Abstract
Promoters and enhancers are cis-acting elements that control gene transcription via complex networks of protein–DNA and protein–protein interactions. Whereas promoters deal with putting in place the RNA polymerase, both enhancers and promoters can control transcriptional initiation and elongation. We have previously shown that promoter structure modulates alternative splicing, strengthening the concept of a physical and functional coupling between transcription and splicing. Here we report that the promoter effect is due to the control of RNA pol II elongation. We found that the simian virus 40 (SV40) transcriptional enhancer, inserted in fibronectin (FN) minigene constructs transfected into mammalian cells, controls alternative splicing by inhibiting inclusion of the FN extra domain I (EDI) exon into mature mRNA. Deletion analysis of enhancer subdomains and competitions in vivo with excess of specific enhancer DNA subfragments demonstrate that the “minimal” enhancer, consisting of two 72-bp repeats, is responsible for the splicing effect. The 72-bp repeat region has been reported to promote RNA pol II elongation. When transcription is driven by the α-globin promoter linked to the SV40 enhancer, basal EDI inclusion and activation by the SR (Ser–Arg-rich) protein SF2/ASF are much lower than with other promoters. Deletion of only one of the two 72-bp repeats not only provokes higher EDI inclusion levels but allows responsiveness to SF2/ASF. These effects are the consequence of a decrease in RNA pol II elongation evidenced both by an increase in the proportions of shorter proximal over full length transcripts and by higher pol II densities upstream of the alternative exon detected by chromatin immunoprecipitation.
Keywords: transcription‖RNA processing‖coupling
Eukaryotic transcription and pre-mRNA processing are coupled. In fact, all three processing reactions (capping, splicing, and cleavage/polyadenylation) occur in intimate association with the elongating RNA polymerase II (for reviews see refs. 1–5). Transcriptional activation of pol II genes provokes association of SR (Ser–Arg-rich) proteins like SF2/ASF to sites of transcription (6). This relocalization does not occur if pol II has a truncated C-terminal domain (7), which is consistent with the fact that truncation of the C-terminal domain causes defects in cleavage/polyadenylation and splicing (8). We have previously demonstrated that differences in promoter structure lead to differences in alternative splicing of the transcript (9). The system analyzed involved transfection of mammalian cells with minigenes carrying the EDI exon (also named EDA or EIIIA), which encodes a facultative repeat of fibronectin (FN) (10, 11). EDI contains an exonic splicing enhancer (12), which is targeted by the SR proteins SF2/ASF and 9G8. Overexpression of SF2/ASF and 9G8 markedly stimulates EDI inclusion, but the effect of these proteins is modulated by the promoter (13). For instance, if transcription is driven by the α-globin (α-gb) promoter, there is little inclusion of EDI and little response to SF2/ASF activation. However, if transcription is driven by the FN or the cytomegalovirus (CMV) promoters, basal EDI inclusion and stimulation by SF2/ASF are 10-fold higher. These effects are not the trivial consequence of different mRNA levels produced by each promoter (promoter strength) but depend on some qualitative properties conferred by promoters to the transcription/RNA processing machinery. A possible mechanism that would explain these observations is that the promoter itself is responsible for recruiting splicing factors, such as SR family proteins, to the site of transcription, possibly through transcription factors that bind the promoter or the transcriptional enhancers. Several candidates have been proposed to mediate such recruitment (2, 14). An alternative, but not exclusive, model suggests that promoters might control alternative splicing via the regulation of pol II elongation or processivity. Low pol II processivity or internal pauses for elongation would favor the inclusion of alternative exons governed by an exon skipping mechanism, whereas a highly elongating pol II, or the absence of internal pauses, would favor exclusion of these kinds of exons. This idea is supported by recent findings that two transcriptional activators with opposite effects on pol II elongation, SV40 T antigen and herpesvirus VP16, have opposite effects on alternative splicing (15).
We report here that the differential behavior of the α-gb compared to the FN or CMV promoters on alternative splicing correlates with different transcriptional processivities. Furthermore, we found that the SV40 transcriptional enhancer, a cis-acting element present in the minigene constructs as a common heterologous component of both kinds of promoters, plays a key role in the promoter effect, by inhibiting EDI inclusion. This is achieved through the binding of transacting factors to the 72-bp repeat region, responsible of stimulating pol II elongation.
Experimental Procedures
Plasmid Constructs.
The references for plasmids are: pSVEDATot (α-gb promoter; ref. 12), pSVEDA/CMV (CMV promoter), and pSVEDA/FN (FN promoter; ref. 9). These constructs contain a SV40 enhancer/origin (e/o) located approximately at −600 bp with respect of the transcriptional start site. To delete this region, the minigene constructs were digested with EcoRI and SfiI and the resulting ends were rendered blunt by treatment with T4 DNA polymerase and Klenow and then religated. The resulting constructs contained a single SspI site, which was used to reinsert, either in the original or in an opposite orientation, a 600-bp segment containing the SV40 e/o in an asymmetric position. The SV40 e/o resulted then positioned at −350, when reinserted in the original orientation, and at −550 when reinserted in the opposite orientation. The use of the SF2 expression plasmid (g10 SF2/ASF) was reported before (13).
Subenhancer Mutants.
pSVEDATot short (15) was digested with EcoRI and SacII to eliminate the whole SV40 e/o and used as vector to clone a series of PCR products, obtained from pSVEDATot as template, encompassing different internal segments of the SV40 e/o and containing EcoRI and SacII sites at their ends. The primers used to generate the segments are: three GC-rich repeats plus ori (131 bp): 5′-CGGAATTCCGAGTCAGCAACCATAGTCC-3′ and 5′-TCCCCGCGGGGACTGCGTTAGCAATTTAACTG-3′; two 72-bp repeats plus three GC-rich repeats (287 bp): 5′-TCCCCGCGGGGAAATTAGTCAGCCATGGGG-3′ and 5′-CGGAATTCCGATAGGCGTATCACGAGGC-3′, and two 72-bp repeats (205 bp): 5′-TCCCCGCGGGAGGGACTATGGTTGCTGAC-3′ and 5′-CGGAATTCCGATAGGCGTATCACGAGGC-3′. To generate the mutant containing one 72-bp repeat plus three GC-rich repeats plus ori, pSVEDATot was digested with SphI and the resulting large fragment was religated. pBS72bp, which harbors two 72-bp repeats, was obtained by cloning the 205-bp PCR product mentioned above between the SacII and EcoRI sites of pBSKS+ (Stratagene).
Transfections and Alternative Splicing Assay.
Transfections with lipofectamine (Life Technologies), RNA preparation (16), and radioactive reverse transcription (RT)-PCR amplification of splicing isoforms by using specific primers (12) have been described (9, 13).
RNase Protection Assay (RPA).
The design of distal and proximal riboprobes and RPA conditions have been described (15).
RNA Pol II Densities on Transfected Templates.
Chromatin immunoprecipitation assay (ChIP).
ChIPs were performed as described in the Upstate Biotechnology (Lake Placid, NY) protocol with minor modifications. Protein–DNA cross-linking was performed by incubating the transfected cell monolayers with 1% (vol/vol) formaldehyde for 10 min 37°C. Glycine (0.125 M) was added to quench the reaction. Cells were scraped, collected by centrifugation, washed with PBS containing proteinases inhibitors, and lysed in 1 ml of 1% SDS, 10 mM EDTA, and 50 mM Tris⋅HCl (pH 8.1) for 10 min at room temperature. The lysate was sonicated with 10 pulses of 20 sec each at 40% of maximum power of a Sonic Dismembrer Digital MDL500 (Fisher) equipped with a microtip, to yield chromatin fragments of an average size of <500 bp. The debris was removed by centrifugation at 14,000 rpm for 10 min at 4°C. The supernatant was diluted 10-fold in SDS lysis buffer (Upstate Biotechnology). Soluble chromatin was precleared by incubation for 2 h at 4°C with 300 μl of protein A-agarose pretreated with 100 μg/ml salmon sperm DNA. An aliquot of precleared chromatin was removed (input) and used in the subsequent PCR analysis. The remainder of the chromatin was incubated with 3 μg of Ab against pol II (N-20 sc-899, Santa Cruz Biotechnology) or rabbit preimmune serum overnight at 4°C. Immune complexes were purified by treatment with Protein A-agarose, followed by washings and elution as specified in the Upstate Biotechnology protocol. Cross-links were reversed by incubation for 4–5 h at 65°C. Samples were digested with 0.24 mg/ml Proteinase K, and DNA was purified by phenol/chloroform extraction followed by ethanol precipitation.
PCR analysis.
Immunoprecipitated (or input) DNAs were used as templates in a quantitative real-time PCR assay performed on the Corbett Research Rotor-Gene 2000 real-time cycler. The PCR mixture contained Platinum Taq polymerase (LifeTechnologies), optimized concentrations of Cybergreen and the following primers: upstream (U) region: 5′-TGGACCCGGTCAACTTCAAG3-′ and 5′-CTCTCTCCGCTTGGATTCTG-3′; downstream (D) region: 5′-AGCTATTCCTGCACCAACTG-3′ and 5′-GCTCCAGCTTAACGGTATTTG-3′, and control (C) region: 5′-AGCCTAGGCCTCCAAAAAGC-3′ and 5′-TGCCACCTGACGTCTAAGAAAC-3′. Cycling parameters were 95°C for 3 min, followed by 40 cycles of 95°C for 30 sec, 57°C for 45 sec, and 72°C for 45 sec. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer.
Results and Discussion
Promoters, Alternative Splicing, and Pol II Processivity.
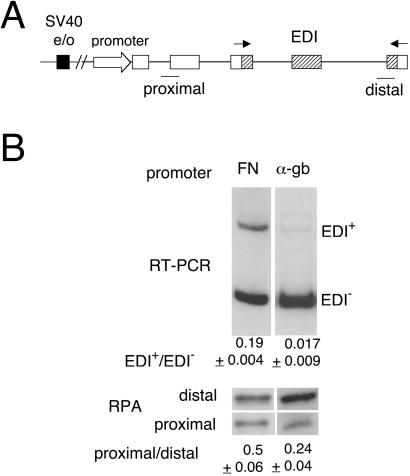
Transcriptional processivity is defined as the ability to elongate through sites where polymerase (in our case pol II) is prone to pause or terminate prematurely. Nonprocessive polymerases are released from the template at some positions within the transcription unit, generating higher proportions of shorter over longer transcripts in the steady–state. Short transcripts also are generated upon total RNA extraction by stalled pol II molecules. Relative proportions of these transcripts can be estimated by quantitative RPA performed with two different probes, one proximal and the other distal to the transcription start site. This assay is based on various observations that prematurely terminated RNAs are stable (17–19). Promoters have been implicated in the control of pol II elongation in several cases (refs. 17 and 20, and references therein). Consistently, we found that constructs carrying different promoters elicit different transcriptional processivities, which correlate inversely with their ability to promote EDI exon inclusion. Minigene constructs (Fig. 1A) with either the FN or the α-gb promoters were transiently transfected into human hepatoma Hep3B cells. Total RNA extracted from the transfected cells was used to assess EDI splicing by radioactive RT-PCR and to quantify expressed mRNA levels with proximal and distal probes by RPA. The FN promoter, which provokes EDI inclusion levels 10 times higher than those of the α-gb promoter, promotes a less processive transcription as evidenced by proximal/distal mRNA levels that are 2-fold higher than those elicited by the α-gb promoter (Fig. 1B).
Figure 1.
FN and α-gb promoters elicit different alternative splicing ratios and pol II processivities. (A) Scheme of the minigenes transfected to assess alternative splicing. Open exons, human α-gb; dashed exons, human FN; black box, SV40 e/o; arrows, primers used to amplify the mRNA splicing variants by RT-PCR, and lines, proximal and distal probes used for RPA. (B Upper) Hep3B cells were transfected with 600 ng of pSVEDA/FN (FN promoter) or pSVEDATot (α-gb promoter) plus 400 ng of pCMVβgal. RNA splicing variants were detected by radioactive RT-PCR and analyzed in 6% native polyacrylamide gels. Ratios between radioactivity in EDI+ bands and radioactivity in EDI− bands are shown under each lane. (Lower) RPA with proximal and distal probes shown in A, to measure levels of short and long transcripts of transfected Hep3B cells. RT-PCR and RPA ratios correspond to at least three independent transfection experiments.
Deletion of the SV40 Enhancer Stimulates Inclusion of the EDI Alternative Exon.
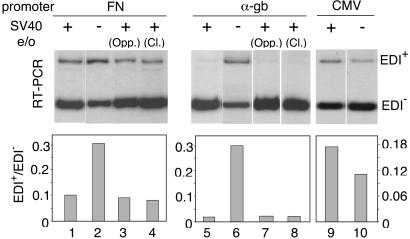
All minigene constructs carrying different promoters used previously by our group (9, 13, 15), including those of Fig. 1, contain the strong SV40 transcriptional enhancer located ≈600 bp upstream of the transcription start site. Yankulov et al. (20) demonstrated that the presence of transcriptional enhancers increases the processivity of transcription. To investigate further if changes in processivity determine changes in splicing, we compared EDI inclusion elicited by three minigenes in the presence or absence of the SV40 e/o. Deletion of the enhancer from the FN or α-gb promoter constructs increased their respective EDI+/EDI− ratios by ≈3- and 10-fold, respectively (Fig. 2, lanes 1, 2, 5, and 6). However, this activating effect was not observed with the CMV promoter construct (Fig. 2, lanes 9 and 10), although the CMV and FN promoters are equally able to elicit higher levels of EDI inclusion than the α-gb promoter. Reinsertion of the SV40 e/o sequences into the minigene plasmids, either in opposite orientation near their original sites, or in the original orientation but 300 bp closer to the start site reestablished the levels of EDI inclusion characteristic of each promoter (Fig. 2, lanes 3, 4, 7, and 8). This indicates that the observed effects correspond of those of bona fide enhancers, which act independently of position and orientation with respect to proximal promoter elements.
Figure 2.
Effects of SV40 e/o deletion on alternative splicing of the EDI exon. Hep3B cells were transfected with 600 ng of minigene constructs carrying FN (lanes 1–4), α-gb (lanes 5–8), or CMV promoters (lanes 9 and 10) plus 400 ng of pCMVβgal. Transfections with variants of these constructs that lack the SV40 e/o are shown in lanes 2, 6, and 10. In the case of the FN and α-gb promoter constructs, variants in which the SV40 e/o was reinserted in opposite orientation near the original site (Opp.) (lanes 3 and 7) or in the original orientation but 300 bp closer to the transcription start site (Cl.) (lanes 4 and 8) are shown. Similar results were obtained in Cos-7 and HeLa cells.
Internal Deletion Analysis Localizes the Enhancer Effect to the 72-bp Repeats.
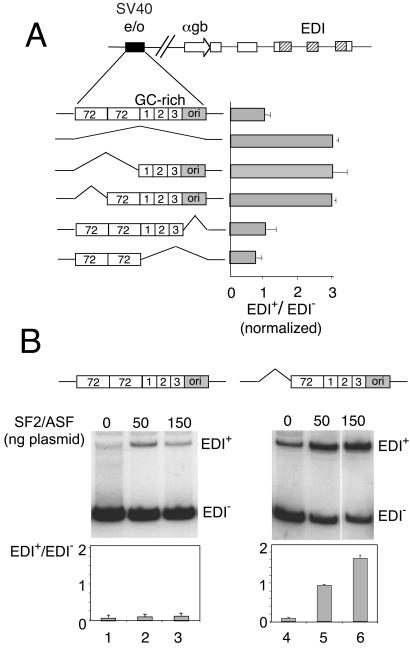
From 5′ to 3′, the SV40 e/o region consists of two tandem copies of a 72-bp repeat, three copies of a 21-bp repeat, a TATA box marking the SV40 early promoter, and the origin of replication (Fig. 3A). Each 21-bp repeat has two copies of a GC-hexanucleotide motif that has been shown to bind the transcription factor Sp1. The minimal enhancer has been localized to the distal 72-bp repeat and 5′-flanking region (21). To identify the cis-acting elements of the SV40 e/o responsible for the inhibition of EDI inclusion, we prepared a series of α-gb promoter minigenes with specific deletions of SV40 e/o subdomains and transfected Hep3B cells to assess alternative splicing ratios. Fig. 3A shows that deletions of the ori segment alone or together with the three 21-bp GC-rich repeats do not affect alternative splicing. However, deletions of either both or only one copies of the 72-bp repeats stimulate EDI inclusion to the same extent as the elimination of the whole SV40 e/o. This indicates that the minimal enhancer is sufficient to control alternative splicing.
Figure 3.
(A) Deletion analysis of the SV40 e/o with respect to alternative splicing of the EDI exon. Horizontal bars indicate normalized EDI+/EDI− ratios of Hep3B cells transfected with a series of α-gb promoter constructs carrying different internal deletions of the SV40 e/o. Results correspond to the mean ± SD of at least three independent transfection experiments. (B) Deletion of only one 72-bp repeat confers responsiveness to SF2/ASF to the α-gb promoter construct. Hep3B cells were transfected with pSVEDATot (lanes 1–3) or a variant lacking the distal 72-bp repeat of the SV40 enhancer (lanes 4–6) and cotransfected with the indicated amounts of a plasmid expressing SF2/ASF (13). Transfections in lanes 1 and 4 contained 150 ng of empty DNA vector. Similar results were obtained in Cos-7 and HeLa cells.
Deletion of One 72-bp Repeat Confers Responsiveness to SF2/ASF.
Minigenes with the α-gb promoter linked to the SV40 e/o display very low response to activation of EDI inclusion by overexpression of the SR protein SF2/ASF (13). Most surprisingly, this lack of response to SF2/ASF is reverted when one of the 72-bp repeats is deleted. Fig. 3B shows that if the SV40 e/o is present in the α-gb promoter construct, SF2/ASF provokes only a 2-fold increase in EDI inclusion. However, in the absence of one 72-bp repeat, not only basal EDI+/EDI− ratios increase but responsiveness to SF2/ASF goes up to 16-fold. This increase in responsiveness to SF2/ASF was also observed when the whole SV40 e/o was deleted and was abolished when the enhancer was reinserted in similar or opposite orientations like in the experiments of Fig. 2 (not shown).
The Enhancer Effect on Splicing Is Independent from Transcription Levels.
In the experiments of Figs. 2 and 3, deletion of the SV40 e/o provokes an important decrease in the total mRNA amounts produced by the transfected minigenes (between 50- and 100-fold, not shown). This opens the possibility that the increase in EDI inclusion is the trivial consequence of extremely low levels of premRNA for which the available concentration of SF2/ASF become less limiting. To rule out this hypothesis, we developed a different strategy, consisting in titrating out the factors that bind to the enhancer sequences by introducing a molar excess of 72-bp DNA into cells transfected with the minigenes carrying the wild-type SV40 e/o. Each 72-bp unit is composed of at least two distinct elements, A and B, which show practically no enhancer activity individually but can cooperate to activate transcription through the synergistic binding of a variety of ubiquitous and cell-specific factors (22–28). These factors, including transcription enhancer factors 1 and 2 (TEF-1 and TEF-2), TC-IIA/NF-κB and TC-IIB/KBF1 heterodimers, the octamer-binding transcription factor, p62 and AP1 (cJun/cFos), participate in the assembly of a multimolecular protein–DNA/protein–protein network known as enhanceosome (29). The relative abundance of these factors is not known; although it is conceivable that some of them might be more limiting than others. Introduction of an excess of 72-bp sequences into the same nucleus where the enhancer is working would compete more efficiently for the limiting factors than for those which are in excess. Competition with a 10 molar excess of plasmid pBS72bp provokes a 4-fold stimulation of EDI inclusion elicited by the α-gb promoter minigene (Fig. 4A, lanes 1 and 2), similar to (or slightly higher than) the effect caused by the deletion of the enhancer in cis. By contrast, transcription is not inhibited: The total amount of transcript increased by 20% (RPA). Control experiments showed similar results in competitions with a plasmid carrying the whole SV40 e/o and no effect in competitions with plasmids carrying subfragments lacking the 72-bp repeats (not shown). We therefore interpret that competition with pBS72bp has titrated out those factors that are important for the transcriptional control of splicing (i.e., processivity factors, see below) but has not inhibited overall transcription.
Figure 4.
(A) Effects of competition with a molar excess of 72-bp DNA on EDI alternative splicing. Hep3B cells were transfected with 600 ng of α-gb (lanes 1 and 2) or CMV (lanes 3–6) promoter constructs, having (lanes 1–4) or lacking (lanes 5 and 6) the SV40 e/o. The same cells were cotransfected with 2 μg of pBS72bp (see Experimental Procedures) (lanes 2, 4, and 6) or an equivalent amount of empty vector (pBSKS+, Stratagene) (lanes 1, 3, and 5). The amount of pBS72bp transfected represented an ≈10-fold molar excess of plasmid with respect to the minigene constructs. RT-PCR and RPA results are shown. Histograms correspond to EDI+/EDI− ratios calculated from RT-PCR results. Similar results were obtained in Cos-7 and HeLa cells. (B) Proximal/distal ratios of RPAs corresponding to Hep3B cells transfected with 600 ng of pSVEDATot (α-gb promoter) and 2 μg of pBS72bp (+) or the same amount of empty vector (−). Ratios correspond to the mean ± SD of at least three independent transfection experiments. (C) The CMV promoter and the SV40 e/o contribute equally to the inhibition of EDI exon inclusion. The model explains why deletion of the SV40 e/o releases inhibition of exon inclusion when transcription is driven by the α-gb promoter but not when it is driven by the CMV promoter. At the same time, the model explains why exon inclusion is stimulated by competition with an excess of 72-bp DNA with both promoter constructs. EPF, elongation promoting factor.
The CMV Promoter Behaves Like the SV40 Enhancer in the Control of Alternative Splicing.
As shown in Fig. 2, deletion of the SV40 e/o has no stimulatory effect on EDI inclusion when transcription is driven by the CMV promoter. However, competition with pBS72bp provokes an important increase (10-fold) in EDI inclusion (Fig. 4A, lanes 3 and 4). This contradictory behavior could be explained if the CMV promoter itself had an SV40-enhancer-like activity, able to promote pol II elongation or, in other words, if this promoter were able to bind factors that also bind to the SV40 minimal enhancer. In fact, the CMV segment used here contains sequences corresponding to the CMV major immediate early enhancer, located upstream of the transcription initiation site, between nucleotides −118 and −524, which were found to functionally substitute for the 72-bp repeats of the SV40 enhancer (30, 31). Thus, competition with pBS72bp would prevent binding of elongation promoting factors from both the SV40 e/o and the CMV promoter and subsequently stimulate exon inclusion. Accordingly, competition over a CMV minigene lacking the SV40 e/o still provokes a significant increase (8-fold) in EDI inclusion (Fig. 4A, lanes 5 and 6), whereas competition over an α-gb minigene lacking the SV40 e/o has no effect at all (not shown). Fig. 4C shows a model illustrating the differential behavior of the α-gb and CMV promoter constructs with respect to deletion and competition experiments involving the SV40 enhancer.
Stimulation of EDI Inclusion by Competition with 72-bp Repeats DNA Correlates with a Decrease in Pol II Processivity.
The fact that elimination of either the whole enhancer or one of its 72-bp repeats alone, as well as competition with 72-bp DNA, have similar effects on splicing is highly supportive for a mechanism involving transcriptional elongation because the 72-bp repeats have been implicated in promoting pol II processivity (20). Processivity analysis of mRNAs produced by α-gb minigenes with or without competition by pBS72bp (Fig. 4B) confirms this speculation: proximal/distal ratios increase by ≈3-fold after competition, which is consistent with lower processivity being associated to higher EDI inclusion. The observation of a 3-fold decrease in processivity accompanied by practically no change in overall transcription (only 20% increase) is consistent with the absence of correlation between promoter strength and elongation capacity, already reported for the c-myc promoter (20) and for the effects of VP16 and SV40 T antigen on transcription of the minigenes used in this report (15). This also is consistent with our previous observations that the promoter effect on alternative splicing was due to promoter quality and not to promoter strength (9, 13).
ChIPs with an Ab to Pol II Confirm Changes in Processivity.
The RPA of Figs. 1 and 4B probe steady–state RNA levels, which are affected by RNA synthesis and degradation. A more direct approach is provided by determining differences in RNA pol II densities along the transfected minigenes by using the chromatin immunoprecipitation technique with an Ab to pol II. Two regions, one U and the other D of the alternative exon were analyzed with respect to a third C region localized outside the transcriptional unit (Fig. 5A). In agreement with RPA results, conditions that favor EDI inclusion display higher pol II densities in the U region compared to the D region, reflecting stalling of the polymerase. Fig. 5B shows that minigenes with the FN and CMV promoters transfected into Cos-7 cells display Ur/Dr ratios (45.5 ± 9.0 and 20.9 ± 2.9) significantly higher than the α-gb promoter (7.7 ± 2.1). Furthermore, in vivo competition of the α-gb construct with the plasmid carrying the SV40 e/o increases pol II density at the U region by >10-fold with a Ur/Dr ratio of 109.4 ± 32.0. Transfections performed in HeLa cells gave essentially similar results, with the following Ur/Dr ratios (defined in legend to Fig. 5): α-gb promoter, 5.3 ± 3.0; FN promoter, 51.0 ± 5.3; CMV promoter, 18.0 ± 1.9; and α-gb competed with SV40 enhancer, 44.0 ± 6.1.
Figure 5.
ChIP with an Ab to RNA pol II. (A) Scheme of the minigenes transfected to assess pol II densities. Arrows indicate the pairs of primers used in real time PCRs to quantitatively amplify DNA that is bound to the immunoprecipitated pol II, at two regions mapping U, D of the EDI alternative exon, and at a third C region outside of the transcription unit. (B) Cells were transfected with α-gb, FN, or CMV promoter constructs and, where indicated, cotransfected with a 10-fold molar excess of a competitor plasmid carrying the SV40 e/o. After 48 h, cells were fixed with formaldehyde and treated for ChIP and real time PCR analysis as described in Experimental Procedures. Ur = Uim/Uin; Cr = Cim/Cin; Dr = Dim/Din where Uim, Cim, and Dim are the template DNA amounts recovered after immunoprecipitation by anti-pol II, and Uin, Cin, and Din are the input DNA amounts, all estimated by real time PCR at regions U, C, and D, respectively. Results correspond to a representative transfection experiment of Cos-7 cells and show the mean ± SD of three real time PCR determinations.
Models for Enhancer Action and Alternative Splicing.
What are the molecular bases for the control of elongation, and concomitantly of alternative splicing, by the SV40 enhancer? One model for enhancer action postulates that enhancer-bound factors synergize with upstream activators, bound in the vicinity of the TATA box, to recruit pol II complexes to the transcriptional start site (32). An alternative model sees that enhancers may function to relieve chromatin-mediated repression of genes (33), a task accomplished by enhancer-mediated recruitment of chromatin-remodeling factors, such as p300, CREB-binding protein or PCAF (p300/CREB-binding protein-associated factor), with acetyltransferase activities. Acetylation of the core histones would facilitate accessibility of transcription factors to DNA elements, otherwise impeded by tight nucleosomal arrays (34–36). The action of enhancers at long-distance might not only facilitate binding of transcription factors to promoters but also affect chromatin structure at internal regions of a gene and therefore the effectiveness of chromatin as template for RNA synthesis (37, 38), that could become particularly critical to facilitate the passage of the transcribing polymerase. The finding that actively transcribing pol II piggybacks a histone acetyltransferase activity (39) has led to the proposal of chromatin opening mediated by DNA tracking (2) to explain why transcription occurs efficiently in vivo on a chromatinized template, whereas in vitro, the ability of purified pol II to transcribe a chromatin template is poor compared to the efficiency in transcribing naked DNA. The viral activator VP16 acts similarly to the SV40 enhancer in promoting transcriptional elongation (17) and in inhibiting EDI exon inclusion (15). The mechanism by which VP16 activates elongation also seems to involve the promotion of histone acetylation far downstream of promoters (40, 41).
Recent evidence, of independent nature, highlights the importance of coupling between splicing and transcriptional elongation. Fong and Zhou (19) found that spliceosomal U small nuclear ribonucleoproteins (UsnRNPs) interact with the human transcription elongation factor TAT-SF1 and strongly stimulate pol II elongation, probably via the binding of TAT-SF1 to the elongation factor P-TEFb. Because the TAT-SF1-UsnRNP complex also stimulates splicing in vitro (19), these results not only reveal that splicing factors function directly to promote transcriptional elongation but that reciprocal interactions exist in the coupling process.
The mechanism by which elongation affects EDI splicing is conditioned by premRNA sequence constraints as suggested before (42, 43). EDI exon skipping occurs because the 3′ splice site of the upstream intron is suboptimal compared to the 3′ splice site of the downstream intron. If the polymerase pauses anywhere between these two sites, only elimination of the upstream intron can take place. Once the pause is passed or the polymerase proceeds, there is no option for the splicing machinery but to eliminate the downstream intron, which leads to exon inclusion. A highly processive elongating pol II, or the absence of internal pauses, would favor the simultaneous presentation of both introns to the splicing machinery, a situation in which the stronger 3′ splice site of the downstream intron outcompetes the weaker 3′ splice site of the upstream intron, resulting in exon exclusion. A less processive pol II would increase the probability of capture of free splicing factors by the nascent transcript and/or facilitate interactions of splicing factors that use the C-terminal domain as landing path with the transcript. This might explain the higher sensitivity to SF2/ASF observed when the SV40 e/o is deleted (Fig. 3B).
Until recently it was assumed that complex patterns of gene expression were mainly achieved through the differential turning on and off of a large number of genes. The realization that the human genome contains a smaller number of genes than foreseen enhances the contribution of alternative splicing to the observed complexity. Results reported here add a novel component to the various and complex network that controls alternative splicing, which not only includes relative abundance, tissue distribution, and posttranslational modifications of splicing factors but also the protein-recruiting and elongating properties of the transcription machinery.
Acknowledgments
This paper is dedicated to the memory of Dr. César Milstein, whose life and scientific achievements remain an inspiration to Argentine scientists. We thank P. Cramer, A. Srebrow, O. Coso, K. Abruzzi, S. Lacadie, and D. Chowdhury for their help. This work was supported by grants from the Fundación Antorchas, the International Center for Genetic Engineering and Biotechnology, and the Agencia Nacional de Promoción de Ciencia y Tecnología (ANPCyT) (to A.R.K.), and National Institutes of Health Grant GM 33205 (to M.R.). S.K. is the recipient of a fellowship from the University of Buenos Aires. A.R.K. is a Howard Hughes Medical Institute International Research Scholar and a career investigator of the Consejo Nacional de Investigaciones Cientiíficas y Técnicas (CONICET) of Argentina.
Abbreviations
- e/o
enhancer/origin
- α-gb
α-globin
- FN
fibronectin
- SV40
simian virus 40
- CMV
cytomegalovirus
- ChIP
chromatin immunoprecipitation assay
- U
upstream
- D
downstream
- C
control
- RT
reverse transcription
- EDI
extra domain I
Footnotes
Deceased March 24, 2002.
Date revised version received by PNAS office.
References
- 1.Bentley D. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 2.Travers A. Proc Natl Acad Sci USA. 1999;96:13634–13637. doi: 10.1073/pnas.96.24.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maniatis T, Reed R. Nature (London) 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 4.Proudfoot N J, Furger A, Dye M J. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 5.Cramer P, Srebrow A, Kadener S, Werbajh S, de la Mata M, Melen G, Nogués G, Kornblihtt A R. FEBS Lett. 2001;498:179–182. doi: 10.1016/s0014-5793(01)02485-1. [DOI] [PubMed] [Google Scholar]
- 6.Misteli T, Cáceres J F, Spector D L. Nature (London) 1997;387:523–526. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 7.Misteli T, Spector D L. Mol Cell. 1999;3:697–705. doi: 10.1016/s1097-2765(01)80002-2. [DOI] [PubMed] [Google Scholar]
- 8.Mc Cracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D. Nature (London) 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 9.Cramer P, Pesce C G, Baralle F E, Kornblihtt A R. Proc Natl Acad Sci USA. 1997;94:11456–11460. doi: 10.1073/pnas.94.21.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kornblihtt A R, Vibe-Pedersen K, Baralle F E. EMBO J. 1984;3:221–226. doi: 10.1002/j.1460-2075.1984.tb01787.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornblihtt A R, Pesce C G, Alonso C R, Cramer P, Srebrow A, Werbajh S E, Muro A F. FASEB J. 1996;10:248–257. doi: 10.1096/fasebj.10.2.8641558. [DOI] [PubMed] [Google Scholar]
- 12.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cramer P, Cáceres J F, Cazalla D, Kadener S, Muro A F, Baralle F E, Kornblihtt A R. Mol Cell. 1999;4:251–258. doi: 10.1016/s1097-2765(00)80372-x. [DOI] [PubMed] [Google Scholar]
- 14.Cáceres J F, Kornblihtt A R. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 15.Kadener S, Cramer P, Nogués G, Cazalla D, de la Mata M, Fededa J P, Werbajh S, Srebrow A, Kornblihtt A R. EMBO J. 2001;20:5759–5768. doi: 10.1093/emboj/20.20.5759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Blau J, Xiao H, McCracken S, O'Hare P, Greenblatt J, Bentley D. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nahreini P, Mathews M B. J Virol. 1995;69:1296–1301. doi: 10.1128/jvi.69.2.1296-1301.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fong Y W, Zhou Q. Nature (London) 2002;414:929–933. doi: 10.1038/414929a. [DOI] [PubMed] [Google Scholar]
- 20.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 21.Zenke M, Grundström T, Matthes H, Wintzerith M, Schatz C, Wildeman A, Chambon P. EMBO J. 1986;5:387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao J H, Davidson I, Matthes H, Garnier J-M, Chambon P. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 23.Xiao J-H, Davidson I, Macchi M, Rosales R, Vigneron M, Staub A, Chambon P. Genes Dev. 1987;1:794–807. doi: 10.1101/gad.1.8.794. [DOI] [PubMed] [Google Scholar]
- 24.Lee W, Mitchell P, Tjian R. Cell. 1987;49:741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- 25.Rosales R, Vigneron M, Macchi M, Davidson I, Xiao J-H, Chambon P. EMBO J. 1987;6:3015–3025. doi: 10.1002/j.1460-2075.1987.tb02607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanno M, Fromental C, Staub A, Ruffenach F, Davidson I, Chambon P. EMBO J. 1989;8:4205–4214. doi: 10.1002/j.1460-2075.1989.tb08606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macchi M, Bornert J-M, Davidson I, Kanno M, Rosales R, Vigneron M, Xiao J-H, Fromental C, Chambon P. EMBO J. 1989;8:4215–4227. doi: 10.1002/j.1460-2075.1989.tb08607.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rachubinski R A, Marcus S L, Capone J P. J Biol Chem. 1999;274:18278–18284. doi: 10.1074/jbc.274.26.18278. [DOI] [PubMed] [Google Scholar]
- 29.Carey M. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 30.Boshart M, Weber F, Jahn G, Dorsch-Hasler K, Fleckenstein B, Schaffner W. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- 31.Fickenscher H, Stamminger T, Ruger R, Fleckenstein B. J Gen Virol. 1989;70:107–123. doi: 10.1099/0022-1317-70-1-107. [DOI] [PubMed] [Google Scholar]
- 32.Treisman R, Maniatis T. Nature (London) 1985;315:73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- 33.Krumm A, Madisen L, Yang X J, Goodman R, Nakatani Y, Groudine M. Proc Natl Acad Sci USA. 1998;95:13501–13506. doi: 10.1073/pnas.95.23.13501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckner R, Ewen M E, Newsome D, Gerded M, De Caprio J A, Lawrence J B, Livingston D M. Genes Dev. 1994;8:869–884. doi: 10.1101/gad.8.8.869. [DOI] [PubMed] [Google Scholar]
- 35.Yang X J, Ogryzko V V, Nishikawa J, Howard B H, Nakatani Y. Nature (London) 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 36.Borelli E R, Hen R, Chambon P. Nature (London) 1984;312:608–612. doi: 10.1038/312608a0. [DOI] [PubMed] [Google Scholar]
- 37.Orphanides G, Reinberg D. Nature (London) 2000;407:471–475. doi: 10.1038/35035000. [DOI] [PubMed] [Google Scholar]
- 38.Madisen L, Krumm A, Hebbes T R, Groudine M. Mol Cell Biol. 1998;18:6281–6292. doi: 10.1128/mcb.18.11.6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittschieben B, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 40.Vignali M, Steger D J, Neely K E, Workman J L. EMBO J. 2000;19:2629–2640. doi: 10.1093/emboj/19.11.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerber H P, Hagmann M, Seipel K, Georgiev O, West M A, Litingtung Y, Schaffner W, Corden J L. Nature (London) 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- 42.Eperon L P, Graham I R, Griffiths A D, Eperon I C. Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 43.Roberts G C, Gooding C, Mak H Y, Proudfoot N J, Smith C W J. Nucleic Acids Res. 1998;26:5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]