Abstract
The GGAs (Golgi-localizing, γ-adaptin ear homology domain, ARF-binding proteins) are a family of proteins implicated in protein trafficking from the Golgi to endosomes/lysosomes. These proteins have modular structures with an N-terminal VHS (VPS-27, Hrs, and STAM) domain followed by a GAT (GGA and TOM1) domain, a connecting hinge segment, and a C-terminal GAE (γ-adaptin ear) domain. Isolated VHS domains have been shown to bind specifically to acidic cluster (AC)-dileucine motifs present in the cytoplasmic tails of the mannose 6-phosphate receptors. Here we report that full-length cytoplasmic GGA1 and GGA3 but not GGA2 bind the cation-independent mannose 6-phosphate receptor very poorly because of autoinhibition. This inhibition is caused by the binding of an AC-LL sequence present in the hinge segment to the ligand-binding site in the VHS domain. The inhibition depends on the phosphorylation of a serine located three residues upstream of the AC-LL motif. The serine is phosphorylated by casein kinase 2 in in vitro assays. Substitution of the GGA1 inhibitory sequence into the analogous location in GGA2, which lacks the AC-LL motif, results in autoinhibition of the latter protein. These data indicate that the activity of GGA1 and GGA3 is regulated by cycles of phosphorylation/dephosphorylation.
The GGAs (Golgi-localizing, γ-adaptin ear homology domain, ARF-binding proteins) are a recently described family of proteins involved in trafficking between the Golgi and endosomes (1–5). Three GGAs have been identified in humans (GGA1, GGA2, and GGA3). These proteins have modular structures consisting of an N-terminal VHS (VPS-27, Hrs, and STAM) domain followed by a GAT (GGA and TOM1) domain, a connecting hinge segment, and a C-terminal GAE domain, which has homology to the ear domain of γ-adaptin (1–5). The VHS and GAT domains are highly conserved among the three mammalian GGAs with 60–75% identity across the N-terminal 300 aa (4). The GAT domain binds to the GTP-bound form of the ADP-ribosylation factor (ARF) family of proteins that serve to recruit the GGAs from the cytosol to the Golgi complex (4, 6, 7). The VHS domain then interacts specifically with the acidic cluster (AC)-dileucine motif found in the cytoplasmic tails of sorting receptors such as sortilin, the cation-independent mannose 6-phosphate receptor (CI-MPR), and the cation-dependent mannose 6-phosphate receptor (CD-MPR), as well as the low-density lipoprotein receptor-related protein 3 (LRP3) (8–13). Although the precise function of the GAE domain is not clear, it has been shown to interact with γ-synergin, rabaptin-5, and at least in the case of GGA1 with clathrin (3, 5, 6, 9).
The hinge regions of the mammalian GGAs range in length from 150 to 280 aa and are divergent from one another, with little sequence identity to each other or to other known proteins (4). The hinge regions of all three GGAs bind clathrin (6, 9). Two conserved clathrin-box motifs are present in the GGA2 hinge (LIDLE and LLDLL), whereas only one slightly imperfect motif has been identified in the GGA1 hinge (LLDDE). The clathrin-binding sequence of GGA3 is uncertain at this time.
We now report that the GGA1 and GGA3 hinge regions contain an internal autoinhibitory AC-LL motif that binds to their VHS domains in an intramolecular fashion, thereby precluding ligand binding. This autoinhibitory sequence, however, is not present in GGA2. We present evidence that the autoinhibition depends on phosphorylation of a proximal serine residue. In addition, the GGA1 internal AC-LL sequence when transplanted into GGA2 also autoinhibits GGA2. These data demonstrate that the activity of GGA1 and GGA3 is regulated by phosphorylation of these proteins, which results in autoinhibition of their VHS domains through the intramolecular binding of their internal AC-LL motifs.
Materials and Methods
Antibodies and Plasmid Construction.
The anti-Myc mAb was purchased from Santa Cruz Biotechnology. The anti-hemagglutinin (HA) mAb was from Covance (Berkeley, CA). The anti-clathrin heavy chain (CHC) mAb TD.1 was provided generously by Frances Brodsky (University of California, San Francisco).
The construct for the glutathione S-transferase (GST) fusion encoding the wild-type bovine CI-MPR 163-aa cytoplasmic tail was made by PCR and ligation into the vector pGEX6P-1 (Amersham Pharmacia) as described (9). Residues 1–145 of the CI-MPR tail were subsequently deleted out by designing primers and using the QuickChange system (Stratagene) to generate a GST-CI-MPR peptide fusion incorporating only the C-terminal 18 aa of the cytoplasmic tail. The two GST-GGA1 peptide fusion constructs (long and short forms) were generated by annealing sense and antisense oligonucleotides corresponding to residues 342–367 and 351–367, respectively, of the human GGA1 hinge region and ligating the double-stranded products into EcoRI/XhoI-digested pGEX-5X-3 (Amersham Pharmacia). Mutagenesis of the GST GGA1 peptide fusion was performed by using primers incorporating the desired mutations with the QuickChange system.
Myc-GGA1pCR3.1 and GGA3pCR3.1 were generous gifts from Juan Bonifacino (National Institutes of Health). GGA2-HApRK5 was provided kindly by Veli-Pekka Lehto (University of Oulu, Oulu, Finland). To construct Myc-GGA1 and GGA2-HA in the pFB1vector (Invitrogen) for expression in Sf9 insect cells, Myc-GGA1pCR3.1 and GGA2-HApRK5 were double-digested with BamHI/NotI, and the cDNA inserts were ligated with BamHI/NotI-digested pFB1 to generate Myc-GGA1pFB1 and GGA2-HApFB1. Myc-GGA3pFB1 was constructed in two steps as follows: a SalI restriction site was engineered into Myc-GGA1pFB1 by substituting the codons for L10 and E11 of the GGA1 cDNA by the QuickChange method. Similarly, a SalI site was engineered into GGA3pCR3.1 by substituting the codons for E7 and S8 of the GGA3 cDNA. Myc-GGA3pFB1 was subsequently constructed by substituting the SalI/NotI fragment of Myc-GGA1pFB1 encoding the GGA1 cDNA with the corresponding SalI/NotI fragment encoding the GGA3 cDNA. The various truncation and switch mutants were generated by using primers incorporating a stop codon or the desired substitution(s) with the QuickChange system. Mutations in Myc-GGA1pCR3.1 and GGA2-HApRK5 were introduced by using the same primer sets as previously described with the QuickChange method. All constructs and mutations were confirmed to be correct by dideoxynucleotide sequencing.
Protein Expression.
The various GST-CI-MPR and GST-GGA1 peptide fusion proteins were expressed in the Escherichia coli strain BL21(RIL) (Stratagene) essentially as described (14). Cells from 1 liter of culture were lysed into 20 ml of CellLytic B reagent (Sigma), sonicated briefly, and centrifuged at 27,000 × g at 4°C for 15 min to remove insoluble material. The clarified lysate then was mixed by tumbling at 4°C for 4 h with glutathione-Sepharose 4B (Amersham Pharmacia) preequilibrated with 20 mM Tris⋅Cl, pH 7.5, containing 0.1% Triton X-100. After four washes with the 20 mM Tris/0.1% Triton X-100 buffer and a single wash with detergent-free 50 mM Tris⋅Cl, pH 8.0, the GST-fusion proteins were eluted competitively with 10 mM reduced glutathione in 50 mM Tris⋅Cl, pH 8.0. Proteins eluted with reduced glutathione were dialyzed overnight against PBS before use in pull-down experiments.
Myc-GGA1pFB1, GGA2-HApFB1, and Myc-GGA3pFB1 were transformed into E. coli DH10Bac-competent cells to generate recombinant bacmids per manufacturer protocol (Invitrogen). Bacmid DNAs were transfected into Sf9 insect cells to produce recombinant baculoviruses that were amplified and used to express the various GGAs in the insect cells. Insect cells expressing the GGA proteins were routinely harvested 48 h postinfection, lysed into cold buffer A (25 mM Hepes-KOH, pH 7.2/125 mM potassium acetate/2.5 mM magnesium acetate/1 mM DTT/0.4% Triton X-100) containing protease-inhibitor mixture (Roche, Indianapolis, IN) by sonication, and centrifuged at 20,000 × g for 10 min. The supernatant containing the GGA protein was stored at −80°C for use in the binding assays. Lysates were clarified by centrifugation at 20,000 × g immediately before use in pull-down experiments.
Transient Cell Transfection.
COS 7 cells were transfected with plasmid DNA by using Lipofectamine 2000 according to manufacturer instructions (Invitrogen). Cells were harvested 48 h posttransfection and lysed into cold buffer A as described for the insect cells. After centrifugation at 20,000 × g, the supernatant containing the GGA protein was stored at −80°C for use in the binding assays. Lysates were clarified by centrifugation at 20,000 × g immediately before use in pull-down experiments.
Binding Assays.
The binding of the various GST fusions with the GGA proteins was assayed in buffer A in a final volume of 300 μl in 1.5 ml of presiliconized microcentrifuge tubes (Midwest Scientific, Valley Park, MO). Routinely, the GST-fusion proteins were immobilized first at room temperature on 30 μl of packed glutathione-Sepharose to concentrations of 3–6 mg/ml. The beads with bound proteins were pelleted by centrifugation at 750 × g for 1 min, the beads were washed once with cold buffer B (buffer A with 0.1% Triton X-100), and 300 μl of the Sf9 cell lysate in buffer A at a final concentration of 2–5 mg/ml was added to the washed beads. To determine binding to clathrin, 300 μl of bovine adrenal cytosol (8 mg/ml) in buffer B was added to the washed beads. The binding reactions were allowed to proceed for 3 h at 4°C with tumbling, after which the samples were subjected to centrifugation at 750 × g for 1 min. An aliquot of the supernatant was saved, and the pellets were washed four times each by resuspension in 1 ml of cold buffer B by centrifugation at 750 × g. The washed pellets were resuspended in SDS sample buffer and heated at 100°C for 5 min. Unless indicated otherwise, 1% of each pellet and supernatant fraction were loaded on SDS gels for pull-downs with insect cell-expressed proteins or 20% for COS 7 cell-expressed proteins.
Electrophoresis and Immunoblotting.
Proteins were resolved on 10% SDS-polyacrylamide gels and transferred to nitrocellulose. Blots were blocked with TBST (100 mM Tris⋅Cl, pH 7.5/150 mM NaCl/0.1% Tween 20) containing 5% nonfat milk for 1 h at room temperature. The blots then were probed with primary antibodies as indicated in the individual figure legends, followed by horseradish peroxidase-conjugated anti-mouse IgG. The immunoreactive bands were visualized on x-ray films by using enhanced chemiluminescence (Amersham Pharmacia).
In Vivo and in Vitro Phosphorylation of GGA Proteins.
For in vivo phosphorylation of GGA proteins, COS 7 cells in six-well plates were transfected with plasmid DNAs to transiently express Myc-GGA1 wild-type and switch or GGA2-HA wild-type and switch in duplicates. Cells were rinsed 48 h posttransfection with either methionine/cysteine-free or phosphate-free DMEM for 35S and 32P labeling, respectively. The cells then were labeled for 4 h with 1 ml of 1 mCi/ml (1 Ci = 37 GBq) Tran35S-label (ICN) in methionine/cysteine-free DMEM with 10% FBS or with 0.5 mCi/ml [32P]orthophosphate (Amersham Pharmacia) in 1 μM okadaic acid (Sigma)/phosphate-free DMEM with 10% FBS. The cells were washed twice with cold PBS and lysed by sonication into 500 μl of buffer A containing protease-inhibitor mixture, 10 μM okadaic acid, and 100 μM sodium orthovanadate (Sigma). After centrifugation at 14,000 × g, the supernatants were immunoprecipitated overnight at 4°C with 2 μg of anti-c-Myc antibody or anti-HA antibody. The samples were washed five times with cold buffer C (100 mM Tris⋅Cl, pH 8.0/100 mM NaCl/1% Triton X-100/0.2% SDS/0.5% sodium deoxycholate/10 μM okadaic acid/100 μM sodium orthovanadate), resuspended in SDS sample buffer, and run on 8% SDS gel. The gel was treated with EN3HANCE (NEN) for 1 h, dried, and exposed to x-ray film at −80°C.
For the in vitro phosphorylation of full-length Myc-GGA1 or Myc-GGA1 switch and full-length GGA2-HA or GGA2-HA switch, Sf9 lysates were used directly as substrate for recombinant human casein kinase 2 (CK2, New England Biolabs). Sf9 lysate (50 μg) in CK2 buffer (100 mM Tris⋅Cl, pH 8.0/100 mM NaCl/50 mM KCl/20 mM Mg2+/100 μM sodium orthovanadate/10 μM okadaic acid) and 20 units of CK2 were combined in a total volume of 20 μl. The reactions were initiated by the addition of 1.5 mM [γ-32P]ATP (8 μCi/mM) at 30°C. After a 30-min incubation, the reactions were terminated by the addition of cold phosphate. The reaction volumes were brought up to 500 μl by the addition of CK2 buffer, and immunoprecipitations carried out as described for in vivo phosphorylation.
For the in vitro phosphorylation of GST-CI-MPR and GST-GGA1 peptides, 5 μg of purified fusion protein from bacteria were combined with 10 units of CK2 in CK2 buffer in a total volume of 20 μl. Manganese was added at the indicated concentrations as noted. The phosphorylation reactions were carried out as described above for full-length GGA1 and GGA2 in cytosol. The reactions were terminated by the addition of SDS sample buffer and boiling for 5 min. The samples were resolved on a 12% SDS gel and exposed to x-ray film after drying the gel.
Results
Full-Length GGA1 and GGA3 Bind Poorly to GST-CI-MPR Peptide.
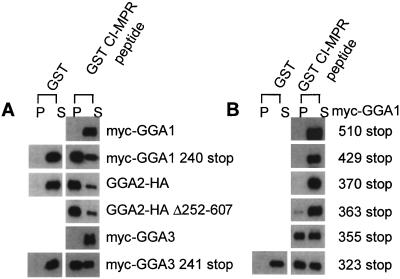
Previous reports have established that the VHS domains of all three GGAs bind to AC-LL motifs (8–13). However, the studies with GGA1 and GGA3 used truncated forms of the proteins rather than full-length molecules. To analyze the interaction of full-length GGAs with AC-LL motifs, the three GGAs were expressed in Sf9 cells and tested for binding to a GST-CI-MPR peptide encoding the C-terminal 18 aa of the bovine CI-MPR cytoplasmic tail in pull-down experiments. Contrary to the results obtained with VHS domains, the full-length Myc-GGA1 and Myc-GGA3 bound extremely poorly to the GST-CI-MPR peptide (Fig. 1A). When truncated forms of these proteins (Myc-GGA1 240 stop and Myc-GGA3 241 stop) were expressed and analyzed, specific binding to the GST-CI-MPR peptide was observed, indicating that GGA1 and GGA3 contain inhibitory sequences distal to residues 240 and 241, respectively (Fig. 1A). GGA2 appeared to lack these inhibitory sequences, because full-length GGA2 and a mutant GGA2 (with residues 252–607 deleted, leaving only six C-terminal amino acids in addition to the HA tag) bound equally well to the GST-CI-MPR peptide.
Figure 1.
GGA1 and GGA3 but not GGA2 exhibit autoinhibition in binding to a GST-CI-MPR AC-LL peptide. (A) Full-length and truncated GGAs 1–3 were expressed in Sf9 cells, and cell extracts containing the various GGAs were used directly in GST pull-down experiments. Proteins were resolved on 8% SDS gels, transferred to nitrocellulose, and probed with either anti-Myc or anti-HA mAbs as described in Materials and Methods. (B) Various GGA1 truncation mutants expressed in Sf9 cells were used to localize the region of GGA1 responsible for the autoinhibition. P, pellet; S, supernatant.
The Hinge Region of GGA1 Contains an Inhibitory Sequence.
To localize the inhibitory sequence of GGA1, a series of truncation mutants were constructed and expressed in Sf9 cells. Fig. 1B shows the results of the pull-down experiments with the various Myc-GGA1 mutants. The Myc-GGA1 510, 429, and 370 stop mutants did not bind to the GST-CI-MPR peptide. Myc-GGA1 363 stop exhibited trace binding, whereas both Myc-GGA1 355 and 323 stop mutants bound about as well as the 240 stop mutant. These results indicated that the inhibitory sequence was located between residues 355 and 363 in the hinge of GGA1.
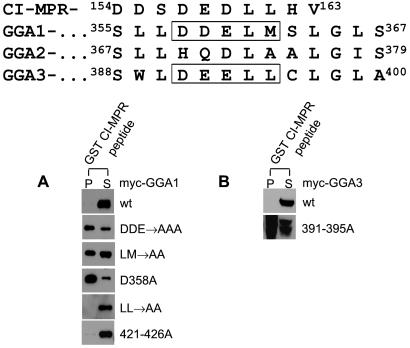
An examination of the amino acid sequence in this region of GGA1 revealed an internal AC-LL motif (358DDELM362; Fig. 2 Upper). Mutation of the three acidic residues to alanines (the DDE → AAA construct) or the leucine and methionine residues to alanines (the LM → AA construct) in the context of the full-length GGA1 resulted in binding to the GST-CI-MPR peptide (Fig. 2A). Strong binding was obtained also with a construct containing a D358A point mutation, indicating that this residue is critical for the inhibitory effect. Substitution of the dileucine sequence at position 356–357 to alanines did not relieve the inhibition. Similarly, mutation of residues 421DDLDLL426 that resemble an AC-LL motif to alanines failed to abolish the autoinhibition. Taken together, these data demonstrate that the 358DDELM362 sequence inhibits binding of GGA1 to the CI-MPR AC-LL motif, most likely by competing for the binding site in the VHS domain.
Figure 2.
GGA1 and GGA3 have internal AC-LL motifs. Sequence alignment of GGAs 1, 2, and 3 in the hinge region is as described in ref. 4. (A and B) Hinge mutants of GGA1 and GGA3 expressed in Sf9 cells were tested for binding to the GST-CI-MPR peptide in pull-down experiments as described in Materials and Methods. Immunoblots were probed with the anti-Myc mAb. P, pellet; S, supernatant; wt, wild type.
An alignment of GGA1, GGA2, and GGA3 amino acid sequences as described by Boman et al. (4) shows that GGA3 also possesses an AC-LL motif (391DEELL395), whereas GGA2 lacks this motif (Fig. 2 Upper). Substitution of residues 391–395 of GGA3 to alanines in the full-length molecule relieved the inhibition of binding to the GST-CI-MPR peptide just as observed with mutation of the equivalent residues of GGA1 (Fig. 2B).
The GGA1 Inhibitory Sequence Is Transplantable.
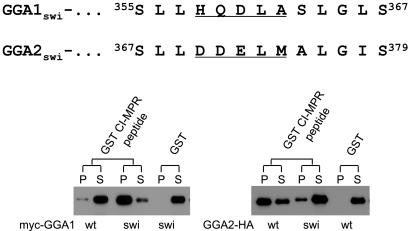
Because the GGA2 hinge lacks the AC-LL motif, we asked whether transplantation of the GGA1 AC-LM motif to the equivalent position into GGA2 would result in inhibition of binding to the GST-CI-MPR peptide. Fig. 3 shows that substitution of the GGA1 358DDELM362 sequence for the 370HQDLA374 sequence in GGA2 results in decreased binding to the GST-CI-MPR peptide. The reciprocal construct (GGA1swi) exhibited strong binding to the CI-MPR peptide, indicating that substitution of the GGA2 sequence into GGA1 relieves the inhibition. These data show that GGA2 is not subject to autoinhibition because of the lack of an AC-LL motif in its hinge region.
Figure 3.
Transplantation of GGA1 AC-LL motif into GGA2 confers autoinhibition of GGA2. Residues 358–362 of GGA1 were substituted for the corresponding residues (370–374) in GGA2 and vice versa to give GGA1 switch (GGA1swi) and GGA2 switch (GGA2swi), respectively. The Myc-GGA1swi and GGA2-HAswi mutants expressed in Sf9 cells were tested for binding to the GST-CI-MPR in pull-down assays as described in Materials and Methods. Immunoblots were probed with either anti-Myc or anti-HA mAbs. P, pellet; S, supernatant; wt, wild type.
Binding of the GGA1 Internal AC-LM Motif to the VHS Domain Depends on Phosphorylation of Serine-355.
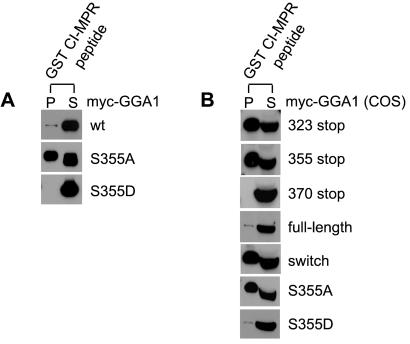
Examination of the amino acid sequences neighboring the internal AC-LL motifs of GGA1 and GGA3 reveals a serine three residues upstream of the acidic residues in both instances (Fig. 2 Upper). This sequence has the potential to serve as a CK2 substrate, raising the possibility that phosphorylation/dephosphorylation of the serine might regulate interaction of the internal AC-LL motifs with the VHS binding site. To test this possibility, serine-355 of GGA1 was mutated to an alanine or aspartate in the full-length molecule, and the resultant proteins were tested for binding to the GST-CI-MPR peptide. As shown in Fig. 4A, GGA1 with the S355A mutation bound well to the GST-CI-MPR peptide, whereas the construct with the S355D mutation failed to bind. Because the substitution of a serine with an aspartate mimics a phosphorylated serine residue (15), these results suggest that serine-355 must be phosphorylated for the autoinhibition to occur.
Figure 4.
Serine-355 of GGA1 regulates its autoinhibition. (A) Serine-355 of GGA1 was mutated to either an alanine (S355A) or aspartate (S355D), and the mutants were expressed in Sf9 cells and compared with the wild-type (wt) GGA1 for binding in GST pull-down experiments. (B) Several GGA1 truncations, GGAswi, and both GGA1 serine-355 mutants were expressed in COS 7 cells by transient transfection. Cell lysates were used directly in GST pull-down experiments to determine binding to the GST-CI-MPR peptide. Immunoblots were probed with the anti-Myc mAb. P, pellet; S, supernatant.
To exclude the possibility that the autoinhibition observed with GGA1 and GGA3 was somehow the result of expressing these proteins in insect cells, full-length GGA1 and several of the mutants were expressed in COS 7 cells and tested for binding to the GST-CI-MPR peptide in pull-down assays. As seen in Fig. 4B, similar results were obtained with COS 7 cell-produced proteins as observed with the insect-cell proteins, which establishes that the autoinhibition is not a peculiarity associated with insect cell-produced proteins.
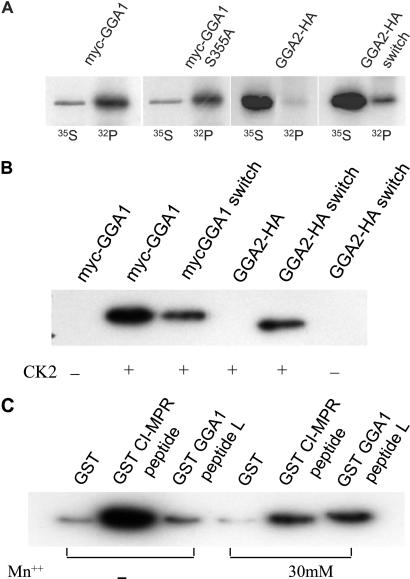
Because wild-type GGA1 present in cytosol is autoinhibited, we predicted that the cytosolic form of the protein is phosphorylated at serine-355. This prediction was examined by expressing Myc-GGA1 and Myc-GGA1 S355A in COS 7 cells, labeling these cells with either [35S]Met/Cys or 32P and then immunoprecipitating the GGAs and determining whether they were phosphorylated. GGA2, which has a serine at the equivalent position but lacks the downstream acidic residues (see Fig. 2 Upper) served as a control. Fig. 5A shows that GGA1 but not GGA2 was phosphorylated in the COS 7 cells. The GGA1 S355A mutant was phosphorylated also, although to a lesser extent, suggesting the presence of multiple phosphorylation sites in the GGA1 molecule. In contrast to GGA2, the GGA2 switch mutant was phosphorylated, indicating that the substitution of the GGA1 AC-LM sequence into GGA2 may have resulted in the phosphorylation of serine-367 of GGA2. Similar results were obtained in in vitro assays when insect cell cytosols containing the various GGAs were incubated with CK2 and [γ-32P]ATP (Fig. 5B). The GGA1 switch exhibited a partial loss of phosphorylation, whereas the GGA2 switch, but not GGA2, was phosphorylated. The phosphorylation was detected only when CK2 was added.
Figure 5.
Serine-355 of GGA1 is phosphorylated in vivo and in vitro. (A) In vivo phosphorylation was performed after transient transfection of COS 7 cells with plasmids encoding Myc-GGA1, Myc-GGA1 S355A, GGA2-HA, and GGA2 switch. Transfections were performed in duplicate in six-well tissue-culture plates. One well was labeled with methionine/cysteine-free DMEM containing 1 mCi/ml Tran35S-label (35S), and the other was labeled with phosphate-free DMEM containing 0.5 mCi/ml [32P]orthophosphate (32P) as described in Materials and Methods. Immunoprecipitates were subject to SDS/PAGE and exposed to film for autoradiography. (B) In vitro phosphorylation of full-length Myc-GGA1 and Myc-GGA1 switch and full-length GGA2-HA and GGA2-HA switch was performed by using Sf9 lysates containing the various GGAs and recombinant CK2 as described in Materials and Methods. Immunoprecipitates were subject to SDS/PAGE and autoradiography. (C) In vitro phosphorylation of GST-CI-MPR and GGA1 (long form) peptides was performed by using recombinant CK2 with and without 30 mM Mn++ as indicated. After 30 min at 30°C, the reactions were terminated by boiling in SDS sample buffer, and the immunoprecipitates were subjected to SDS/PAGE and autoradiography.
CK2 also phosphorylated a GST-GGA1 peptide encoding residues 342–367 (Fig. 5C). The extent of phosphorylation was much less than that observed with a GST-CI-MPR peptide encoding a canonical CK2 site (SDEDLL). However, in the presence of 30 mM Mn++, the phosphorylation of the GST-GGA1 peptide was stimulated strongly, whereas that of the GST-CI-MPR peptide was reduced, resulting in equivalent phosphorylation. This result is similar to the finding that phosphorylation of the Gα-interacting protein by CK2 is Mn++-dependent (16).
The GGA1 Internal AC-LM Motif Binds Directly to the VHS Domain.
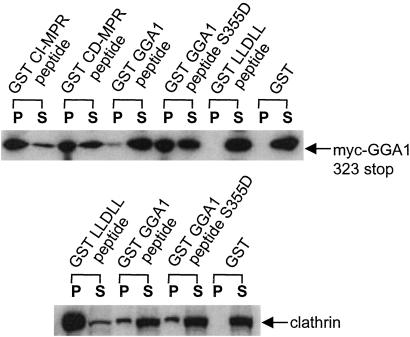
We next determined whether a GST-peptide fusion encoding serine-355 and the AC-LM motif of GGA1 could interact with its own VHS domain in solution. Because the GST-GGA1 peptide expressed and purified from bacteria is not phosphorylated, a S355D mutant was also made to mimic a phosphorylated serine. Fig. 6 Upper shows that the GST-GGA1 peptide S355D mutant bound Myc-GGA1 323 stop as strongly as the GST-CD-MPR peptide, although both of these peptides bound the truncated GGA1 less efficiently than the GST-CI-MPR peptide (note that the pellet fraction of the GST-CI-MPR peptide represents only a 10th of the loading compared with the rest of the GST-fusion proteins or GST). In contrast, the GST-GGA1 peptide with the wild-type sequence bound Myc-GGA1 323 stop very poorly, demonstrating the importance of phosphorylation of serine-355 in interaction with the VHS domain. Neither GST nor the GST-LLDLL peptide bound the Myc-GGA1 323 stop mutant. Because the GST-GGA1 peptide fusions included the LLDDE sequence reported by Puertollano et al. (6) to be the clathrin-binding motif of GGA1, we tested the wild-type peptide and the S355D mutant to determine whether clathrin binding was affected also by phosphorylation of serine-355. As shown in Fig. 6 Lower, both the wild-type and the S355D GGA1 peptides bound clathrin to an equal extent, and both are much weaker clathrin-binding motifs when compared with the AP-1 γ-LLDLL sequence (17).
Figure 6.
GST-GGA1 hinge peptide binds to GGA1 VHS domain. (Upper) The GST GGA1 peptides (short form) encode either wild-type sequence from residues 351–367 of GGA1 or the serine-355 mutated to an aspartate. The GST-CI-MPR and -CD-MPR peptides encode their AC-LL motifs. The GST-LLDLL peptide encodes residues 626–633 of mouse γ-adaptin, which is the clathrin-binding motif of the AP-1 γ-subunit. The various GST fusions were used in pull-down experiments with an Sf9 cell extract containing the Myc-GGA1 323 stop mutant, and the immunoblot was probed with the anti-Myc mAb. Twenty percent of each pellet (P) was analyzed except for the GST-CI-MPR pellet, from which 2% percent was loaded. (Lower) GST pull-down was performed with 10 mg/ml bovine adrenal cytosol, and the immunoblot was probed with the anti-clathrin TD.1 mAb. S, supernatant; P, pellet.
The GGA1 and CI-MPR AC-LL Motifs Bind to the Same Site in the GGA1 VHS Domain.
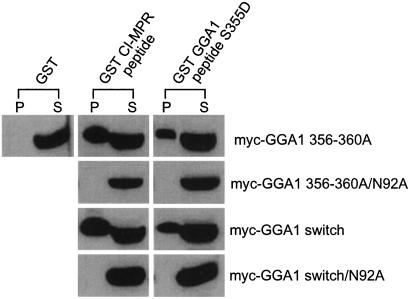
Recently, two groups reported the crystal structure of the VHS domains of human GGA1 and GGA3 complexed with the AC-LL motifs of the MPRs (12, 13). The structures reveal that asparagine at position 92 is critical for the binding of the MPR AC-LL motifs to the VHS domain of GGA1. Thus, we mutated asparagine-92 to an alanine in the Myc-GGA1 356–360Ala and the Myc-GGA1 switch mutants to determine whether the GGA1 internal AC-LM motif bound to the same site as the MPR AC-LL motifs. Fig. 7 shows the results of the pull-down assays with the various GGA1 proteins expressed in COS 7 cells. Both the GST-CI-MPR peptide and GST-GGA1 S355D peptide (short form) lost all binding to the GGA1 variants when asparagine-92 in the VHS domain was changed to an alanine, indicating that both peptides are binding to the same site within the VHS domain.
Figure 7.
The GGA1 and CI-MPR AC-LL motifs bind to the same site in the GGA1 VHS domain. Asparagine-92 was mutated to an alanine in the context of Myc-GGA1 356–360Ala or Myc-GGA1 switch to determine whether the GGA1 and CI-MPR AC-LL motifs compete for the same site in the GGA1 VHS domain. Both mutants were expressed in COS 7 cells by transient transfection, and cell lysates were used directly for binding with either the GST-CI-MPR peptide or the GST-GGA1 peptide. The immunoblot was probed with the anti-Myc mAb. P, pellet; S, supernatant.
Discussion
The data presented in this study establish that an internal AC-LL motif present in the hinge region of GGA1 and GGA3 binds to the ligand-binding site in the VHS domain and thereby blocks interaction with the AC-LL motifs of cargo molecules. Because the GGAs have been reported to be monomeric in the cytoplasm (3), we assume that this autoinhibition results from the intramolecular binding of the internal AC-LL motif to the VHS domain rather then intermolecular binding. GGA2 lacks the AC-LL motif and is not subject to this form of autoinhibition. The previous studies of GGA1 and GGA3 binding to the MPRs used VHS domains rather than full-length molecules, accounting for the failure to detect the autoinhibition (8, 11).
A key finding is that the autoinhibition depends on the phosphorylation of a serine residue located three residues upstream of the AC. Mutation of this serine to an alanine completely relieved the autoinhibition, whereas mutation to an aspartate, which mimics phosphorylation, maintained the inhibition. This requirement for serine phosphorylation to effect autoinhibition may relate to the internal location of the AC-LL motifs of GGA1 and GGA3. We and others have shown that the binding of the MPR AC-LL motifs to the VHS domains requires a precise spacing between the dileucines and the free C terminus of the receptors (12, 18). The addition of alanines to the MPR C terminus greatly inhibited binding to the VHS domain. In view of this finding, it was surprising that an internal AC-LL motif as found in GGA1 and GGA3 could compete effectively with the MPR ligand for VHS binding. One factor that likely facilitates binding of the internal AC-LL motifs is their molecular proximity to the VHS domains, which could increase the effective local concentration by several orders of magnitude (19). However, this process by itself apparently is unable to provide the strong binding needed to give autoinhibition. We postulate that the phosphorylated serine also binds to the VHS domain and that this additional interaction increases the avidity of the hinge segment to the point at which strong interaction occurs.
Several lines of evidence indicate that the serines are phosphorylated by a CK2-type enzyme. First, CK2 is known to act on serines positioned three residues upstream of an acidic residue, which occurs in GGA1 and GGA3 (20). Further, recombinant CK2 phosphorylated full-length GGA1 as well as a peptide encoding residues 342–367 of the hinge. By contrast, CK2 did not phosphorylate GGA2, which has the relevant serine but lacks the acidic residue. Substitution of an acidic residue at the third position downstream of the serine in GGA2 allowed the molecule to be phosphorylated by CK2. Although the actual kinase that phosphorylates GGA1 and GGA3 in cells has yet to be identified, it should be noted that a CK2-type enzyme has been shown to be associated with clathrin-coated vesicles (16, 21–24). In preliminary experiments, we found that an activity associated with purified clathrin-coated vesicles phosphorylates the GGA1 peptide.
The finding that GGA1 and GGA3 are subject to autoinhibition that is regulated by the state of phosphorylation of the proteins suggests that this process may have an important physiologic role. Because the cytoplasmic forms of these GGAs are phosphorylated, it is likely that after recruitment onto the Golgi by ARF–GTP, the GGAs become dephosphorylated, which would relieve the autoinhibition and allow the proteins to bind their cargo molecules. Several groups have documented that a portion of the GGAs on the Golgi are localized to clathrin-coated buds and vesicles, making it likely that the GGAs with their bound cargo next associate with forming clathrin-coated structures (2, 25). It is at this point that we speculate that GGA1 and GGA3 may be rephosphorylated by a clathrin-coated vesicle-associated CK2-type enzyme. If this indeed occurs, we would predict that the GGAs would release their cargo molecules and perhaps return to the cytoplasm as phosphoproteins, which could explain why isolated clathrin-coated vesicles have undetectable amounts of the GGAs (3). But what happens to the cargo molecule? One possibility is that the MPRs pass from the GGAs to AP-1 for final packing into the clathrin-coated vesicles. In this scenario, the GGA–MPR complex would serve as a transient intermediate to the formation of the AP-1–MPR complex that is found in clathrin-coated vesicles. This process would be similar to the role of phosphofurin acidic cluster-sorting protein (PACS1) in presenting the MPRs to AP-1 in endosomes (26). This mechanism does not preclude an independent role for the GGAs in packaging the MPRs into clathrin-coated vesicles in the Golgi, as suggested by several groups (25, 27, 28). In fact, this process could be regulated by controlling the phosphorylation of the GGAs. Finally, it is important to note that because GGA2 is not subject to this form of autoinhibition, it may have a different role in sorting.
Acknowledgments
We thank Linton Traub, Dan Ory, Rosalind Kornfeld, and members of the Kornfeld lab for reading of the manuscript and helpful comments. B.D. is the recipient of a postdoctoral fellowship from the Singapore National Science and Technology Board. This work was supported by National Institutes of Health Grant CA08759 (to S.A.K.).
Abbreviations
- GGA
Golgi-localizing, γ-adaptin ear homology domain, ARF-binding protein
- VHS
VPS-27, Hrs, and STAM
- GAT
GGA and TOM1
- AC
acidic cluster
- CI-MPR
cation-independent mannose 6-phosphate receptor
- CD-MPR
cation-dependent mannose 6-phosphate receptor
- HA
hemagglutinin
- GST
glutathione S-transferase
- CK2
casein kinase 2
References
- 1.Poussu A, Lohi O, Lehto V P. J Biol Chem. 2000;275:7176–7183. doi: 10.1074/jbc.275.10.7176. [DOI] [PubMed] [Google Scholar]
- 2.Dell'Angelica E C, Puertollano R, Mullins C, Aguilar R C, Vargas J D, Hartnell L M, Bonifacino J S. J Cell Biol. 2000;149:81–94. doi: 10.1083/jcb.149.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirst J, Lui W W, Bright N A, Totty N, Seaman M N, Robinson M S. J Cell Biol. 2000;149:67–80. doi: 10.1083/jcb.149.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boman A L, Zhang C J, Zhu X, Kahn R A. Mol Biol Cell. 2000;11:1241–1255. doi: 10.1091/mbc.11.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takatsu H, Yoshino K, Nakayama K. Biochem Biophys Res Commun. 2000;271:719–725. doi: 10.1006/bbrc.2000.2700. [DOI] [PubMed] [Google Scholar]
- 6.Puertollano R, Randazzo P A, Presley J F, Hartnell L M, Bonifacino J S. Cell. 2001;105:93–102. doi: 10.1016/s0092-8674(01)00299-9. [DOI] [PubMed] [Google Scholar]
- 7.Zhdankina O, Strand N L, Redmond J M, Boman A L. Yeast. 2001;18:1–18. doi: 10.1002/1097-0061(200101)18:1<1::AID-YEA644>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 8.Puertollano R, Aguilar R C, Gorshkova I, Crouch R J, Bonifacino J S. Science. 2001;292:1712–1716. doi: 10.1126/science.1060750. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Doray B, Poussu A, Lehto V P, Kornfeld S. Science. 2001;292:1716–1718. doi: 10.1126/science.1060896. [DOI] [PubMed] [Google Scholar]
- 10.Nielsen M S, Madsen P, Christensen E I, Nykjaer A, Gliemann J, Kasper D, Pohlmann R, Petersen C M. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takatsu H, Katoh Y, Shiba Y, Nakayama K. J Biol Chem. 2001;276:28541–28545. doi: 10.1074/jbc.C100218200. [DOI] [PubMed] [Google Scholar]
- 12.Misra S, Puertollano R, Kato Y, Bonifacino J S, Hurley J H. Nature (London) 2002;415:933–937. doi: 10.1038/415933a. [DOI] [PubMed] [Google Scholar]
- 13.Shiba T, Takatsu H, Nogi T, Matsugaki N, Kawasaki M, Igarashi N, Suzuki M, Kato R, Earnest T, Nakayama K, Wakatsuki S. Nature (London) 2002;415:937–941. doi: 10.1038/415937a. [DOI] [PubMed] [Google Scholar]
- 14.Drake M T, Downs M A, Traub L M. J Biol Chem. 2000;275:6479–6489. doi: 10.1074/jbc.275.9.6479. [DOI] [PubMed] [Google Scholar]
- 15.Jones B G, Thomas L, Molloy S S, Thulin C D, Fry M D, Walsh K A, Thomas G. EMBO J. 1995;14:5869–5883. doi: 10.1002/j.1460-2075.1995.tb00275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer T, Elenko E, Wan L, Thomas G, Farquhar M G. Proc Natl Acad Sci USA. 2000;97:4040–4045. doi: 10.1073/pnas.97.8.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doray B, Kornfeld S. Mol Biol Cell. 2001;12:1925–1935. doi: 10.1091/mbc.12.7.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doray B, Bruns K, Ghosh P, Kornfeld S. J Biol Chem. 2002;277:18477–18482. doi: 10.1074/jbc.M201879200. [DOI] [PubMed] [Google Scholar]
- 19.Deshaies R J, Ferrell J E. Cell. 2001;107:819–822. doi: 10.1016/s0092-8674(01)00620-1. [DOI] [PubMed] [Google Scholar]
- 20.Allende J E, Allende C C. FASEB J. 1995;9:313–323. doi: 10.1096/fasebj.9.5.7896000. [DOI] [PubMed] [Google Scholar]
- 21.Schook W J, Puszkin S. Proc Natl Acad Sci USA. 1985;82:8039–8043. doi: 10.1073/pnas.82.23.8039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bar-Zvi D, Levin A E, Branton D. J Biol Chem. 1987;262:17719–17723. [PubMed] [Google Scholar]
- 23.Meresse S, Ludwig T, Frank R, Hoflack B. J Biol Chem. 1990;265:18833–18842. [PubMed] [Google Scholar]
- 24.Morris S A, Mann A, Ungewickell E. J Biol Chem. 1990;265:3354–3357. [PubMed] [Google Scholar]
- 25.Hirst J, Lindsay M R, Robinson M S. Mol Biol Cell. 2001;12:3573–3588. doi: 10.1091/mbc.12.11.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan L, Molloy S S, Thomas L, Liu G, Xiang Y, Rybak S L, Thomas G. Cell. 1998;94:205–216. doi: 10.1016/s0092-8674(00)81420-8. [DOI] [PubMed] [Google Scholar]
- 27.Black M W, Pelham H R B. J Cell Biol. 2000;151:587–600. doi: 10.1083/jcb.151.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costaguta G, Stefan C J, Bensen E S, Emr S D, Payne G S. Mol Biol Cell. 2001;12:1885–1896. doi: 10.1091/mbc.12.6.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]