of the nπ*-transition (B), respectively. In A, the curve for azobenzene is plotted on two different scales to depict its overall appearance and the matching behavior at delay times past 1 ps. A logarithmic scale is applied to the delay time.
The transient absorption changes at 527 nm in Fig. 2B probe the nπ*-transition and, therefore, can also indicate relaxation processes in the trans ground state S*0 (steps 2 and 3). An additional 15-ps kinetics is apparent both for the linear APB-peptide (blue curve) and free azobenzene (green curve). For azobenzene, it has been observed and attributed (19) to the cooling of S*0 (step 2). In contrast, the cyclic compound (red curve) exhibits additional and much longer-lived intermediates. Because of their duration, they have to be assigned to the ongoing conformational relaxation of the peptide still exerting strains on the chromophore (step 3). As we have seen in Fig. 3A, that dynamics is not yet complete after 1 ns.
These results are corroborated by Fig. 3B, which summarizes the kinetic differences between the cyclic and linear APB-peptides over the whole spectral range. For its construction, the respective transient spectra have been subtracted. Negative values (dark blue) appearing in the first picosecond after excitation at wavelengths above 450 nm demonstrate that the ballistic photoisomerization is slightly faster (270 fs) in the cyclic than in the linear molecule (380 fs). Here, positive values at subsequent delay times mark the long-lived intermediate conformations of the cyclic peptide. Positive values are also seen near 370 nm. They extend over the whole time range and indicate that the quantum yield of cis/trans-photoisomerization is larger in the cyclic than in the linear peptide.
The ππ*-absorption bands of conjugated dyes like APB are known to show distinct spectral shifts on twists within the π-electron system and on solvation changes. In particular, the spectral location of the ππ*-band of trans-APB near 370 nm or, equivalently, the vertical excitation energy ΔE of the second excited singlet state S2, should sensitively respond to distortions of the diazene bridge connecting the benzene rings. Such distortions can derive from strains exerted on trans-APB by an incompletely relaxed peptide cycle and are expected to affect the energy of S0 more strongly than that of S2 (31). Therefore, in addition to the vibrational relaxation of the chromophore ground state S0, the temporal behavior of ΔE should reflect also its torsional relaxation. As noted above, corresponding band shifts are evident in Fig. 3A.
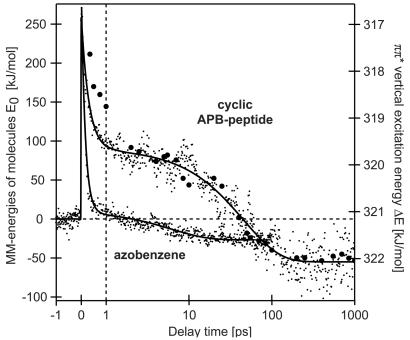
For a clear-cut presentation of this observable, Fig. 4 displays ΔE (large black dots) as a function of delay time. Apparently, the relaxation kinetics of ΔE reflects dynamical processes occurring in the cyclic APB-peptide on its way from the cis to the trans conformation. Fig. 4 compares the temporal changes of ΔE with those of the total energy E0 of the cyclic APB-peptide in solution extracted from MD simulations (small dots and fit curves). Because ΔE measures relative effects localized at APB, whereas E0 pertains to the whole APB-peptide, different energy scales apply. The computer experiments (see Materials and Methods) simulate the ballistic cis/trans-photoisomerization by switching on a model potential, which injects an energy of 270 kJ/mol into the cis-chromophore and mechanically drives the isomerization. In this way, the calculations account for the photon energy absorbed by the chromophore and for the S1→S*0 transition. Furthermore, the explicit treatment of the solvent allows to describe the dissipation of the injected energy through solvation changes as well as vibrational and conformational relaxation.
Figure 4.
Comparison of the total energies (small dots) of the cis-APB-peptide and of azobenzene during six 1-ns MD simulations on light-induced cis/trans-isomerization (see Materials and Methods). The elapsed time is shown on the x axis on a linear/logarithmic scale, with the origin depicting the electronic excitation. Fits to the temporal evolution of the energies with biexponential decays are represented by solid lines. They are compared with experimental data (large dots) in the form of vertical excitation energies ΔE of the trans ππ*-transition, derived from Gaussian fits to peaks of the ππ*-absorption near 370 nm in the transient spectra depicted in Fig. 3A.
Simulation results and observations are seen to closely match at delay times larger than about 3 ps. In particular, a 45-ps kinetics is revealed by the point of inflection in the fit-curve, reflecting identical time dependencies of peptide conformational relaxation in experiment and theory.
To enable a discussion of the physical processes covered by the simulations and to distinguish the effects confined to the chromophore from those caused by the peptide, we have also simulated the cis/trans-photoisomerization of azobenzene in solution and have included the corresponding results into Fig. 4. Here, a large fraction of the energy initially deposited at the dye is dissipated to the surrounding solvent within 150 fs, which is close to the 170-fs component observed for azobenzene in the same solvent, DMSO (by construction, the mechanical model potential driving isomerization cannot account for the 2-ps component of azobenzene; in the case of the APB-peptide, the corresponding shortcoming explains the deviation of the black dots from the theoretical fit curve in the time range between 300 fs and 3 ps). After 150 fs, a small fraction of the initial energy remains at trans-azobenzene in the form of vibrations and solvation energy. Subsequently, with a time constant of 6 ps, thermal equilibrium is reached by vibrational relaxation and solvation. During this process, the energy becomes negative, because the cis/trans-isomerization removes an intramolecular steric hindrance, which is characteristic for the cis-isomer and places it energetically above the trans-isomer. As to be expected from the overestimate of specific heats, generic to classical mechanics descriptions of nuclear motion in condensed matter, the 6-ps relaxation time underestimates the 15-ps kinetics experimentally observed for cooling (19).
In the case of the cyclic APB-peptide, only two-thirds of the initial energy is dissipated within 300 fs of APB to the solvent (spectroscopy, 230 fs), whereas the remainder is used for building up a strain within the cycle. The trajectories (data not shown) reveal a ballistic change of the APB chromophore to a trans-like geometry on that fast time scale. Isomerization of the chromophore is followed by slow (45-ps) strain-driven relaxations, on which about 150 kJ/mol of energy (see Fig. 4) is dissipated from the cyclic APB-peptide into the solvent. Unfortunately, the conformational changes related to these relaxations cannot be readily extracted from the MD trajectories. Such changes involve concerted motions of many degrees of freedom. Two of us (H.C. and P.T., unpublished work) are currently developing tools that will serve to identify these collective coordinates in the space of dihedral angles. Preliminary results indicate that after 1 ns, the relaxation of the peptide backbone toward its trans-conformation is about halfway complete. This agrees with Fig. 3A, which has demonstrated that the static spectrum is not yet reached after 1 ns.
Conclusion
We have reported a real-time observation of elementary events in peptide conformational dynamics on its intrinsic subnanosecond time scale by ultrafast spectroscopy on cyclic model compounds with built-in light switches. The remarkably good agreement between spectroscopic and computer experiments leads us to assume that the virtual reality of atomic motions generated by our simulations faithfully models important molecular processes monitored by spectroscopy. In particular, the simulations allow association of ballistic cis/trans-isomerization motions, solvation and vibrational cooling processes, as well as conformational peptide relaxations to the observations. They show that substantial conformational transitions proceed on the quite rapid time scale of 50 ps in the cyclic peptide. In general, our integrated approach paves the way for studying the effects of chemical composition on peptide conformational dynamics and improving the quality of MD techniques.
Acknowledgments
We are grateful to D. Oesterhelt for stimulating discussions. This study was supported by the Sonderforschungsbereich 533 of the Deutsche Forschungsgemeinschaft.
Abbreviations
- APB
(4-aminophenyl)azobenzoic acid
- MD
molecular dynamics
References
- 1.Frauenfelder H, Sligar S G, Wolynes P G. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 2.Grubmüller H, Tavan P. J Chem Phys. 1994;101:5047–5057. [Google Scholar]
- 3.Dill K A, Chan H S. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 4.Guo Z Y, Brooks C L, Boczko E M. Proc Natl Acad Sci USA. 1997;94:10161–10166. doi: 10.1073/pnas.94.19.10161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duan Y, Kollman P A. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 6.Daura X, Jaun B, Seebach D, van Gunsteren W F, Mark A E. J Mol Biol. 1998;280:925–932. doi: 10.1006/jmbi.1998.1885. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y Q, Karplus M. Nature (London) 1999;401:400–403. doi: 10.1038/43937. [DOI] [PubMed] [Google Scholar]
- 8.Williams S, Causgrove T P, Gilmanshin R, Fang K S, Callender R H, Woodruff W H, Dyer R B. Biochemistry. 1996;35:691–697. doi: 10.1021/bi952217p. [DOI] [PubMed] [Google Scholar]
- 9.Thompson P A, Eaton W A, Hofrichter J. Biochemistry. 1997;36:9200–9210. doi: 10.1021/bi9704764. [DOI] [PubMed] [Google Scholar]
- 10.Lednev I K, Karnoup A S, Sparrow M C, Asher S A. J Am Chem Soc. 1999;121:8074–8086. doi: 10.1021/ja003381p. [DOI] [PubMed] [Google Scholar]
- 11.Bieri O, Wirz J, Hellrung B, Schutkowski M, Drewello M, Kiefhaber T. Proc Natl Acad Sci USA. 1999;96:9597–9601. doi: 10.1073/pnas.96.17.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren Z, Perman B, Srajer V, Teng T Y, Pradervand C, Bourgeois D, Schotte F, Ursby T, Kort R, Wulff M, Moffat K. Biochemistry. 2001;40:13788–13801. doi: 10.1021/bi0107142. [DOI] [PubMed] [Google Scholar]
- 13.Woutersen S, Mu Y G, Stock G, Hamm P. Proc Natl Acad Sci USA. 2001;98:11254–11258. doi: 10.1073/pnas.201169498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong D P, Zewail A H. Proc Natl Acad Sci USA. 2001;98:11867–11872. doi: 10.1073/pnas.211440398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dobler J, Zinth W, Kaiser W, Oesterhelt D. Chem Phys Lett. 1988;144:215–220. [Google Scholar]
- 16.Holzapfel W, Finkele U, Kaiser W, Oesterhelt D, Scheer H, Stilz H U, Zinth W. Proc Natl Acad Sci USA. 1990;87:5168–5172. doi: 10.1073/pnas.87.13.5168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrendt R, Renner C, Schenk M, Wang F Q, Wachtveitl J, Oesterhelt D, Moroder L. Angew Chem Int Ed. 1999;38:2771–2774. doi: 10.1002/(sici)1521-3773(19990917)38:18<2771::aid-anie2771>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 18.Behrendt R, Schenk M, Musiol H J, Moroder L. J Pept Sci. 1999;5:519–529. doi: 10.1002/(SICI)1099-1387(199911)5:11<519::AID-PSC223>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 19.Nägele T, Hoche R, Zinth W, Wachtveitl J. Chem Phys Lett. 1997;272:489–495. [Google Scholar]
- 20.Renner C, Behrendt R, Spörlein S, Wachtveitl J, Moroder L. Biopolymers. 2000;54:489–500. doi: 10.1002/1097-0282(200012)54:7<489::AID-BIP20>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 21.Seel M, Wildermuth E, Zinth W. Meas Sci Technol. 1997;8:449–452. [Google Scholar]
- 22.Wilhelm T, Piel J, Riedle E. Opt Lett. 1997;22:1494–1496. doi: 10.1364/ol.22.001494. [DOI] [PubMed] [Google Scholar]
- 23.Huber R, Satzger H, Zinth W, Wachtveitl J. Opt Commun. 2001;194:443–448. [Google Scholar]
- 24.Brooks B R, Bruccoleri R E, Olafson B D, States D J, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 25.Eichinger M, Grubmüller H, Heller H, Tavan P. J Comput Chem. 1997;18:1729–1749. [Google Scholar]
- 26.Niedermeier C, Tavan P. J Chem Phys. 1994;101:734–748. [Google Scholar]
- 27.Tironi I G, Sperb R, Smith P E, van Gunsteren W F. J Chem Phys. 1995;102:5451–5459. [Google Scholar]
- 28.Frisch M J, Trucks G W, Schlegel H B, Scuseria G E, Robb M A, Cheeseman J R, Zakrzewski V G, Montgomery J A, Jr, Stratmann R E, Burant J C, et al. GAUSSIAN98. Pittsburgh: Gaussian; 1998. [Google Scholar]
- 29.Eichinger M, Tavan P, Hutter J, Parrinello M. J Chem Phys. 1999;110:10452–10467. [Google Scholar]
- 30.Ryckaert J-P, Ciccotti G, Berendsen H J C. J Comp Phys. 1977;23:327–330. [Google Scholar]
- 31.Monti S, Orlandi G, Palmieri P. Chem Phys. 1982;71:87–99. [Google Scholar]