Abstract
Acute promyelocytic leukemia (APL) cells invariably express aberrant fusion proteins involving the retinoic acid receptor α (RARα). The most common fusion partner is promyelocytic leukemia protein (PML), which is fused to RARα in the balanced reciprocal chromosomal translocation, t(15;17)(q22:q11). Expression of PML/RARα from the cathepsin G promoter in transgenic mice causes a nonfatal myeloproliferative syndrome in all mice; about 15% go on to develop APL after a long latent period, suggesting that additional mutations are required for the development of APL. A candidate target gene for a second mutation is FLT3, because it is mutated in approximately 40% of human APL cases. Activating mutations in FLT3, including internal tandem duplication (ITD) in the juxtamembrane domain, transform hematopoietic cell lines to factor independent growth. FLT3-ITDs also induce a myeloproliferative disease in a murine bone marrow transplant model, but are not sufficient to cause AML. Here, we test the hypothesis that PML/RARα can cooperate with FLT3-ITD to induce an APL-like disease in the mouse. Retroviral transduction of FLT3-ITD into bone marrow cells obtained from PML/RARα transgenic mice results in a short latency APL-like disease with complete penetrance. This disease resembles the APL-like disease that occurs with long latency in the PML/RARα transgenics, suggesting that activating mutations in FLT3 can functionally substitute for the additional mutations that occur during mouse APL progression. The leukemia is transplantable to secondary recipients and is ATRA responsive. These observations document cooperation between PML/RARα and FLT3-ITD in development of the murine APL phenotype.
Acute promyelocytic leukemia (APL) comprises 5–10% of cases of AML and is almost always associated with rearrangement of the retinoic acid receptor α (RARα) gene as a consequence of balanced reciprocal chromosomal translocations. The t(15;17)(q22;q11.2) is the most common such translocation and most APL cells with this translocation express both the promyelocytic leukemia (PML)/RARα and RARα/PML fusion proteins. Mouse models and cell culture assays have demonstrated that the PML/RARα fusion protein contributes to the leukemic phenotype in part by inhibiting differentiation and promoting survival of hematopoietic progenitor cells (1–6).
Data from murine models and analysis of human APL genotypes indicate that the PML/RARα fusion protein is necessary, but not sufficient for the APL disease phenotype. Transgenic mice expressing PML/RARα under the control of the cathepsin G promoter can develop acute myeloid leukemia (AML) with many of the features of APL (1, 2, 7, 8). All transgenic animals expressing this transgene display a myeloproliferative syndrome characterized by elevated numbers of myeloid cells in the marrow and spleen. After a latency of 6–13 months, 15–20% of animals develop an APL-like disease. However, the penetrance increases to about 60% when both the PML/RARα and the reciprocal RARα/PML cDNAs are expressed in early myeloid cells (1, 7). Progression to leukemia in this model system is often associated with acquisition of additional cytogenetic abnormalities (3, 8). The long latency, incomplete penetrance, and cytogenetic changes accompanying disease progression strongly suggest that additional mutations are required for the development of APL in the murine models.
In human APL, all-trans-retinoic acid (ATRA) directly targets the PML/RARα fusion protein (9–11), but does not induce long-term disease-free survival unless it is combined with intensive induction chemotherapy (10, 12–14). Although there are several possible explanations for the inability of ATRA to cure APL, it is plausible that there are second mutations present that are not ATRA sensitive. Perhaps the most convincing evidence to support the hypothesis that additional mutations contribute to the APL phenotype has been recent reports of a high frequency of activating mutations in the FLT3 receptor tyrosine kinase in APL. Approximately 37% of t(15;17) APL patients have mutations that constitutively activate the FLT3 tyrosine kinases (15).
These genetic and epidemiological data suggest a role for mutant FLT3 as a second event in pathogenesis of APL. Internal tandem duplications (ITDs) in FLT3 (FLT3-ITD) are found in 27% of all AML cases (16–18) and the frequency is even higher (37%) in APL patients (15). FLT3-ITDs are constitutively activated and can confer factor-independent growth to 32D (19, 20) or Ba/F3 cells (21–23). Furthermore, when transduced into primary mouse bone marrow cells, FLT3-ITDs induce a myeloproliferative disease in recipient mice (23) with normal maturation and differentiation of myeloid lineage cells. These data indicate that although FLT3-ITD is not sufficient to cause an AML phenotype in this model, it does cause myeloid cell proliferation or survival.
Based on these considerations, and the high frequency of FLT3-ITD mutations in human APL, we have tested the hypothesis that FLT3-ITD cooperates with PML/RARα by using a murine model. Our experimental approach uses retroviral transduction of FLT3-ITD into bone marrow cells derived from cathepsin G-PML/RARα transgenic mice (1), followed by transplantation of transduced cells into lethally irradiated syngeneic recipient mice to test for development of leukemia. We present data consistent with cooperation between PML/RARα and FLT3-ITD in the development of APL. These findings have important implications for pathogenesis of APL and targeted molecular therapy of APL patients with an activated FLT3 gene.
Methods
Mouse Strains and Bone Marrow Transplantation (BMT).
Cathepsin G-PML/RARα transgenic mice were created on a mixed B6×C3H background and maintained by random interbreeding with each other (1, 7). Mice were genotyped either by Southern analysis for the presence of the transgene as described (1, 7) or PCR. A 360-bp PCR product was obtained from the PML/RARα transgene and a 312-bp product from the RARα/PML transgene by using the primer pairs PF, 5′-AGGAAGGTCATCAAGATGGAGTC, and PR2, 5′-GAAGCCCTTGCAGCCCTCACA, or RF, 5′-CTGGGGCTAGGCGGTCCATCCA, and RR, 5′-GACCACTCTCCAGCACCAGCTTCCAG, respectively. BMT assays were carried out as described (23–25). The FLT3-ITD construct used was FLT3-ITD/51 (23) with enhanced green fluorescent protein (EGFP) expressed from an internal ribosomal entry site (IRES) in the murine stem cell virus (MSCV). EGFP-MSCV was a kind gift from W. Pear, University of Pennsylvania, Philadelphia (26), and transduction efficiency as measured by EGFP fluoresence was between 5% and 10% at 24 and 2 h post-spin infections 1 and 2, respectively. A total of 1 × 106 cells were injected into the lateral tail vein of lethally irradiated (2 × 535 cGy) female (B6×C3H)F1 recipient mice. These recipients were chosen as they will readily accept grafts from either B6 or C3H strains. For secondary transplants, mice were sublethally irradiated (535 cGy) and transplanted with 1 × 106 spleen cells. ATRA pellets (10 mg, 60-day release, Innovative Research of America) were implanted s.c.
Analysis of Transplanted Mice.
Peripheral blood was collected from the retro-orbital plexus by using a heparinized capillary, and blood smears were prepared and total and differential blood counts were determined (ADVIA 120 hematology system, Bayer). The spleen, liver, heart, lungs, intestine, kidneys, lymph nodes, and hindlimb bones were collected in 10% neutral buffered formalin for preparation of histological sections as described (23). Single cell suspensions of spleen and bone marrow were stained with allophycocyanin-conjugated antibodies to Gr-1, Mac-1, or CD4 and phycoerythrin-conjugated antibodies to Mac-1, c-Kit and CD8α as described (23).
Southern Analysis.
DNA (20 μg) was digested with EcoRI and Southern hybridization was performed exactly as described (23). The EGFP probe was a 750-bp fragment isolated by using a NcoI/SalI digest from EGFP-murine stem cell virus. The human cathepsin G probe was a 550-bp fragment spanning exons 2 and 3.
Colony Assays.
Colony assays were performed in RPMI media containing IL-3 (6 ng/ml), IL-6 (10 ng/ml), stem cell factor (10 ng/ml), 10% FCS, and 10% bovine citrated plasma. Cells were diluted in this medium, which was clotted by the addition of 10 units/ml of thrombin per ml and immediately transferred into cell culture dishes. Cells were seeded over a range of 1 × 104–2 × 105 cells/ml, and assays were harvested and analyzed at 5–7 days. Clotted colonies were compressed onto slides through silk mesh and dried with Whatman paper. Colonies were stained with Wright and Giemsa. ATRA diluted in 0.1% EtOH was included at 10−6 M where indicated.
Results
Neither PML/RARα Nor FLT3-ITD Independently Induce Acute Leukemia in a Murine BMT Model.
To assess the potential for cooperative effects between FLT3-ITD and PML/RARα, we used the single PML/RARα transgenic model because it has low disease penetrance and long latency (1). Therefore, any change in disease latency or penetrance caused by FLT3-ITD would be clearly evident. As a control, we initially characterized the phenotype of mice transplanted with bone marrow cells derived from PML/RARα transgenic mice. In the PML/RARα asymptomatic transgenics, the white blood cell count shows a slight elevation in mature myeloid cells caused by elevated promyelocyte counts as described (1). Cells from healthy 10-week-old, age-matched transgenic donors were pooled and transduced with a retroviral vector control containing the EGFP gene expressed from an internal ribosomal entry site. We found that transplantation of EGFP-transduced PML/RARα transgenic bone marrow cells into lethally irradiated (B6×C3H)F1 recipients resulted in a similar disease latency, low penetrance, and APL-like phenotype as the reported transgenic model (1), validating the use of the adoptive transfer model for these experiments. After 100 days APL developed in some animals transplanted with these cells. Thus, when the age of the donor mice was taken into account, disease latency was comparable to >180-day latency observed in unmanipulated PML/RARα transgenic animals. This BMT protocol involves treating donor mice with 5-fluorouracil (5-FU) to induce cycling of hematopoietic progenitor cells, which renders them susceptible to retroviral transduction. Although 5-FU treatment may have effects on progenitor cells in other genetic backgrounds (27), we did not find a statistically significant change in disease latency when recipients of cells from 5-FU-treated and untreated animals were compared (data not shown).
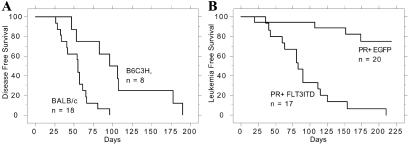
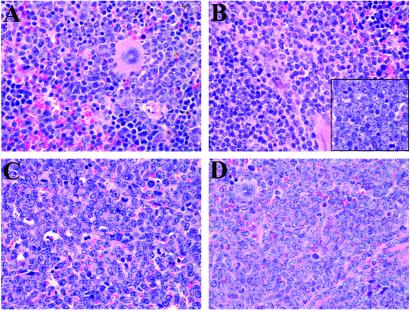
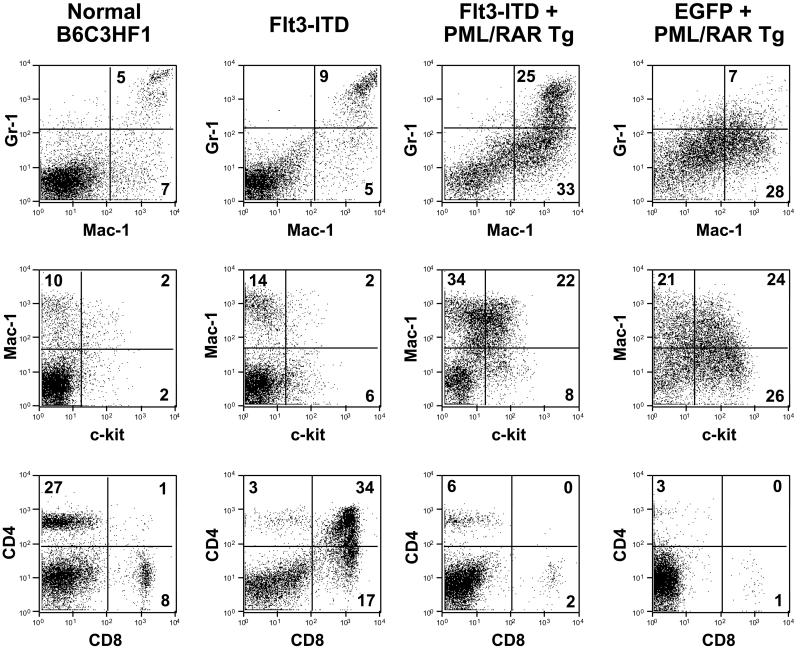
FLT3-ITD mutations alone are also not sufficient to induce acute leukemia in the murine BMT models. FLT3-ITD expression induces a myeloproliferative disease with a median latency of 55 days in BALB/c mice (23). In contrast, we observed that (B6×C3H)F1 recipients of FLT3-ITD-transduced bone marrow (mixed B6/C3H) pooled from wild-type littermates of the transgenics developed features of a T cell lymphoblastic lymphoma with a median latency of 100 days (Fig. 1A). Diseased animals had thymic involvement and associated mediastinal lymphadenopathy, but did not typically display leukocytosis, hepatomegaly, or splenomegaly (Table 1). Histopathologic analysis showed features characteristic of lymphoblastic lymphoma consisting of small to intermediate-sized immature lymphoid cells with dispersed chromatin and scant cytoplasm (Fig. 2B Inset). Immunophenotypic analysis of spleen cells (Fig. 3) and the thymic and mediastinal masses (data not shown) by flow cytometry demonstrated increases in CD4+CD8+ and CD8+ cells. Taken together, these data indicate that in contrast with data from BALB/c backgrounds, in the B6/C3H background FLT3-ITD causes a long latency T cell lymphoblastic lymphoma-like disease, which was not transplantable to secondary recipients with 106 spleen cells (Table 2). Although the basis for the strain-specific difference in latency and phenotype is not known, similar observations of strain-specific influences have been previously reported (28). The phenotype observed with FLT3-ITD alone provides a clear contrast for the comparison of the contribution of the FLT3-ITD allele to PML/RARα leukemic phenotype.
Figure 1.
Kaplan–Meier analysis for leukemia-free survival. The percentage of surviving mice (y axis) is plotted with respect to time in days (x axis). The number of animals per group is indicated. (A) The disease-free survival of BALB/c animals receiving 106 cells transduced with FLT3-ITD compared with the survival of (B6×C3H)F1 animals receiving 106 cells transduced with FLT3-ITD. This plot presents data from four independent experiments. (B) The leukemia-free survival of (B6×C3H)F1 animals receiving PML/RARα transgenic cells (PR) transduced with either EGFP or FLT3-ITD. This plot presents data from two independent experiments.
Table 1.
Gross analysis of PML/RAR+ FLT3-ITD compared to controls
| Survival time, days [median] | White cell count, per μl [median] | Platelet count, per μl [median] | Spleen weight, mg [median] | Liver weight, mg [median] | Enlarged nodes, mg [median] | Histological and immunological phenotype | |
|---|---|---|---|---|---|---|---|
| PML/RAR+ | 40–210 | 6.5 × 103–1.28 × 105 | 35–1,044 | 585–1,500 | 1,800–4,000 | None | AML |
| FLT3-ITD | [80] | [3.5 × 104] | [499] | [1,000] | [2,200] | ||
| n = 14 | n = 12 | n = 15 | n = 13 | ||||
| PML/RAR | 20–314 (n = 7) | 4 × 103 –1 × 105 | 124–614 | 500–718 | 1,600–2,680 | None | AML |
| or PML/ | [173] | [8 × 103] | [334] | [641] | [2,300] | ||
| RAR + EGFP | >330 (n = 7) | n = 12 | n = 12 | n = 10 | n = 6 | ||
| WT+ | 53–190 | 2.4–4 × 103 | 539–798 | 100–570 | No data | 835–1,070 | T lymphoblastic |
| FLT3-ITD | [106] | [3.2 × 103] | [745] | [319] | [1,000] | lymphoma/ | |
| n = 4 | n = 3 | n = 4 | n = 3 | leukemia | |||
| WT +EGFP | >330 | 2–6.8 × 103 | 539–798 | 100–101 | 1,300–1,544 | None | Normal |
| [4.4 × 103] | [745] | [101] | [1,422] | ||||
| n = 2 | n = 3 | n = 2 | n = 2 |
The survival times are taken from the Kaplan–Meier survival plots (Fig. 1). The n value in each field represents the number of animals for which that particular analysis was performed.
Figure 2.
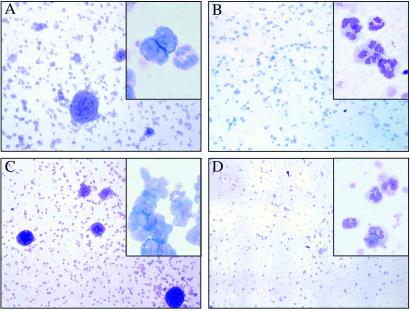
Histopathology of the spleen. Sections of hematoxylin and eosin-stained spleen (×500 magnification) are shown. (A) Wild-type healthy (B6×C3H)F1 mouse spleen transduced with EGFP. (B) Diseased mouse spleen expressing FLT3-ITD on wild-type background, with an Inset showing a section of the lymphoid tumor. (C) Diseased mouse spleen with PML/RARα alone, which developed APL after long latency. (D) Diseased mouse spleen expressing FLT3-ITD on the PML/RARα transgenic background.
Figure 3.
Immunophenotype of spleen cells. The immunophenotypes for normal (B6×C3H)F1 control spleen cells, FLT3-ITD-transduced control spleen cells, FLT3-ITD-transduced PML/RARα transgenic spleen cells, and EGFP-transduced PML/RARα transgenic cells are shown. Spleen cells were stained with allophycocyanin (APC)-anti-Gr-1 and phycoerythrin (PE)-conjugated Mac-1, APC-Mac-1, and PE-c-Kit or APC-CD4 and PE-CD8. The dot plots are gated for live cells based on forward- and side-scatter profiles. The percentages of cells in quadrants of interest are shown.
Table 2.
Transplantability of leukemia cells
| Source of cells | Transplantable | Leukemic mice, # | Median latency |
|---|---|---|---|
| P/R+ITD 1° A | Yes | 6/6 | 37 |
| P/R+ITD 1° B | Yes | 4/4 | 51 |
| P/R+ITD 1° C | Yes | 3/4 | 67 |
| P/R+ITD 2° A | Yes | 4/4 | 37 |
| P/R AML 1° | Yes | 4/4 | 48 |
| P/R asym. 1° | No | 0/4 | — |
| Wt + ITD 1° | No | 0/4 | — |
Animals designated P/R+ITD 1° A, B, C represent different primary mice with AML induced by PML/RARα and FLT3-ITD. P/R+ITD 2° A represents an animal with AML caused by transplantation of cells from P/R+ITD 1° A. P/R AML 1° and P/R asym. 1° represent animals transplanted with PML/RARα transgenic cells that either developed AML or remained asymptomatic, respectively. Wt + ITD 1° represents an animal with lymphoid disease induced by FLT3-ITD in wild-type cells. Cells from the spleen were transplanted in each case.
PML/RARα and FLT3-ITD Cooperate to Induce a Short Latency APL-Like Disease with Complete Penetrance.
PML/RARα transgenic cells were transduced with FLT3-ITD and introduced into lethally irradiated (B6×C3H)F1 recipients to determine the effect of the combination of both oncogenes on the disease phenotype. Control experiments with retroviral transduction of internal ribosomal entry site (IRES)-EGFP into transgenic cells and FLT3-ITD–IRES-EGFP into cells from wild-type littermates were also performed. In three independent experiments, the combination of FLT3-ITD and PML/RARα (F3+PR) induced an APL-like disease in 100% of animals, with a significantly reduced latency compared with EGFP and PML/RARα (EGFP+PR) (Fig. 1B). As expected, a few animals with EGFP+PR eventually developed APL, which was compared with the APL phenotype induced by F3+PR (see below).
Animals that had F3+PR disease displayed leukocytosis, splenomegaly, and hepatomegaly, but did not have mediastinal lymphadenopathy or thymic involvement as was seen with FLT3-ITD in wild-type cells (Table 1). Histopathologic examination of the spleens of F3+PR (Fig. 2) showed a high proportion of immature myeloid cells, in contrast to wild-type or FLT3-ITD-only spleens. Hepatomegaly, associated with infiltration of immature myeloid cells, and bone marrow hyperplasia comprised of immature myeloid lineage cells, was also observed in F3+PR animals. There was also a consistent increase in the proportion of eosinophilic myeloid forms in the spleen, bone marrow, and peripheral blood of F3+PR mice compared with controls. Consistent with the histopathologic analysis, flow cytometric analysis of spleen cells (Fig. 3) demonstrated the presence of an immature population of c-KIT+ (CD117) myeloid cells and an increase in the proportion of Gr-1+ and Mac-1+ cells (Fig. 3), corresponding to the CD34+ Gr-1low population described previously (1).
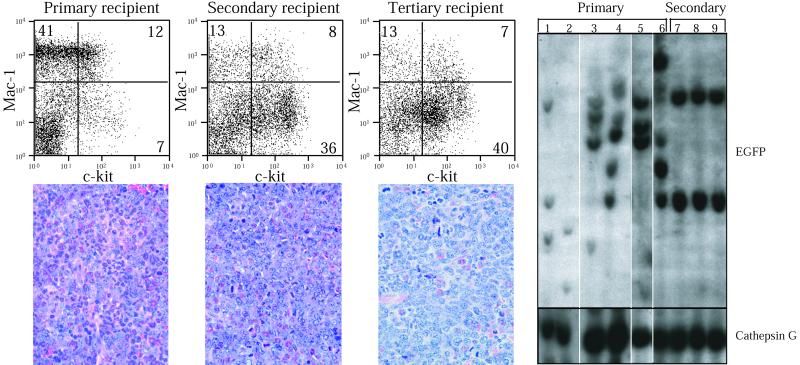
Only a fraction of cells are transduced by retrovirus in the BMT assay, so a critical corollary of the hypothesis that FLT3-ITD mutations cooperate with PML/RARα to cause an APL phenotype is that the FLT3-ITD provirus should be present in leukemic cells. This was demonstrated with Southern blot analysis using a probe for the FLT3-ITD retrovirus (EGFP), where several positive bands were observed in each primary recipient (Fig. 4, lanes 1–6). Southern analysis indicates that the disease is oligoclonal in the primary animals, and clonal selection occurs upon transplanation to secondary recipients. Presence of the PML/RARα transgene was demonstrated with a cathepsin G probe and was also an indication of equivalent DNA loading.
Figure 4.
The FLT3-ITD, PML/RARα-induced disease is transplantable. The c-Kit (allophycocyanin conjugated), Mac-1 (phycoerythrin conjugated) staining profile of spleen cells from primary, secondary, and tertiary recipients of FLT3 and PML/RARα (F3+PR) cells is shown. The percentages of cells in each quadrant is indicated. Below each FACS profile is a histological section of the spleen from a primary, secondary, or tertiary animal, stained with hematoxylin and eosin (×500 magnification). Southern analysis of cells from primary (lanes 1–6) and secondary (lanes 7–9) recipients is shown; each lane represents an individual mouse. The animal represented in lane 6 was the source of primary cells used for secondary transplantation (lanes 7–9), hybridized with EGFP. These data were generated on a single blot. The blot was rehybridized with cathepsin G as a control for DNA loading.
The FLT3 + PML/RARα Disease Is Transplantable to Secondary Recipients.
The APL-like leukemia from F3+PR animals was transplantable into sublethally irradiated secondary recipients (Table 2), as was the less frequently occurring EGFP+PR disease, as previously reported (1). The disease latency in secondary recipients of F3+PR and EGFP+PR leukemic cells (spleen) was comparable; all animals that received 106 cells derived from either leukemic EGFP+PR or F3+PR mice developed disease within 60 days. The disease latency was shorter in tertiary transplant recipients (Table 2), which may indicate selection for initiating cells. Finally, cells from asymptomatic EGFP+PR mice, or cells from FLT3-ITD mice with the lymphoid disease, did not cause leukemia in secondary recipients with a follow-up of 150 days.
Interestingly a slightly greater degree of myeloid maturation, as judged by morphology and surface antigen expression, was noted in primary F3+PR disease compared with the EGFP+PR disease (Figs. 2 and 3), although this distinction was not evident after transfer to 2° or 3° recipients. Examination of the spleen (Fig. 4B) showed the highest proportion of immature myeloid cells in tertiary recipients. Flow cytometry (Fig. 4A) confirmed that the proportion of immature c-Kit+, Mac-1low myeloid cells increased with successive transplantations. These findings are consistent with clonal selection occurring with serial transfer of primary F3+PR cells into secondary recipients. Southern analysis strongly supports this, as dominant leukemic clones emerge in secondary transplants (Fig. 4C, lanes 7–9) from an oligoclonal background seen in the primary transplant mice (Fig. 4C, lane 6).
The FLT3-ITD + PML/RARα Leukemic Cells Respond to ATRA Treatment in Vitro and in Vivo.
F3+PR and EGFP+PR leukemia cells from the spleen were cultured in vitro in clotted plasma in the presence of growth factors and treated with ATRA (10−6 M) as reported (1, 2, 7). ATRA treatment induced differentiation into mature neutrophils but did not result in proliferation (Fig. 5). In contrast, untreated cells continued to grow in compact colonies with a low rate of spontaneous differentiation, as reported (1, 2, 7). The morphology of the cells without ATRA was less differentiated than those growing in the presence of ATRA as seen with higher magnification (Fig. 5 Insets). Both the F3+PR and EGFP+PR cells responded equally well to ATRA, indicating that the F3+PR cells expressed PML/RARα and retained ATRA sensitivity.
Figure 5.
ATRA responsiveness of FLT3-ITD, PML/RARα APL cells. Colony assays with PML/RARα APL cells and FLT3-ITD + PML/RARα APL cells were performed in clotted plasma in the absence or presence of ATRA, dried, and stained with Wright and Giemsa. In each case the colonies are shown at ×100 magnification with an Inset showing the cells at ×500 magnification. (A) PML/RARα cells without ATRA. (B) PML/RARα cells with ATRA 10−6 M. (C) FLT3-ITD + PML/RARα cells without ATRA. (D) FLT3-ITD + PML/RARα cells with ATRA 10−6 M.
Response of the disease to ATRA was also tested in vivo. Mice with a secondary disease resulting from a transplant of either 106 F3+PR or EGFP+PR cells were given 10-mg, 60-day, slow-release ATRA pellets 19 days after transplant (when splenomegaly could be detected by palpation). At 29 days F3+PR animals with ATRA pellets had a marked reduction in splenomegaly (90–100 mg, n = 3) compared with untreated controls (600–1,100 mg, n = 3). Similiarly, at 37 days EGFP+PR animals with ATRA pellets showed a reduction in spleen size (40–55 mg, n = 4) compared with untreated controls (469–564 mg, n = 2). ATRA-treated animals also showed a shift toward normal in the white blood cell differential and a dramatic reduction in histological evidence of leukemia in the spleen (data not shown). This finding further indicates that the F3+PR disease depends on PML/RARα and remains equally responsive to the effects of ATRA.
Discussion
We have tested the potential of the mutant proteins FLT3-ITD and PML/RARα to cooperate to cause APL in a murine BMT assay. The combination of FLT3-ITD and PML/RARα (F3+PR) induces an APL-like leukemia in recipient mice over a period of 7–23 weeks with 100% penetrance. In contrast, the latency is longer and the frequency of disease is lower in recipients of the EGFP-transduced PML/RARα (EGFP+PR) bone marrow and corresponds to the 15–30% penetrance level of APL reported (1) for unmanipulated transgenic mice (Fig. 1A). Alone, FLT3-ITD leads to a lymphoid disease in the mixed B6×C3H background used for these experiments (Fig. 1B). Therefore, we conclude that the observed disease reflects a cooperative rather than an additive effect between FLT3-ITD and PML/RARα.
The APL syndromes induced by either F3+PR or EGFP+PR are very similar. The animals display leukocytosis, splenomegaly, and hepatomegaly (Table 1). Histological and surface marker analysis suggests that the expanded cell populations are comprised of immature myeloid cells (Figs. 2 and 3). The diseases are transplantable to secondary recipients, indicating that they are fully transformed (Table 2). Finally, APL cells from both sets of mice respond in vitro and in vivo to the differentiating effects of ATRA. These data suggest that the FLT3-ITD can functionally substitute for the additional mutations occurring to produce spontaneous leukemias in the PML/RARα transgenic mice.
Although the phenotypes were very similar, primary animals with the F3+PR had a higher proportion of mature cells (Gr-1+, Mac-1+) than EGFP+PR animals. With secondary and tertiary transplants of F3+PR cells, the proportion of immature (c-Kit+, Mac1low) cells increased and the immunophenotype of F3+PR animals was indistinguishable from the EGFP+PR animals. The clonal selection with successive transplants, shown by Southern analysis, may reflect a selection of the clones with enhanced potential to cause leukemia. Clones with differing leukemogenic potential may result from transduction of a mixture of different progenitor cells in the bone marrow of PML/RARα with FLT3-ITD retrovirus or from variation in FLT3-ITD genomic integration sites. Differential cell counts show a consistent increase in neutrophils and eosinophils in the F3+PR peripheral blood, whereas the EGFP+PR samples have more variability, with increased monocytes, eosinophils, or both, often seen in addition to increased neutrophils. This may reflect transduction of FLT3-ITD into a distinct subset of progenitor cells, such as proeosinophils, from total bone marrow of the transgenic mice. It may also reflect differences between the mouse BMT system and the natural evolution of leukemia in humans, as the murine APL model (1, 7), in contrast to human APL, exhibits some differentiation to mature neutrophils. Cumulatively, these data indicate that although there are subtle differences in the phenotype between the F3+PR and EGFP+PR diseases, FLT3-ITD cooperates with PML/RARα to cause APL.
We favor the hypothesis that AML is the consequence of cooperation between at least two classes of mutation, one that impairs hematopoietic differentiation and a second that confers a proliferative and/or survival advantage. Several lines of evidence support this model. A spectrum of AML patients have been identified where two gene rearrangements were observed involving mutation of a transcription factor and constitutive activation of a tyrosine kinase. Examples include AML/ETO or NUP98/HOXA9 translocations in combination with TEL/PDGFβR or BCR/ABL, respectively (29, 30). Avian model systems also have demonstrated mutations in both cell surface receptors and nuclear factors in leukemia development (31, 32). In addition, although activated kinases and altered function of hematopoietic transcription factors are associated with leukemia (33), neither mutation alone is sufficient to induce AML in mouse BMT models (24, 34, 35). Cooperativity of AML1/EVI1 or NUP98/HOXA9 with BCR/ABL in leukemia progression has been recently reported in mouse models (36) (A. Dash and D.G.G., unpublished observation). The long latency APL that spontaneously develops in the PML/RARα transgenics (1) indicates that additional mutations are required for disease progression. In mice, the chromosomal changes observed with APL progression are varied but not random, suggesting that one or several of a particular subset of target genes must be mutated for progression to APL (8). The BMT model described here bypasses this acquisition step and demonstrates that FLT3-ITD can serve as a second hit in the development of APL in the mouse. The experiments presented here demonstrate cooperativity between FLT3-ITD and PML/RARα in the induction of APL, as evidenced by a decrease in latency, an increase in penetrance, and a change in disease phenotype compared with the controls with only PML/RARα or FLT3-ITD. The histological and immunological phenotype of the APL observed with FLT3-ITD and PML/RARα is comparable to PML/RARα transgenic animals with a spontaneous APL. Additional experiments to assess the consequence of the reciprocal RARα/PML in this model system where FLT3-ITD was transduced into PML/RARα and RARα/PML double transgenic cells demonstrated no change in disease latency or phenotype when compared with FLT3-ITD- and PML/RARα-induced disease (data not shown). Mutations in RAS have also been associated with human APLs and interestingly, do not coexist with FLT3 mutations (15, 37, 38). These data suggest that there may be other activating mutations in tyrosine kinases or their downstream effectors in the remainder of APL cases.
Our model provides a useful tool for comparing the gene expression profiles between an APL caused by FLT3 and PML/RARα and spontaneous AML in the PML/RARα transgenic and for studying the disease at a molecular level. Because PML/RARα is a well-characterized protein, it may be possible to understand some of the molecular events that occur in its cooperation with FLT3-ITD. Several promising FLT3 inhibitors are currently being developed (39, 40) (C.L.B., L.M.K., and D.G.G., unpublished observations). This model can also be used to test combinations of ATRA and specific FLT3 inhibitors in mice and may aid in the design of clinical trials in humans. Despite the success with treating APL patients with ATRA, ≈30% of patients will succumb to their disease or complications of therapy. In addition, ATRA therapy must currently be used in combination with anthracycline-based induction chemotherapy for prolonged disease-free survival. Depending on genotype, use of FLT3 inhibitors may improve the prognosis of APL patients who are refractory to ATRA treatment, or they may be useful in targeting or sensitizing the proliferating leukemia stem cells that may escape the effects of ATRA.
Acknowledgments
We thank L. Seaton for administrative assistance and L. Urwiller for expert preparation of the histological samples. We also thank members of the Gilliland and Tenen laboratories for valuable discussions and Dr. J. Growney for helpful comments on the manuscript. This work was supported in part by National Institutes of Health Grants CA66996 and DK50654. L.M.K. is an Associate and D.G.G. is an Associate Investigator of the Howard Hughes Medical Institute. L.M.K. is a fellow of the Leukemia and Lymphoma Society.
Abbreviations
- APL
acute promyelocytic leukemia
- RARα
retinoic acid receptor α
- PML
promyelocytic leukemia
- AML
acute myeloid leukemia
- ATRA
all-trans-retinoic acid
- ITD
internal tandem duplication
- EGFP
enhanced green fluorescent protein
- BMT
bone marrow transplantation
References
- 1.Grisolano J L, Wesselschmidt R L, Pelicci P G, Ley T J. Blood. 1997;89:376–387. [PubMed] [Google Scholar]
- 2.He L Z, Tribioli C, Rivi R, Peruzzi D, Pelicci P G, Soares V, Cattoretti G, Pandolfi P P. Proc Natl Acad Sci USA. 1997;94:5302–5307. doi: 10.1073/pnas.94.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown D, Kogan S, Lagasse E, Wiessman I, Alcalay M, Pelicci P G, Atwater S, Bishop J M. Proc Natl Acad Sci USA. 1997;94:2551–2556. doi: 10.1073/pnas.94.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrucci P F, Grignani F, Pearson M, Fagioli M, Nicoletti I, Pelicci P G. Proc Natl Acad Sci USA. 1997;94:10901–10906. doi: 10.1073/pnas.94.20.10901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grignani F, Ferrucci P F, Testa U, Talamo G, Fagioli M, Alcalay M, Mencarelli A, Srignani F, Peschle C, Nicoletti I, et al. Cell. 1993;74:423–431. doi: 10.1016/0092-8674(93)80044-f. [DOI] [PubMed] [Google Scholar]
- 6.Lavau C, Heard J M, Danos O, Dejean A. Exp Hematol. 1996;24:544–551. [PubMed] [Google Scholar]
- 7.Pollock J L, Westervelt P, Kurichety A K, Pelicci P G, Grisolano J L, Ley T J. Proc Natl Acad Sci USA. 1999;96:15103–15108. doi: 10.1073/pnas.96.26.15103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zimonjic D B, Pollock J L, Westervelt P, Popescu N C, Ley T J. Proc Natl Acad Sci USA. 2000;97:13306–13311. doi: 10.1073/pnas.97.24.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raelson J V, Nervi C, Rosenauer A, Benedetti L, Monczak Y, Pearson M, Pelicci P G, Miller W H., Jr Blood. 1996;88:2826–2832. [PubMed] [Google Scholar]
- 10.Warrell R P, Jr, Maslak P, Eardley A, Heller G, Miller W H, Jr, Frankel S R. Leukemia. 1994;8:929–933. [PubMed] [Google Scholar]
- 11.Ding W, Li Y P, Nobile L M, Grills G, Carrera I, Paietta E, Tallman M S, Wiernik P H, Gallagher R E. Blood. 1998;92:1172–1183. [PubMed] [Google Scholar]
- 12.Fenaux P, Chastang C, Chevret S, Sanz M, Dombret H, Archimbaud E, Fey M, Rayon C, Huguet F, Sotto J J, et al. Blood. 1999;94:1192–1200. [PubMed] [Google Scholar]
- 13.Kanamaru A, Takemoto Y, Tanimoto M, Murakami H, Asou N, Kobayashi T, Kuriyama K, Ohmoto E, Sakamaki H, Tsubaki K, et al. Blood. 1995;85:1202–1206. [PubMed] [Google Scholar]
- 14.Tallman M S, Nabhan C, Feusner J H, Rowe J M. Blood. 2002;99:759–767. doi: 10.1182/blood.v99.3.759. [DOI] [PubMed] [Google Scholar]
- 15.Kottaridis P D, Gale R E, Frew M E, Harrison G, Langabeer S E, Belton A A, Walker H, Wheatley K, Bowen D T, Burnett A K, et al. Blood. 2001;98:1752–1759. doi: 10.1182/blood.v98.6.1752. [DOI] [PubMed] [Google Scholar]
- 16.Yokota S, Kiyoi H, Nakao M, Iwai T, Misawa S, Okuda T, Sonoda Y, Abe T, Kahsima K, Matsuo Y, Naoe T. Leukemia. 1997;11:1605–1609. doi: 10.1038/sj.leu.2400812. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, et al. Blood. 2001;97:2434–2439. doi: 10.1182/blood.v97.8.2434. [DOI] [PubMed] [Google Scholar]
- 18.Stirewalt D L, Kopecky K J, Meshinchi S, Appelbaum F R, Slovak M L, Willman C L, Radich J P. Blood. 2001;97:3589–3595. doi: 10.1182/blood.v97.11.3589. [DOI] [PubMed] [Google Scholar]
- 19.Fenski R, Flesch K, Serve S, Mizuki M, Oelmann E, Kratz-Albers K, Kienst J, Leo R, Schwartz S, Berdel W E, Serve H. Br J Haematol. 2000;108:322–330. doi: 10.1046/j.1365-2141.2000.01831.x. [DOI] [PubMed] [Google Scholar]
- 20.Mizuki M, Fenski R, Halfter H, Matsumura I, Schmidt R, Muller C, Gruning W, Kratz-Albers K, Serve S, Steur C, et al. Blood. 2000;96:3907–3914. [PubMed] [Google Scholar]
- 21.Hayakawa F, Towatari M, Kiyoi H, Tanimoto M, Kitamura T, Saito H, Naoe T. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- 22.Tse K-F, Mukherjee G, Small D. Leukemia. 2000;14:1766–1776. doi: 10.1038/sj.leu.2401905. [DOI] [PubMed] [Google Scholar]
- 23.Kelly L M, Liu Q, Kutok J L, Williams I R, Boulton C L, Gilliland D G. Blood. 2001;99:310–318. doi: 10.1182/blood.v99.1.310. [DOI] [PubMed] [Google Scholar]
- 24.Schwaller J, Frantsve J, Tomasson M, Aster J, Williams I, Van Rompey L, Marynen P, Van Etten R, Ilaria R, Gilliland D G. EMBO J. 1998;17:5321–5333. doi: 10.1093/emboj/17.18.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Q, Schwaller J, Kutok J, Cain D, Aster J C, Williams I R, Gilliland D G. EMBO J. 2000;19:1827–1838. doi: 10.1093/emboj/19.8.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Persons D A, Allay J A, Allay E R, Smeyne R J, Ashmun R A, Sorrentino B P, Nienhuis A W. Blood. 1997;90:1777–1786. [PubMed] [Google Scholar]
- 27.Schwaller J, Parganas E, Wang D, Cain D, Aster J C, Williams I R, Lee C K, Gerthner R, Kitamura T, Frantsve J, et al. Mol Cell. 2000;6:693–704. doi: 10.1016/s1097-2765(00)00067-8. [DOI] [PubMed] [Google Scholar]
- 28.Szymanska H, Sitarz M, Krysiak E, Piskorowska J, Czarnomska A, Skurzak H, Hart A A, de Jong D, Demant P. Int J Cancer. 1999;83:674–678. doi: 10.1002/(sici)1097-0215(19991126)83:5<674::aid-ijc18>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 29.Golub T R, Barker G F, Lovett M, Gilliland D G. Cell. 1994;77:307–316. doi: 10.1016/0092-8674(94)90322-0. [DOI] [PubMed] [Google Scholar]
- 30.Ahuja H G, Popplewell L, Tcheurekdjian L, Slovak M L. Genes Chromosomes Cancer. 2001;30:410–415. doi: 10.1002/1098-2264(2001)9999:9999<::aid-gcc1108>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 31.Graf T, von Weizsaecker F, Grieser S, Coll J, Stehelin D, Patschinsky T, Bister K, Bechade C, Calothy G, Leutz A. Cell. 1986;45:357–364. doi: 10.1016/0092-8674(86)90321-1. [DOI] [PubMed] [Google Scholar]
- 32.Kahn P, Frykberg L, Brady C, Stanley I, Beug H, Vennstrom B, Graf T. Cell. 1986;45:349–356. doi: 10.1016/0092-8674(86)90320-x. [DOI] [PubMed] [Google Scholar]
- 33.Lacronique V, Boureux A, Valle V D, Poirel H, Quang C T, Mauchauffe M, Berthou C, Lessard M, Berger R, Ghysdael J, Bernard O A. Science. 1997;278:1309–1312. doi: 10.1126/science.278.5341.1309. [DOI] [PubMed] [Google Scholar]
- 34.Tomasson M H, Williams I R, Hasserjian R, Udomsakdi C, McGrath S M, Schwaller J, Druker B, Gilliland D G. Blood. 1999;93:1707–1714. [PubMed] [Google Scholar]
- 35.Castilla L H, Garrett L, Adya N, Orlic D, Dutra A, Anderson S, Owens J, Eckhaus M, Bodine D, Liu P P. Nat Genet. 1999;2:144–146. doi: 10.1038/13776. [DOI] [PubMed] [Google Scholar]
- 36.Cuenco G M, Ren R. Oncogene. 2001;20:8236–8248. doi: 10.1038/sj.onc.1205095. [DOI] [PubMed] [Google Scholar]
- 37.Kiyoi H, Towatari M, Yokota S, Hamaguchi M, Ohno R, Saito H, Naoe T. Leukemia. 1998;12:1333–1337. doi: 10.1038/sj.leu.2401130. [DOI] [PubMed] [Google Scholar]
- 38.Kiyoi H, Naoe T, Nakano Y, Yokota S, Minami S, Miyawaki S, Asou N, Kuriyama K, Jinnai I, Shimazaki C, et al. Blood. 1999;93:3074–3080. [PubMed] [Google Scholar]
- 39.Levis M, Tse K, Smith D, Garrett E, Small D. Blood. 2001;98:885–888. doi: 10.1182/blood.v98.3.885. [DOI] [PubMed] [Google Scholar]
- 40.Zhao M, Kiyoi H, Yamamoto Y, Ito M, Towatari M, Omura S, Kitamura T, Ueda R, Saito H, Naoe T. Leukemia. 2000;14:374–378. doi: 10.1038/sj.leu.2401680. [DOI] [PubMed] [Google Scholar]