Abstract
The envelope glycoprotein (Env) of HIV-1 is incorporated into virions that bud from the cell surface of infected T cells. With immunofluorescence microscopy and subcellular membrane fractionation techniques, the intracellular fate of Env in the secretory pathway of HIV-1-infected T cells was evaluated. Rather than trafficking constitutively from the Golgi to the cell surface, Env is directed to intracellular CTLA-4-containing granules, whose recruitment to the cell surface is regulated. The use of the regulated pathway for intracellular Env storage before virion assembly holds implications for the staging of Env exposure at the cell surface of infected cells and of coordinating HIV virion assembly.
The envelope glycoprotein (Env) of the HIV type 1 (HIV-1) is crucial to viral infectivity for binding to the CD4 and chemokine receptors present on T cells and for driving membrane fusion (1–4). The HIV-1 Env gene product consists of a complex of two subunits, gp120 and gp41. They are synthesized in the endoplasmic reticulum (ER) as the gp160 precursor protein, which is folded into trimers before exit from the ER (5, 6). The gp160 trimers are then transported to the Golgi apparatus where further oligosaccharide modifications take place (7). During its transport through the secretory pathway of the host T cell, the precursor is proteolytically cleaved by the PC6 protease of the subtilisin-like pro-protein convertase family, to yield the mature gp120 and gp41 (refs. 8–13 and Z. Hu, G. Pott, L.R.M., Q. Wang, Y. Lu, J. Schaack, X. Zhang, H. J. Choi, R. T. Schooley, J. W. M. Creemers, J. van de Loo, N. Seidah, K. Nakayama, and A.F., unpublished data). The cleaved Env is assembled together with other viral components for virion budding from the cell surface (14–17).
Several regulatory steps in the intracellular itinerary of HIV-1 Env remain to be resolved, such as whether Env traffics to the surface of T cells via the constitutive or regulated secretory pathway. The regulated branch of the post-Golgi secretory pathway of T cells contains specialized membrane compartments known collectively as regulated secretory granules, secretory lysosomes, or lytic granules (18). One function of this compartment is to direct the delivery of molecules used for killing tumors or virally infected cells. Stored molecules include perforin and granzymes, Fas ligand, and CTLA-4 (19–21). CTLA-4 (CD152) is an important T cell regulatory protein that acts as a negative regulator of the immune response (for review see refs. 22–24). Cell surface expression of CTLA-4 is tightly controlled. Before T cell activation, CTLA-4 traffics through the secretory pathway to the cell surface then is rapidly internalized by endocytosis, and delivered to the intracellular regulated secretory granules (25–27). Interestingly, the transport of Env to the cell surface seems to be tightly regulated as well. Although the HIV buds from the cell surface (28, 29), most of the mature Env is found or “stored” in an unidentified intracellular compartment (ref. 30 and A.F., unpublished data).
Understanding the proper itinerary for Env would potentially reveal new information about the regulation of its cell surface expression and the coordination of events for virion budding. In this report, we have discovered that the Env traffics directly to the intracellular CTLA-4-containing granules. These results suggest the timing and delivery of Env to the surface of HIV-infected T cells may be controlled through utilization of the regulated secretory pathway.
Materials and Methods
Cell Lines.
The human T cell line H9 was obtained from the American Type Culture Collection. Reagents were obtained from Sigma unless otherwise indicated. Fresh blood from healthy adult donors was used to isolate CD4+ cells from peripheral blood mononuclear cells with CD4 MicroBeads (Miltenyi Biotec, Auburn, CA), as described by the manufacturer. H9 cells were grown in DMEM containing 10% FBS (Gemini Biological Products, Woodland, CA), 10 μg/ml of gentamicin (GIBCO/BRL), and 41.4 μg/ml of 2-mercaptoethanol. Recombinant human IL-2 (10 units/ml; Roche Molecular Biochemicals) and 3 μg/ml of phytohemagglutinin were added to the human CD4+ cells 3 days before HIV-1 infection. This treatment was also used to increase the levels of expression of the endogenous CTLA-4 (31).
Generation of H9 Cells Stably Expressing CTLA-4.
Full-length mouse CTLA-4 gene fused directly to green fluorescent protein (GFP) at the C terminus under the control of the human ubiquitin promoter was engineered in plasmid pUp. H9 cells (8 × 106) in serum-free DMEM were mixed with 30 μg of DNA in a 4-mm gap cuvette and electroporated in a BTX electroporator (Genetronics, San Diego) that was set for 500 V capacitance/resistance mode, 1,050 μF capacitance, 720 ohms resistance, and 260 V charging voltage. Cells were incubated 48 h in DMEM after which Geneticin (GIBCO/BRL) was added to 800 μg/ml. H9 cells stably expressing CTLA-4-GFP were obtained from three rounds of fluorescence activated cell sorting.
Viruses and Abs.
The D47 (gp120) and D61 (gp41) hybridomas (32, 33), as well as the recombinant vaccinia viruses vPE16 (34), expressing the gp160 precursor were kindly provided by P. Earl (National Institutes of Health). Ascites for these Abs were produced in BALB/c mice as described (35). The 902 mAb against gp120 was obtained from the National Institutes of Health AIDS Research and Reference Program. HIV-1 strain IIIB was purchased from Advanced Biotechnologies (Columbia, MD). A mAb against GFP (CLONTECH), a polyclonal Ab against MG160 (from K. Howell, Univ. of Colorado Health Sciences Center), a mAb against human CTLA-4 (J. Bluestone, Univ. of Illinois, Chicago), polyclonal Abs against human CD4 receptor, Fgr kinase, calreticulin, and the extracellular domain of mouse CTLA-4 (Santa Cruz Biotechnology) were used in these studies. Phycoerythrin-conjugated anti-mouse CTLA-4 Ab was purchased from BD Biosciences–PharMingen. Fluorescent-conjugated secondary Abs were obtained from Jackson ImmunoResearch.
Virus Infections.
For infection with the recombinant vaccinia virus vPE16, H9 cells were infected at a multiplicity of infection (moi) of 30 plaque-forming units per cell for 3 h (37°C, 5% CO2), after which cells were washed and incubated with fresh medium. At 14 h after infection, cells were processed for immunological analysis. In experiments involving HIV-1, CD4+ cells isolated from peripheral blood mononuclear cells, as well as H9 cells, were infected at an moi of 0.5 tissue culture 50% infective dose (TCID50) per cell for 3 h. Cells were then extensively washed with fresh medium and incubated for an additional 72 h (37°C, 5% CO2). Cells were pelleted, washed, and processed for further experiments.
Imunocytochemistry, Cell Imaging Analysis, and Flow Cytometry.
For immunofluorescence, 5 × 104 HIV-infected T cells were bound to washed, poly-d-lysine-coated cover slips (3 mg/ml), fixed for 30 min with 3% paraformaldehyde/3% sucrose (Fisher Scientific), and quenched 10 min in 50 mM NH4Cl (Fisher Scientific). Cells were permeabilized 4 min with 0.1% Triton X-100 in PBS and blocked 5 min with 0.2% gelatin in PBS. Cells were incubated with Abs and then observed with a Zeiss Axiovert M100 epifluorescence microscope. Visual data were acquired with a Cooke Corporation (Auburn Hills, MI) SensiCam charge-coupled device camera and were digitally deconvolved with a nearest-neighbors algorithm with slidebook software (Intelligent Imaging, Denver). In addition, colocalization analysis was performed with the masking and statistics capabilities of slidebook. Initially, stacks of images were acquired in 0.2-μm steps throughout the cell volume. Stacks were deconvolved with a constrained iterative algorithm.
For the Ab uptake experiments, 3 × 104 cells were incubated 45 min on ice with 1 μg of D47 or mouse CTLA-4 (extracellular domain) Ab. After washing 5 times with PBS, cells were resuspended in DMEM and incubated for 45 min at 37°C, 5% CO2 to allow Ab internalization. After fixation, cells were incubated with the corresponding secondary Abs conjugated to biotin, followed by streptavidin-Cy3.
For flow cytometry analysis of CTLA-4 surface expression, HIV-1-infected H9-CTLA-4-GFP cells were washed 3 times in ice-cold PBS and incubated with saturating concentration of the phycoerythrin-conjugated CTLA-4 Ab for 2 h on ice. Cells were fixed, then washed 5 times with ice-cold PBS and analyzed in a Becton Dickinson FACScan.
Metabolic Labeling, Immunoprecipitations, and Western Blots.
For labeling of Env expressed by recombinant vaccinia viruses, at 14 h after infection, 3 × 106 cells were washed and then starved 30 min in methionine-free DMEM with 5% dialyzed FBS. Cells were pulsed 40 min in the presence of 100 μCi of [35S]methionine (ICN) and chased for different time points. To immunoprecipitate Env, gradient fractions of lysed cells were incubated 10 min in lysis buffer [PBS containing 1% TX-100, 0.4% deoxycholic acid, 100 mM PMSF, and 1× Complete protease inhibitor mixture (Roche Molecular Biochemicals)], after which the lysate was clarified by centrifugation at 14,000 × g for 10 min at 4°C. Typically, 1 μl of ascites fluid or 1 μg of polyclonal Ab was used per immunoprecipitation. Incubations for at least 3 h at 4°C were followed by addition of protein-A Sepharose for at least 2 h. Complexes were washed 4 times with lysis buffer and eluted from beads by SDS-loading buffer. Proteins were separated by SDS/PAGE (8%) and visualized by autoradiography.
For Western blot analyses, infected cells were washed with PBS and the pellet treated with 100 μl of lysis buffer. The lysate was clarified by centrifugation (14,000 × g), and SDS-loading buffer was added to 100 μg of the clarified supernatant. Total proteins from the gradient fractions were precipitated by methanol/chloroform (4:1) extraction, and the pellet washed with methanol. Proteins were then subjected to SDS/PAGE (8%) and transferred (16 h) onto Immobilon membranes (Millipore), which were incubated with the primary Ab, followed by peroxidase-conjugated secondary Ab, and revealed by enhanced chemiluminescence (NEN).
Density Gradient Fractionation.
Separation of granules from other membranes was performed as described (36) with a few modifications. After infection of H9 cells with vaccinia virus vPE16 for 14 h, cells were radiolabeled as indicated previously, rinsed with PBS, and harvested in 0.25 M sucrose/5 mM Tris⋅HCl (pH 7.3)/1 mM EGTA/2 mM PMSF/1X protease inhibitor mixture. Cells were homogenized by multiple passage through a 22.5-gauge needle on an insulin syringe. After 60–80% cell lysis (as judged by phase-contrast microscopy), the homogenates were sedimented twice at 1,000 × g for 10 min to pellet nuclei and unbroken cells. The resulting postnuclear supernatant was layered onto linear 0.3–1.6 M sucrose gradient and sedimented to equilibrium at 154,325 × g (RCFmax) for 16 h in an SW41 rotor at 4°C. Fractions were collected from the top. Membrane fractionations were analyzed by immunoblots of the fractions by using Abs for calreticulin (ER marker protein), MG160 (Golgi marker protein), CD4 (cell surface/endosome marker protein), and CTLA-4-GFP (regulated granule marker protein). Immunoprecipitation of proteins from the sucrose fractions was achieved by adding 10X lysis buffer before the addition of Abs and protein A-Sepharose.
Results
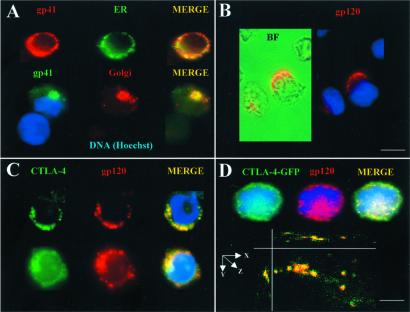
Indirect immunofluorescence was performed to detect the intracellular compartments where Env was targeted in the T cell secretory pathway. Representative images for Env localization in HIV-1-infected CD4+ cells isolated from peripheral blood mononuclear cells are shown in Fig. 1. The expected appearance of Env in ER and Golgi organelles was observed as reported (30, 32, 37). In the present study, ∼40% cells showed a pattern of Env localization in the ER and ∼25% showed labeling associated with the Golgi (Fig. 1A). About 25% of Env-expressing cells revealed cell surface labeling (Figs. 1B and 2B). A significant fraction of HIV-1-infected cells (10%) showed an intracellular punctate pattern of Env localization. Double-labeling with the granular marker CTLA-4 identified the punctate staining as intracellular regulated secretory granules (Fig. 1C). The granule localization of Env was also observed with Abs to gp41 (data not shown), as well as with either human or mouse T cells infected with Env-expressing vaccinia virus vPE16 (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). The localization of Env in the regulated secretory granules was confirmed with HIV-infected human H9 T cells that stably express a CTLA-4-GFP fusion protein (Fig. 1D). The colocalization of CTLA-4-GFP and Env in the same intracellular granules was verified by three-dimensional analysis of multiple focal planes with digital deconvolution fluorescence microscopy (Fig. 1D). The results from immunofluorescence microscopy suggested that Env normally traffics to CTLA-4-containing regulated secretory granules as part of its intracellular trafficking itinerary after HIV-1 infection of human T cells.
Figure 1.
Env targeting to the CTLA-4-containing granules is part of the HIV-1 life cycle. The inventory of Env localization in representative cells that had been infected with HIV-1 is shown. (A) HIV-1-infected CD4+ cells were double-stained for immunofluorescence with an anti-gp41 Ab (red) and calreticulin (ER marker protein, green), or anti-gp41 (green) and anti-Fgr kinase (Golgi marker protein, red). Colocalization of the two markers in the cell is revealed by yellow. (B) Staining with an anti-gp120 Ab (red) shows surface staining on some cells. The same field is shown by bright field microscopy (BF) to reveal the contours of the cell. (C) Double staining for immunofluorescence with anti-gp120 Ab (red) and anti-CTLA-4 Ab (green). The punctate staining shows the localization of the regulated secretory granules. Colocalization of the two markers in the cell is revealed by yellow. The staining pattern in two separate cells is shown (Upper and Lower). (D) HIV-1-infected H9-CTLA-4-GFP cells labeled with anti-gp120 Ab (red). Colocalization of the two markers in the cell is revealed by yellow. The staining pattern in two separate cells is shown (Upper and Lower). The bottom image shows colocalization of both molecules within the same compartment in a three-dimensional view of the granules. The frequency of these patterns was assessed by examining approximately 90 cells for the staining of each compartment. [Bars = 10 (A–D Upper) and 3 μm (D Lower).]
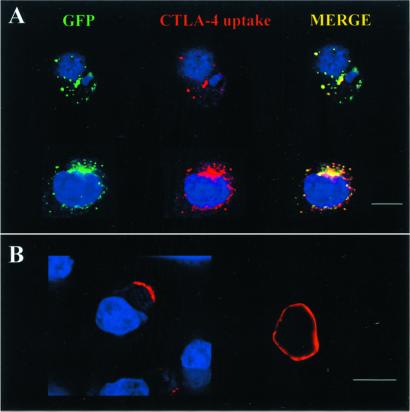
Figure 2.
CTLA-4, but not Env, is endocytosed from the surface of HIV-infected T cells. (A) An Ab against the CTLA-4 luminal domain was incubated with H9-CTLA-4-GFP cells in culture before fixation, permeabilization, and staining with a Cy3-conjugated second Ab (red). The anti-CTLA-4 Ab was internalized to the same granules as those containing CTLA-4-GFP at steady state (GFP, green). (B) Three days after infection by HIV-1 (multiplicity of infection, 0.5), H9-CTLA-4-GFP cells were incubated on ice with anti-gp120 Ab. Cells were then washed and gp120 Ab internalization was allowed at 37°C before fixation, permeabilization, and staining with a secondary Cy3-conjugated Ab (red). Although gp120 was detected at the cell surface in some of the HIV-infected cells, endocytosis of the anti-gp120 was not detected in any of the cells. [Bars = 10 μm.]
HIV-1 Env Traffics Directly from Golgi to CTLA-4-Containing Granules.
The steady-state localization of CTLA-4 in the intracellular granules is achieved by rapid endocytosis from the cell surface (27). Ab uptake experiments were performed to determine whether Env was similarly directed to the intracellular granules by endocytosis from the cell surface. As reported (27), we confirmed that Abs against the extracellular domain of CTLA-4 were internalized to the same intracellular granules as those harboring CTLA-4 at steady state in the H9-CTLA-4-GFP cell line (Fig. 2A). In contrast, no endocytic uptake of Abs to the extracellular domain of Env was detected in HIV-infected H9 cells, even though these Abs label Env protein at the surface of some cells (Fig. 2B). These experiments suggest that, unlike CTLA-4, Env localization to the intracellular granules does not use the pathway of endocytosis from the cell surface.
In studying Env trafficking, the exit of Env from the ER is slow, most likely a result of complications in protein folding, trimerization, and a result of the potential for binding CD4 proteins in the ER of human T cells (6, 38). A significant fraction of nascently synthesized Env proteins is degraded by ER quality control mechanisms. Because Env protein “trickles” out of the ER, post-ER trafficking of Env can be effectively monitored. To dissect the intracellular trafficking itinerary of Env, pulse-chase radiolabeled H9 T cells were analyzed by membrane fractionation in sucrose density gradients. This approach resolved the key organelles of the secretory pathway involved in Env trafficking, i.e., ER, Golgi, plasma membrane, and the CTLA-4-containing granules (Fig. 3D).
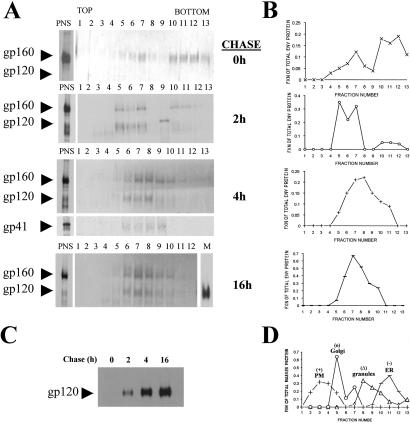
Figure 3.
HIV-1 Env traffics directly from Golgi to CTLA-4-containing granules. Lysates of radiolabeled H9-CTLA-4-GFP cells infected with vaccinia virus vPE16 (encoding HIV-1 Env) were resolved by density sucrose gradient centrifugation. (A) The images represent a 30-min radiolabeled pulse followed by 0-, 2-, 4-, or 16-h chase periods. An aliquot from the total postnuclear supernatant before resolution on the gradient is shown on the left side and immunoprecipitated for gp120 Env. Each fraction from the gradients was analyzed for the presence of radiolabeled gp120 Env by immunoprecipitation. In addition, the distribution of gp41 in the gradients at the 4-h chase is shown. (B) The autoradiographs from A were digitized and quantified with NIH IMAGE software. The fraction of total Env protein (gp120 + gp160) loaded onto the gradient and recovered from each gradient fraction is shown. (C) Immunoprecipitates of gp120 protein shed into the media from the pulse-chase experiments are shown on the autoradiograph. Equal volumes of culture supernatant were used from each time point. (D) The resolution of marker proteins of specific organelles on the gradient was analyzed by immunoblots. The resolved fractions were blotted for the marker proteins calreticulin (ER), MG160 (Golgi), CTLA-4 (regulated secretory granules), and CD4 (plasma membrane and endosomes). For each fraction, the amount of the marker is expressed as a percentage of total immunoreactivity across the gradient.
At different chase times, the organelles from lysates of H9-CTLA-4-GFP cells infected with vaccinia virus vPE16 were resolved by density gradient fractionation. At the start of the chase experiment (after the 30-min pulse), there was no detectable cleavage of HIV-gp160, and most of Env was found in dense fractions 10–12 (Fig. 3 A and B), corresponding to ER membranes (Fig. 3D). A small proportion of HIV-gp160 was also found in lighter gradient fractions correlating with the Golgi marker protein (Fig. 3 A, B, and D). After a 2-h chase period, the proportion of HIV-gp160 in the Golgi fractions increased, whereas the fraction of Env in the ER diminished (Fig. 3 A, B, and D). The Env cleavage products were evident in fractions 5–7 (Fig. 3 A and B), corresponding to Golgi compartments (Fig. 3D). The appearance of gp120 shed into the medium starting at the 2-h chase time point is consistent with partial Env distribution in post-Golgi compartments (Fig. 3C).
A shift in intracellular Env localization began to be apparent by the 4-h chase. The peak of HIV-gp160 and the mature gp120-gp41 products were recovered in fractions 6–10 (Fig. 3 A and B), which overlaps with the distribution of CTLA-4-containing granules (Fig. 3D). At even longer chase times, i.e., the 16-h chase, most of Env remained in fractions 6–10. Interestingly, Env was not detected in more buoyant membrane fractions containing plasma membrane marker proteins (Fig. 3D), even though a fraction of the total gp120 pool was shed into the media (Fig. 3A, 16-h chase, lane M). Immunoprecipitation analysis of gp120 from the media started as early as 2 h after the chase yet was greatly elevated by 4–16 h (Fig. 3C). The release of gp120 into the medium would imply that gp41 would be present to a significant degree at the cell surface. Nevertheless, Env fractionation experiments performed with a gp41 Ab (Fig. 3A, 4-h chase, Lower) failed to detect Env in cell surface fractions yet they show overlap with gp120-containing membranes. This result suggests that the granule compartment constitutes the major post-Golgi fraction of Env protein in the cell. At longer chase times, gp120 continued to be shed even though most of the intracellular Env pool was localized to CTLA-4-containing granules. Taken together, these results showed that Env transited the secretory pathway from ER to Golgi (as expected) yet was directly delivered to and retained in CTLA-4-containing granules rather than traffic to the cell surface. Shedding of gp120 into the medium seems to represent the outcome of Env trafficking in a minor population of cells, whereas trafficking to and residence in the regulated secretory granules is the principal itinerary of Env before virion assembly.
Cell Surface Recruitment of the CTLA-4 Granule During HIV Infection.
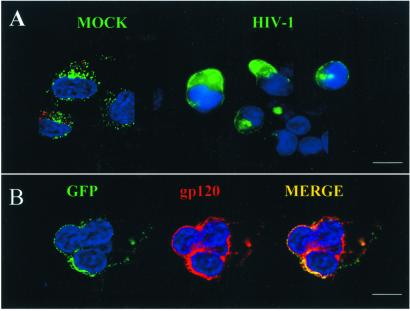
HIV-1 infection of H9-CTLA-4-GFP cells triggered a visible alteration in the immunofluorescent pattern for CTLA-4 and for Env. In place of the typical punctate staining of the granules, the majority of cells incubated with HIV now revealed CTLA-4 and Env localization at the cell surface (Fig. 4 A and B). However, no change in surface CTLA-4 expression was detected in cells that solely expressed Env by vaccinia virus (Fig. 5). The fluorescent intensity of CTLA-4 and the number of cells expressing CTLA-4 at the cell surface significantly increased after HIV infection (Fig. 6, which is published as supporting information on the PNAS web site). The concomitant expression of CTLA-4 and gp120 at the cell surface strongly suggested that the CTLA-4-containing granules were recruited to the cell surface after HIV-1 infection.
Figure 4.
HIV-1 infection drives cell surface expression of CTLA-4. The impact of HIV infection on expression of CTLA-4 at the cell surface of H9-CTLA-4-GFP cells is shown. (A) GFP fluorescence reveals the localization of CTLA-4 protein in mock (Left) and HIV-infected cells (Right). (B) Double staining by immunofluorescence of CTLA-4 (green) and Env (red) shows concomitant expression at the cell surface (yellow). [Bars =1 0 μm.]
Discussion
The intracellular trafficking of HIV-1 Env in CD4+ cells is crucial to the assembly and budding of infectious virus. Although HIV buds from the surface of infected cells, here we show that Env is not constitutively delivered to the cell surface as has been previously assumed. Instead, Env traffics from the Golgi to intracellular granules of the regulated secretory pathway of human T cells. These intracellular granules contain the immunomodulatory protein, CTLA-4, which is normally recruited to the cell surface after T cell activation (25, 26). Delivery of Env to the cell surface of HIV-infected cells is accompanied by CTLA-4 surface expression, as would be expected if both molecules reside in the same intracellular granule. The results from this study suggest that control of Env trafficking to the cell surface may provide an additional regulatory step in HIV virion assembly.
The observation that Env does not constitutively reside at the cell surface of HIV-infected T cells has been described (30). Furthermore, our studies do not contradict previous reports on HIV-1 Env trafficking yet provide added definition of the intracellular compartments where Env is found during HIV-1 infection. This added definition is possible because most studies of intracellular Env trafficking have been performed in cell types, such as fibroblasts, which are not normal targets for HIV infection (6, 37, 39). The organization of the secretory pathway branches in T cells and macrophages contributes to the distinctive itinerary of Env trafficking in those cells. Hence, defining the intermediates in the trafficking pattern of Env in HIV-infected T cells is crucial to understanding the regulation of the assembly and budding of nascent HIV virions.
Rather than being delivered directly from the Golgi to the cell surface, the Env protein is diverted into granules of the regulated branch of the secretory pathway (Fig. 3). This interpretation is supported by observations that gp160 cleavage does not depend on the furin protease, which functions in the constitutive branch of the secretory pathway (40, 41). Rather, Env is cleaved by the PC6 protease that inhabits the regulated secretory pathway (Z. Hu, G. Pott, L.R.M., Q. Wang, Y. Lu, J. Schaack, X. Zhang, H. J. Choi, R. T. Schooley, J. W. M. Creemers, J. van de Loo, N. Seidah, K. Nakayama, and A.F., unpublished data). Therefore, Env is sorted into the regulated pathway, where its delivery to the cell surface would depend on specific signals, which influences the timing of viral assembly and budding.
The results of this study indicate that Env transits from the ER to the Golgi and then transits directly to CTLA-4-containing granules, without first trafficking to the cell surface. This conclusion is based on the failure to uptake gp120 Ab from the cell surface (Fig. 2), and the absence of Env in membrane fractions containing endosomes and plasma membrane (Fig. 3). In this respect, Env trafficking to the CTLA-4-containing granules resembles that of Fas ligand, which is transported directly to these granules without transiting to the cell surface (42). The trafficking itineraries of Env and Fas ligand are therefore distinct from that of CTLA-4, which is endocytosed from the cell surface to the intracellular granules (21, 25–27, 42, 43). Sorting of Env from the Golgi to the CTLA-4-containing granules seems to be an inherent property of the protein, because identical results were observed whether Env was expressed by recombinant vaccinia virus or by HIV-1. Hence, more than one pathway exists for the delivery of molecules to the intracellular CTLA-4-containing granules.
What events during HIV-1 infection trigger Env delivery to the cell surface? Cell surface expression of CTLA-4 increases during HIV protein production (ref. 31 and supporting information), suggesting that CTLA-4-containing granules are apparently signaled to translocate to the plasma membrane at some late stage of HIV virus assembly. Because of their shared residence in granules, CTLA-4 and Env are recruited concomitantly to the cell surface (Fig. 4). The trigger that drives CTLA-4-containing granules to translocate to the cell surface is not provided solely by the Env protein, because increased CTLA-4 expression at the cell surface is not observed when Env is the only HIV protein expressed by recombinant vaccinia virus (see supporting information). Nevertheless, T cell activation seems to be sufficient to induce cell surface expression of CTLA-4 (26, 33) and gp120 (see supporting information). Furthermore, when Env is finally delivered to the cell surface, its distribution is clustered in focused regions on the plasma membrane (Fig. 2), consistent with reported observations on the intracellular localization of the matrix subunit of HIV-1 Gag protein (44). Because viral assembly depends on the recruitment of HIV-1 Gag protein, the binding of Gag protein to Env on the cytosolic surface of these intracellular CTLA-4-containing granules is predicted to initiate the translocation of the granules to the cell surface (45).
We hypothesize that intracellular Env storage in regulated secretory granules provides two benefits in support of HIV proliferation. First, a prolonged residence of Env at the cell surface would prematurely alert the immune system before productive virions would be assembled and released for proliferation. HIV-1 has apparently developed multiple mechanisms for escaping detection by the immune system. One example is the Nef-mediated down-regulation of MHC-I proteins to prevent viral antigen presentation and cytotoxic T cell-mediated destruction of HIV-infected cells (46–48). It would be similarly advantageous to reduce the probability of Ab-mediated detection of HIV-infected cells by limiting exposure time of Env at the cell surface until productive viral progeny can be rapidly assembled.
The second predicted benefit for Env retention in the regulated granules relates to the role of CTLA-4 in attenuating activation signals of the infected T cells (22, 24). Because HIV proliferation depends on T cell activation, the cellular machinery coopted to produce HIV virions may depend on maintaining cells in an activated state. Therefore, it may be important to prevent CTLA-4 from attenuating T cell activation during HIV protein production. The timing of CTLA-4 translocation to the cell surface may be delayed by events related directly or indirectly to the presence of Env in the intracellular granules. However, because other regulatory molecules of the immune system may be present in the CTLA-4-containing granules, further studies must be conducted to determine which molecules are involved in controlling HIV proliferation and/or evasion of immune surveillance. The results provided by this study therefore suggest new avenues for research into the potential regulation of Env trafficking to the cell surface and of HIV virion assembly.
Supplementary Material
Acknowledgments
We are grateful to Hannah Kupfer, Dr. Ben Freiberg, Dr. Alex Sorkin, and Dr. Ani Banerjee for their assistance with digital deconvolution fluorescence microscopy; to Drs. Patricia Earl and Bernard Moss (National Institutes of Health) for monoclonal Env Abs; to Drs. Tom Campbell, Elizabeth Connick, and Dan Kuritzkes of the Infectious Diseases Division at the University of Colorado Health Sciences Center for use of the BL3 facility; to members of the Franzusoff lab for their contributions to this project; to Dr. Elaine del Nery Santos for her work on cell lines used in this project; and to Dr. Sandra Meech [Univ. of Colorado Health Sciences Center (UCHSC)] for providing the H9 T cell line, Dr. Peter Angel (DKFZ, Heidelberg, Germany) for the Ubiquitin C promoter fragment, and to David Shugarts (UCHSC) for supplying human peripheral blood mononuclear cells for this study. We thank Dr. Greg Pott for critical reading of the manuscript. This research was supported by National Institute of Allergy and Infectious Diseases/National Institutes of Health Grant AI-34747 (to A.F.), National Institutes of Health Grant AI-23764 (to A.K.), and by a Special Fellowship from the Leukemia and Lymphoma Society (to B.C.S.).
Abbreviations
- Env
envelope glycoprotein
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
Note Added in Proof.
While this manuscript was under review, Leng et al. (49) examined the percentage of CD4+ CTLA4+ cells from peripheral blood mononuclear cells (PBMCs) of HIV-1-infected individuals. They reported that an increased percentage of CD4+ CTLA4+ cells significantly correlated with high viral load in HIV-1-infected individuals, thus extending the results reported by Steiner et al. (31). Our observations may serve to explain the clinical findings by our proposal that CD4+ T cells undergoing active HIV-1 replication exhibit CTLA4 on their surface during the late stages of virion assembly. Therefore, the prediction that follows from our work and that of Leng et al. and Steiner et al. is that sorting for CD4+ CTLA4+ cells may be used to monitor and to isolate individual HIV-1-infected T cells from patient PBMCs.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman J C, Montagnier L. Nature (London) 1984;312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 2.McDougal J S, Kennedy M S, Sligh J M, Cort S P, Mawle A, Nicholson J K. Science. 1986;231:382–385. doi: 10.1126/science.3001934. [DOI] [PubMed] [Google Scholar]
- 3.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 5.Pinter A, Honnen W J, Tilley S A, Bona C, Zaghouani H, Gorny M K, Zolla-Pazner S. J Virol. 1989;63:2674–2679. doi: 10.1128/jvi.63.6.2674-2679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Earl P L, Moss B, Doms R W. J Virol. 1991;65:2047–2055. doi: 10.1128/jvi.65.4.2047-2055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kozarsky K, Penman M, Basiripour L, Haseltine W, Sodroski J, Krieger M. J Acquired Immune Defic Syndr Hum Retrovirol. 1989;2:163–169. [PubMed] [Google Scholar]
- 8.Franzusoff A, Volpe A M, Josse D, Pichuantes S, Wolf J R. J Biol Chem. 1995;270:3154–3159. doi: 10.1074/jbc.270.7.3154. [DOI] [PubMed] [Google Scholar]
- 9.Decroly E, Wouters S, Di Bello C, Lazure C, Ruysschaert J M, Seidah N G. J Biol Chem. 1996;271:30442–30450. doi: 10.1074/jbc.271.48.30442. [DOI] [PubMed] [Google Scholar]
- 10.Miranda L, Wolf J R, Pichuantes S, Duke R, Franzusoff A. Proc Natl Acad Sci USA. 1996;93:7696–7700. doi: 10.1073/pnas.93.15.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vollenweider F, Benjannet S, Decroly E, Savaria D, Lazure C, Thomas G, Chretien M, Seidah N G. Biochem J. 1996;314:521–532. doi: 10.1042/bj3140521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hallenberger S, Moulard M, Sordel M, Klenk H D, Garten W. J Virol. 1997;71:1036–1045. doi: 10.1128/jvi.71.2.1036-1045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Decroly E, Benjannet S, Savaria D, Seidah N G. FEBS Lett. 1997;405:68–72. doi: 10.1016/s0014-5793(97)00156-7. [DOI] [PubMed] [Google Scholar]
- 14.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger H G. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosson P. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 16.Freed E O, Martin M A. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vincent M J, Melsen L R, Martin A S, Compans R W. J Virol. 1999;73:8138–8144. doi: 10.1128/jvi.73.10.8138-8144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrews N W. Trends Cell Biol. 2000;10:316–321. doi: 10.1016/s0962-8924(00)01794-3. [DOI] [PubMed] [Google Scholar]
- 19.Lowin B, Peitsch M C, Tschopp J. Curr Top Microbiol Immunol. 1995;198:1–24. doi: 10.1007/978-3-642-79414-8_1. [DOI] [PubMed] [Google Scholar]
- 20.Bossi G, Griffiths G M. Nat Med. 1999;5:90–96. doi: 10.1038/4779. [DOI] [PubMed] [Google Scholar]
- 21.Iida T, Ohno H, Nakaseko C, Sakuma M, Takeda-Ezaki M, Arase H, Kominami E, Fujisawa T, Saito T. J Immunol. 2000;165:5062–5068. doi: 10.4049/jimmunol.165.9.5062. [DOI] [PubMed] [Google Scholar]
- 22.Lee K M, Chuang E, Griffin M, Khattri R, Hong D K, Zhang W, Straus D, Samelson L E, Thompson C B, Bluestone J A. Science. 1998;282:2263–2266. doi: 10.1126/science.282.5397.2263. [DOI] [PubMed] [Google Scholar]
- 23.Chambers C A, Allison J P. Curr Opin Cell Biol. 1999;11:203–210. doi: 10.1016/s0955-0674(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 24.Oosterwegel M A, Greenwald R J, Mandelbrot D A, Lorsbach R B, Sharpe A H. Curr Opin Immunol. 1999;11:294–300. doi: 10.1016/s0952-7915(99)80047-8. [DOI] [PubMed] [Google Scholar]
- 25.Linsley P S, Bradshaw J, Greene J, Peach R, Bennett K L, Mittler R S. Immunity. 1996;4:535–543. doi: 10.1016/s1074-7613(00)80480-x. [DOI] [PubMed] [Google Scholar]
- 26.Alegre M L, Noel P J, Eisfelder B J, Chuang E, Clark M R, Reiner S L, Thompson C B. J Immunol. 1996;157:4762–4770. [PubMed] [Google Scholar]
- 27.Chuang E, Alegre M L, Duckett C S, Noel P J, VanderHeiden M G, Thompson C B. J Immunol. 1997;159:144–151. [PubMed] [Google Scholar]
- 28.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. J Gen Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 29.Salzwedel K, West J T, Jr, Mulligan M J, Hunter E. J Virol. 1998;72:7523–7531. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steiner K, Waase I, Rau T, Dietrich M, Fleischer B, Broker B M. Clin Exp Immunol. 1999;115:451–457. doi: 10.1046/j.1365-2249.1999.00806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Earl P L, Broder C C, Long D, Lee S A, Peterson J, Chakrabarti S, Doms R W, Moss B. J Virol. 1994;68:3015–3026. doi: 10.1128/jvi.68.5.3015-3026.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Earl P L, Broder C C, Doms R W, Moss B. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Earl P L, Hugin A W, Moss B. J Virol. 1990;64:2448–2451. doi: 10.1128/jvi.64.5.2448-2451.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harlow E D, Lane D. Antibodies: A Laboratory Manual. Plainview, New York: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 36.Waites C L, Mehta A, Tan P K, Thomas G, Edwards R H, Krantz D E. J Cell Biol. 2001;152:1159–1168. doi: 10.1083/jcb.152.6.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.San Jose E, Munoz-Fernandez M A, Alarcon B. Virology. 1997;239:303–314. doi: 10.1006/viro.1997.8872. [DOI] [PubMed] [Google Scholar]
- 38.Raja N U, Vincent M J, Jabbar M A. J Gen Virol. 1993;74:2085–2097. doi: 10.1099/0022-1317-74-10-2085. [DOI] [PubMed] [Google Scholar]
- 39.Otteken A, Earl P L, Moss B. J Virol. 1996;70:3407–3415. doi: 10.1128/jvi.70.6.3407-3415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohnishi Y, Shioda T, Nakayama K, Iwata S, Gotoh B, Hamaguchi M, Nagai Y. J Virol. 1994;68:4075–4079. doi: 10.1128/jvi.68.6.4075-4079.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molloy S S, Anderson E D, Jean F, Thomas G. Trends Cell Biol. 1999;9:28–35. doi: 10.1016/s0962-8924(98)01382-8. [DOI] [PubMed] [Google Scholar]
- 42.Blott E J, Bossi G, Clark R, Zvelebil M, Griffiths G M. J Cell Sci. 2001;114:2405–2416. doi: 10.1242/jcs.114.13.2405. [DOI] [PubMed] [Google Scholar]
- 43.Barrat F J, LeDeist F, Benkerrou M, Bousso P, Feldmann J, Fischer A, de Saint Basile G. Proc Natl Acad Sci USA. 1999;96:8645–8650. doi: 10.1073/pnas.96.15.8645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fais S, Capobianchi M R, Abbate I, Castilletti C, Gentile M, Fei P C, Ameglio F, Dianzani F. AIDS. 1995;9:329–335. [PubMed] [Google Scholar]
- 45.Ono A, Orenstein J M, Freed E O. J Virol. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scheppler J A, Nicholson J K, Swan D C, Ahmed-Ansari A, McDougal J S. J Immunol. 1989;143:2858–2866. [PubMed] [Google Scholar]
- 47.Collins K L, Baltimore D. Immunol Rev. 1999;168:65–74. doi: 10.1111/j.1600-065x.1999.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 48.Piguet V, Wan L, Borel C, Mangasarian A, Demaurex N, Thomas G, Trono D. Nat Cell Biol. 2000;2:163–167. doi: 10.1038/35004038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leng Q, Bentwich Z, Magen E, Kalinkovich A, Borkow G. AIDS. 2002;16:519–529. doi: 10.1097/00002030-200203080-00002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.