Abstract
Previously described animal models for Helicobacter pylori infection have been limited by cumbersome host requirements (e.g., germ-free conditions or unusual species) or are applicable to only special subsets of H. pylori strains (e.g., fresh clinical isolates or animal-adapted derivatives). Here, we report that 5- to 6-day-old outbred CD-1 (ICR) suckling mice support 24-h colonization of all H. pylori strains tested (SS1, 26695 SmR-1, 43504 SmR-1, and G27 SmR-1), including lab-passaged strains that cannot be adapted for colonization of adult animals. Total colony-forming units (cfu) recovered from infection with lab-passaged strains did not differ from those with mouse-adapted SS1. We also tested this model's ability to detect colonization defects in strains carrying mutations in known virulence genes by coinfecting with wild-type H. pylori and measuring differential recovery. This competition assay identified colonization defects in several classes of known attenuated mutants, including those defective in acid resistance (ureA), metabolism (frdA), motility (motB), and chemotaxis (cheY). A mutant defective in copA (copper transporting P-type ATPase) is nonattenuated in adult and infant mice. Possibly because of the limited duration of infection, our model did not identify defects in vacuolating cytotoxin (vacA) or γ-glutamyltranspeptidase (ggt) as attenuating, in contrast to results from other animal models. We also identified a new virulence gene (HP0507) encoding a conserved hypothetical protein, which is important for colonization in our model. The suckling mouse model offers a rapid method to identify colonization defects in any H. pylori strain and may have utility as a new tool for studying immunity to primary infection.
The bacterial pathogen Helicobacter pylori chronically colonizes the gastric mucosa of humans. After decades of exposure to this microbe, humans suffer increased risk for various gastrointestinal (GI) pathologies including GI ulcers, gastritis, and gastric cancer (1, 2). Accordingly, there has been much interest in modeling H. pylori GI colonization in experimental animals in which prophylactic measures, such as vaccines and drug therapies, can be evaluated. Several animal models for H. pylori infection have been developed, including nonhuman primates (3), gnotobiotic piglets (4), beagle dogs (5), Mongolian gerbils (6), guinea pigs (7), rats (8), and mice (9, 10). In general, these animals only support colonization by special H. pylori strains (fresh clinical isolates or animal-adapted derivatives). Some clinical isolates spontaneously colonize certain animal species in high numbers, whereas other strains initially do not, although a hypercolonizing derivative can be selected through an adaptation process (10, 11). Transgenic mice expressing the Lewis-b blood group antigen are capable of being colonized by H. pylori without prior animal adaptation, provided the bacteria express the BabA adhesin (12). To a greater or lesser degree, all of these animal models exhibit progression to a disease that mimics human H. pylori-associated pathology and thus are useful for studying chronic infection, gastric inflammation and injury, and host immune responses.
Despite the usefulness of these models, strain restriction has hindered progress in studying H. pylori pathogenesis. The most widely used animal model involves infection of adult mice with the mouse-adapted strain SS1, which consistently produces chronic disease. Unfortunately, SS1 also exhibits spontaneous loss of infectivity, is notoriously difficult to manipulate genetically, and its genome has not been sequenced. Moreover, because only fresh clinical isolates have been reported to colonize adult animals, several lab-passaged strains that are well characterized and/or easily manipulated genetically have not been usable for animal studies. The piglet-adapted strain 26695 is appealing because its genomic DNA sequence was recently reported (13); however, it cannot be adapted to infect mice, a more convenient and less expensive animal model with better host genetics and immunological tools.
As H. pylori is a highly heterogeneous species, there is clearly a need for a model that can be used to study and compare all strains. Comparison of H. pylori genomes has revealed substantial diversity (14), including differences in vacA alleles (15), cag pathogenicity island status, and restriction-modification systems (16). Interpretation of animal results by using different uncharacterized strains is therefore difficult; conflicting reports in the literature may be because of the use of mutants made in different backgrounds.
Furthermore, the exclusive study of strains that have been selected for their ability to colonize animals chronically may result in an incomplete or inaccurate picture of H. pylori host–pathogen interactions. Such selection of strain subsets might bias results toward over-emphasizing the roles of certain virulence determinants that are important to establishing chronic infection in that particular host (e.g., immune evasion, inflammation, gastric injury). Other H. pylori factors that make critical contributions to the earliest steps in the infection process, perhaps those most important for vaccine development efforts to target, may be overlooked as a result. Finally, focusing on only animal-adapted strains is incompatible with the goal of understanding the role of strain variability in the virulence and epidemiology of this globally distributed human pathogen.
Here, we report the use of 5- to 6-day-old suckling infant mice to assay H. pylori colonization in a short-term infection. We show that infant mice are permissive for colonization by all H. pylori strains tested, including lab-passaged strains, which are not adaptable for chronic infection of adult animals. Moreover, our assay shows no significant difference in recovery between the mouse-adapted strain SS1 and any other tested strains. We validate the model by demonstrating that it can identify colonization defects for known attenuating mutations. Furthermore, we identify a virulence gene that previously has not been tested in animals. Although short-term colonization has its limitations, the infant mouse model is capable of identifying genes critical for early establishment of infection, and it is an affordable, convenient system for rapidly screening large numbers of mutants. Furthermore, by allowing use of any strain, this model promises to provide a means to study subtle properties that vary between strains but may have critical influences on the success of this organism as a human pathogen.
Materials and Methods
Bacterial Strains and Culture.
H. pylori strains were cultured on tryptic soy agar containing 5% (vol/vol) defibrinated horse or sheep blood (HemoStat Labs, Dixon, CA) or 10% (vol/vol) newborn calf serum (GIBCO/BRL). Antibiotics were used at the following concentrations: 5 μg/ml trimethoprim (T), 10 μg/ml vancomycin (V), 8 μg/ml amphotericin B (A), 5 μg/ml cefsulodin (C), 2 μg/ml bacitracin (B), 10 μg/ml nalidixic acid (N), 20 μg/ml streptomycin (Sm), 10 μg/ml chloramphenicol (Cm), 20 μg/ml kanamycin (Km); 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) was used at 80 μg/ml. Combinations of antibiotics are denoted by combining together the above abbreviations. In liquid culture for mouse experiments, TVACSm was used for G27 SmR-1, VABN for AH244 and G27, and TVAC for all other strains. Bacteria were incubated at 37°C in an atmosphere containing 5% O2, 10% CO2, and 85% N2. See Table 1 for a list and descriptions of H. pylori strains used in this study.
Table 1.
H. pylori strains
| Strain | Description or gene function affected | Ref. |
|---|---|---|
| SS1 | Motile, mouse-adapted clinical isolate | 10 |
| AH244 | Motile, mouse virulent clinical isolate | 17 |
| 26695 SmR-1 | Spontaneous Smr derivative of 26695 | 13 |
| 43504 SmR-1 | Motile lab-passaged type strain | ATCC |
| G27 | Motile clinical isolate, not mouse virulent | 18 |
| G27 SmR-1 | Nonmotile, spontaneous Smr derivative of G27 | 19 |
| copA∷cat AH244 | Copper transporting P-type ATPase | 17 |
| frdA∷cat AH244 | Fumarate reductase | 17 |
| motB∷kan G27 | Motility, also nonflagellated | K. M. Ottemann |
| cheY∷kan G27 | Chemotaxis | T. M. Andermann, Y.-T. Chen, and K. M. Ottemann, unpublished work |
| vacA∷kan G27 SmR-1 | Vacuolating cytotoxin | 19 |
| ggt∷kan G27 SmR-1 | γ-glutamyltranspeptidase | This work |
| ureA∷kan G27 SmR-1 | Urease subunit | 19 |
Vibrio cholerae O395-Sm WT and Tcp2, infant mouse colonization-defective (20), were grown in LB containing 100 μg/ml Sm at 37°C. Escherichia coli DH10B (GIBCO/BRL) was used for propagation of plasmids and was grown in LB containing 50 μg/ml Km, 20 μg/ml Cm, and/or 100 μg/ml ampicillin as appropriate.
Plasmid Construction and Generation of Mutants.
H. pylori mutants ggt∷kan and HP0507∷kan were constructed by in vitro mutagenesis as described (19). pBAD-TOPO (Invitrogen) was used for cloning ggt by using primers ggt-F, 5′-CCCATAGCGTTTTGATCAAATAAGCC-3′, and ggt-R, 5′-TCCTTGATCCGTTGAACCATAGA-3′. pMON/HP0508, which contains the 3′ half of HP0507, was used for mutagenizing HP0507. The insert was amplified by using primers Klas-F, 5′-GCGCAGGGCTTGTGGATAAAGCTAA-3′, and Klas-R, 5′-CCCCCCGCTCACGCTCAACGCTCCC-3′.
The ureA, vacA, cheY, motB, and ggt mutants were confirmed as null mutants by urease assay (19), Western blot (19), chemotaxis assay (T. M. Andermann, Y.-T. Chen, and K. M. Ottemann, unpublished work), motility assay (K. M. Ottemann, personal communication), and γ-glutamyltranspeptidase kit assay (Sigma), respectively.
Infant Mouse Colonization Assays.
H. pylori.
ICR mice (5–6 day-old, Charles River Breeding Laboratories) were starved by separation from their mothers for 6 h before intragastric inoculation. H. pylori cultures were diluted from plates into 1 ml of Brucella broth layered over 5 ml per well of agar with TVAC (described above) in six-well culture dishes and shaken overnight. Bacteria were pelleted and resuspended in the same volume of Brucella broth. Gastric tube lavage was used to deliver ≈107–108 organisms in a 50–100 μl volume directly into the stomach. For competitions, equal quantities of mutant and wild-type (WT) were mixed. Unless otherwise indicated, stomachs were homogenized in 5 ml of Brucella broth 20–24 h postinfection (p.i.). One hundred μl was plated to determine output cfus. For G27 SmR-1, homogenates were plated on TVACSm and X-Gal; for AH244 and G27, VABN was used. For competitions of G27 SmR-1 mutants, hopZ∷kanlacZ G27 SmR-1 (18) was used as WT. We have determined that hopZ∷kanlacZ G27 SmR-1 has a less than two-fold defect in competition with G27 SmR-1 (data not shown). Mutant and WT were distinguished by blue/white screening after growth on plates containing X-Gal. For competitions of all other strains, ratios were determined by testing 50–150 colonies per mouse for growth on plates containing Km or Cm.
Vibrio cholerae.
An overnight culture of V. cholerae was diluted 1:2,000 in LB and inoculated at ≈105 organisms (50 μl volume). Twenty to twenty-four hours p.i., stomachs and small intestines were homogenized separately in 5 ml of LB. Dilutions of the homogenates were plated on LB agar containing Sm to determine cfus per animal.
Tissue Sections and Microscopy.
Infected stomachs were dissected and immediately preserved in neutral buffered formalin [3.7% (vol/vol) formaldehyde, 29 mM NaH2PO4, 46 mM Na2HPO4]. Tissue was paraffin-embedded and Warthin Starry silver-stained for detection of H. pylori. Histopathology was performed at the Massachusetts Institute of Technology Division of Comparative Medicine.
Protein Sequence Alignments.
Protein sequences were aligned by using the CLUSTALW program (21).
Results
Several H. pylori Strains Colonize the Infant Mouse Stomach.
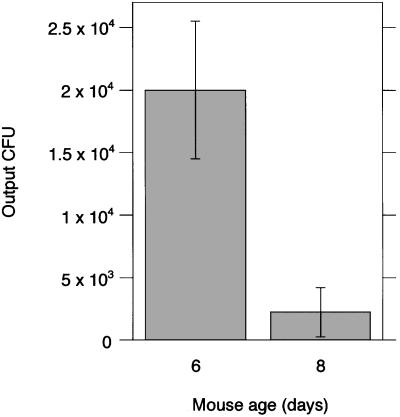
We inoculated ICR infant mice intragastrically with H. pylori strains SS1, G27 SmR-1, 26695 SmR-1, and 43504 SmR-1. SmR-1 strains are spontaneous Smr derivatives of the indicated parent strains. Eight-day-old mice did not support infection as well as 6-day-old mice. Nine-fold fewer organisms were recovered 20–24 h p.i. from 8-day-old mice compared with 6-day-old mice, even with a larger inoculation dose for the former (Fig. 1). To maximize recoverable organisms, 5- to 6-day-old mice were used for all subsequent studies.
Figure 1.
Dependence of mouse age on cfus recovered. 43504 SmR-1 was inoculated at 7.7 × 107 and 1.7 × 108 cfus into 6- and 8-day-old mice, respectively. Results are the averages of cfus recovered from three and seven mice, respectively. SDs are shown.
In contrast with other models, motility does not play a role in colonization in the absence of competition, because the nonmotile G27 SmR-1 colonized as well as motile strains SS1, 26695 SmR-1, and 43504 SmR-1 (Table 2). In addition, Ottemann and Lowenthal (22) analyzed a flagellated, nonmotile mutant of SS1 and found that at high doses, it could colonize FVB/N mice for 2 weeks. One interpretation they present is that some mouse strains are more permissive for colonization by nonmotile H. pylori. It is important to note that strains not adapted for mouse infection (G27 SmR-1, 43504 SmR-1, and 26695 SmR-1), and not capable of being adapted because of a history of lab-passaging (Table 1), colonized as well as mouse-adapted SS1.
Table 2.
Different H. pylori strains colonize the infant mouse stomach
| Strain | No. of mice | cfus input | cfus output | Motility | Strain details |
|---|---|---|---|---|---|
| SS1 | 5 | 1.7 × 108 | 3.3 × 104 ± 1.2 × 104 | + | Mouse-adapted |
| G27 SmR-1 | 4 | 1.4 × 108 | 4.2 × 104 ± 3.6 × 104 | − | Lab-passaged |
| 26695 SmR-1 | 4 | 7.5 × 108 | 1.4 × 104 ± 9.0 × 103 | + | Piglet-adapted |
| 43504 SmR-1 | 3 | 7.7 × 107 | 2.0 × 104 ± 5.5 × 103 | + | Lab-passaged |
Results are the averages of total cfus recovered. SDs are shown.
Microscopically, H. pylori SS1 could be detected in silver-stained, paraffin-embedded tissue sections of infant mouse stomachs 24 h p.i. (data not shown). Low numbers of small (<3 μm), plump, slightly curved rods were detected on the mucosal surface within deep mucosal folds. No organisms were identified within gastric pits, glandular lumens, or epithelium. Tissue did not show any evidence of inflammation. We also attempted to visualize colonization of G27 SmR-1, but no organisms could be found (data not shown). It is possible that because of its lack of motility, G27 SmR-1 does not colonize the same location and was washed away during processing of the tissue for sectioning.
H. pylori Is Better Able to Colonize the Infant Mouse Stomach than the Intestine.
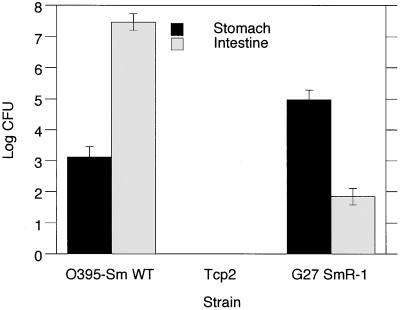
Because the recovered cfus from the stomach were 3–4 logs lower than the inoculating dose, we were concerned that these organisms might represent a fraction of the inoculum that simply had not washed out of the stomach rather than organisms truly colonizing the stomach (Table 2). To address this issue, we tested whether V. cholerae, a bacterium known to colonize and replicate in the small intestine of infant mice, could also colonize the stomach in comparable numbers. V. cholerae O395-Sm WT was inoculated at ≈105 cfu, which is sufficient to result in colonization of the small intestine. A much lower dose of V. cholerae (compared with H. pylori) had to be used, because a dose of 107 is lethal to infant mice. Twenty to twenty-four hours p.i., H. pylori was recovered from stomach and intestine at ≈105 and ≈102 cfu, respectively, whereas V. cholerae O395-Sm WT was recovered from stomach and intestine at ≈104 and ≈107 cfu, respectively (Fig. 2). There is a 3-log difference in cfus recovered from these two GI locations, which is inverted for H. pylori and V. cholerae; this result strongly suggests that these organisms colonize these sites differently. Because V. cholerae does not possess urease and is acid-sensitive, it is likely that the V. cholerae recovered from the stomach were actually present because of reflux from the intestine. We tested this possibility with strain Tcp2, a V. cholerae mutant unable to colonize the intestine, and found that this mutant was not recovered from the stomach, suggesting that clearance mechanisms (e.g., peristalsis, mucus flow) are adequate in the infant mouse to clear all noncolonizing organisms from both the stomach and upper intestine within 20–24 h.
Figure 2.
V. cholerae and H. pylori recovered from stomach and intestine. V. cholerae O395-Sm WT, V. cholerae Tcp2 (intestinal colonization defective), and H. pylori G27 SmR-1 (WT) were inoculated at 2.9 × 106, 1.9 × 105, and 1.3 × 108 cfus, respectively. Results are the averages of total cfus recovered from four, four, and five mice, respectively. SDs are shown.
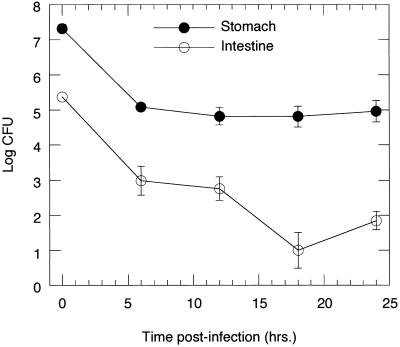
To evaluate the progression of colonization over time in these two tissues, we compared the cfus of H. pylori recovered from the stomach vs. the small intestine (above the cecum) over time at 6-h intervals p.i. In the stomach at 6 h, there was a large drop in organisms recovered compared with the inoculation dose, but the cfus then remained steady at ≈105 between 6 and 24 h (Fig. 3). In the small intestine, the cfus declined steadily from ≈105 to 102. Furthermore, the bacteria in the stomach outnumbered those in the intestine by a factor of 102-104 at all time points. These results suggest that in the stomach, H. pylori are surviving and perhaps replicating, whereas in the upper intestine, they seem to be dying or washing out over time. Because the intestine has a much larger surface area for colonization than the stomach, these data suggest that H. pylori is better able to colonize the stomach. No H. pylori could be recovered 48 h p.i. (data not shown), possibly because of the return of the infants to their mothers and their subsequent feeding on milk, which contains lactoferrin, a substance inhibitory to H. pylori growth (23).
Figure 3.
H. pylori recovered from the stomach and intestine over 24 h. Inoculating dose was 1.3 × 108 cfu. Results shown are the averages of cfus recovered from two to five mice per time point. SDs are shown.
In Vivo Competition Assay Can Identify Attenuated Mutants.
We used a competition assay to determine colonization of H. pylori mutants with previously described in vivo phenotypes, both attenuating and nonattenuating (Table 3). Competitive colonization assays provide greater sensitivity in identifying mildly attenuating mutations as coinfecting with WT provides an internal reference standard. Infant mice were inoculated with roughly equal cfus of mutant and WT bacteria. Twenty to twenty-four hours p.i., organisms were recovered, and ratios of mutant to WT were determined by blue/white screening (G27 SmR-1 strains) or growth after transferring colonies to plates containing the antibiotic to which the mutant is resistant. Competitive indices (C.I.s) were calculated as the ratio of the number of mutants over WT recovered (output) divided by the ratio of the number of mutants over WT inoculated (input).
Table 3.
In vivo competitions
| Strain | Infant mice
|
Previously tested animal models
|
||||
|---|---|---|---|---|---|---|
| Rodents
|
Piglets
|
|||||
| No. of mice | C.I. | Colonization | Ref. | Colonization | Ref. | |
| copA∷cat AH244* | 4 | 0.98 ± 0.19 | + | 17 | NA | |
| vacA∷kan G27 SmR-1 | 5 | 0.98 ± 0.25 | +, +/− | 24, 25 | + | 26 |
| ggt∷kan G27 SmR-1 | 4 | 0.93 ± 0.55 | +/−, − | 27, 28 | +/− | 28 |
| motB∷kan G27† | 6 | 0.33 ± 0.09 | +/−, − | 22, 29, 30 | − | 31 |
| cheY∷kan G27 | 5 | 0.26 ± 0.07 | − | 32 | − | 32 |
| HP0507∷kan G27 SmR-1 | 4 | 0.03 ± 0.02 | NA | NA | ||
| frdA∷cat AH244* | 6 | <4.5 × 10−4 | − | 17 | NA | |
| ureA∷kan G27 SmR-14 | 4 | <4.4 × 10−4 | − | 33 | − | 34 |
Competitive index (C.I.) = (cfu muant/cfu WT) output/(cfu mutant/cfu WT) input. SDs are shown. Mutants with a C.I. <0.5 are considered attenuated. NA, data not available. +, colonization same as WT. +/−, colonization lower than WT. −, no colonization.
Same mutant as used in reference.
† Same phenotype as mutants used in reference(s). All other mutants have an insertion in the same gene, but in a different strain background than used in the reference(s).
Mutants with defects in ureA (urease subunit), frdA (fumarate reductase), cheY (chemotaxis), or motB (motility) were attenuated in this model, with C.I.s of <4.4 × 10−4, < 4.5 × 10−4, 0.26, and 0.33, respectively (Table 3). These results are in agreement with published data. It has been reported that ureA (33) and frdA (17) mutants cannot be recovered from mice, and that motility mutants are defective for colonization of mice (35), gnotobiotic piglets (31), and Mongolian gerbils (30). However, in our model, the motB mutant can be recovered, but competes less well in the presence of WT. We did not test the motB mutant in the absence of WT but expect that no defect would be detectable, which is the case for the spontaneously nonmotile G27 SmR-1 (Table 2).
Mutants defective in copA (copper transporting P-type ATPase), ggt (γ-glutamyltranspeptidase) or vacA (vacuolating cytotoxin) had C.I.s of 0.98, 0.98, and 0.93, respectively, indicating that they are fully competent for the colonization of suckling mice (Table 3). Our result with the copA mutant is in agreement with the report of Ge et al. (17), which also shows no colonization defect of this same mutant in adult mice. However, in some other animal models, ggt mutants (27, 28) and vacA (25) are defective for colonization. The two proposed functions for bacterial γ-glutamyltranspeptidase, protection against oxidative damage and de novo amino acid synthesis, may not be crucial in a short-term infection without inflammation. It was not surprising that the vacA mutant was not colonization-defective in the suckling mouse model. Some investigators have reported that vacA mutants are not attenuated (24, 26), whereas Salama et al. show that a vacA-associated colonization defect is apparent in competition with WT or when the mutant is administered alone at low inoculation doses (25). We did not test lower inoculation doses in our model.
Identification of a New Virulence Gene in the Suckling Mouse Model.
During the course of assaying mutants for another study, we identified HP0507 as a gene that is important for colonization in infant mice. A mutant defective in HP0507, which encodes a conserved hypothetical protein, gave a C.I. of 0.03, indicating it is severely attenuated (Table 3).
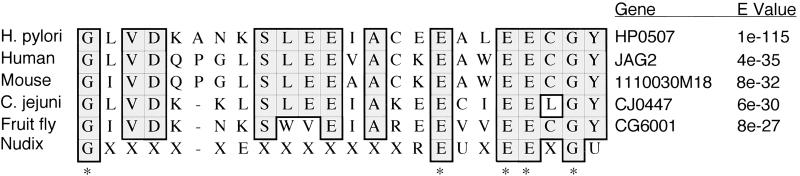
In an attempt to determine a possible function for HP0507, we performed a BLASTP search against known protein sequences (36). Sequences with the highest identities were found in hypothetical proteins from Homo sapiens, Mus musculus, Campylobacter jejuni, and Drosophila melanogaster (Fig. 4). The search also revealed a putative Nudix (nucleoside diphosphate moiety X-linked) motif, which is present in a wide diversity of proteins that hydrolyze substrates composed of a nucleoside diphosphate linked to some other moiety, X (37). Nudix hydrolases are thought to function in cleansing the cell of potentially deleterious metabolites (38). HP0507 protein and its closest homologues show partial identity to the Nudix consensus sequence (39), matching 5 of 9 consensus residues (Fig. 4).
Figure 4.
CLUSTALW protein sequence alignment of the Nudix motif consensus sequence with the Nudix-like domain of H. pylori HP0507 and its homologues in Homo sapiens (human), Mus musculus (mouse), Campylobacter jejuni, and Drosophila melanogaster (fruit fly). Identical residues are shaded. X, any residue; U, residues usually leucine (L), valine (V), or isoleucine (I); *, residues in consensus sequence with matches in HP0507.
Discussion
The immature GI tract of infant mice permits colonization by a variety of microorganisms. For example, infant mice have been used to study V. cholerae (40), Legionella pneumophila (41), Pseudomonas aeruginosa (42), Neisseria meningitidis (43), E. coli (44), and Candida albicans (45). It might be argued that this model is too permissive to provide useful information about virulence properties, but this concern has not proven true for V. cholerae, a pathogen for which infant mice have been used extensively. Indeed, for all strains tested so far, V. cholerae mutants that display colonization defects in the infant mouse show similar or even more severe colonization defects in volunteer human subjects (20). Similarly, a V. cholerae strain with a mutation in a putative colonization factor but that showed no defect in the infant mouse model (46, 47) was eventually found to colonize humans normally as well (48). Thus, infant mice provide a simple and accurate model for studying the interaction of this pathogen with the human small bowel mucosa. Based on the results presented here, infant mice also are useful for studying H. pylori, with the potential of providing a better understanding of its interaction in the human stomach.
Elucidating the mechanisms of H. pylori pathogenesis has been challenging for a number of reasons. In vivo studies necessitate using certain strains that colonize well, but may not have other desirable qualities. The sequenced strain 26695 was previously known to colonize only gnotobiotic piglets, a model that is widely trusted but much less convenient than small animals such as mice. Some strains that colonize mice do not do so consistently. The most commonly used strain, SS1, infects mice consistently but is difficult to transform and can lose infectivity during routine laboratory manipulations such as passaging and electroporation (49). Because of these difficulties, in vitro studies often are performed with the strain of choice, whereas mutants are recreated in an animal-adapted strain for in vivo studies. Because H. pylori are highly diverse, it would be beneficial to be able to use directly or compare any strain(s) in one animal model. The model we present in this article enables researchers to attain consistent infection in infant mice by using any strain of H. pylori.
In 5- to 6-day-old mice, we observed the same level of colonization in the stomach throughout most of the 24-h infection. However, no organisms could be recovered at 48 h (24 h after returning the mice to their mothers; data not shown). Also, older mice (8 days old) were less efficient at supporting colonization, with a 9-fold lower average recovery (Fig. 1). Why older mice are more resistant to H. pylori infection is unknown, but this fact may be because of a better-developed innate immune system, more active mechanical clearance mechanisms, or the establishment of a normal flora relative to their immature counterparts. Colonization is specific for the stomach, with a steady colonization over 24 h and many fewer organisms surviving in the intestine (Figs. 2 and 3). At 20–24 h, ≈105 organisms per 100 mg of tissue (infant mouse stomach is ≈10 mg) were routinely recovered (Table 2), similar to 105-106 cfu per 100 mg of tissue (10) and more than 103-104 cfu per 100 mg of mucus (11) reported for infection of mouse-adapted strains in adult mice. Unlike in other models, nonmotile strains inoculated alone were able to colonize infant mice (Table 2), although a nonmotile mutant, defective in motB, was less efficient at colonizing in competition with the isogenic WT (Table 3). Although the infection period for this model is extremely short, we were able to demonstrate that we could identify colonization defects associated with mutations in genes of varied function, including acid resistance, central metabolism, motility, and chemotaxis, that had been shown to be attenuated in other animal models. Moreover, we used this model to identify a virulence gene, HP0507, that encodes a conserved hypothetical protein with a putative Nudix domain (Fig. 4). Mutation of HP0507 produced a highly attenuated phenotype in infant mice (Table 3).
Our model did not detect mutations in ggt or vacA as attenuating, perhaps because of the use of different strains from those used in published reports. Alternatively, our model may be able to identify only a subset of the mutations that would be attenuating in a chronic infection of adult animals. This idea is supported by the fact that in the case of vacA, the reported difference in infection was very subtle and only discernible when infecting with much lower doses than routinely used by most researchers (25). Its inability to detect some mutants as attenuated may be caused by the short infection time, different resident microflora, or the limited immune system of infant mice.
Because of the brief infection period, the mutants identified will likely be those important early in the establishment of infection. With the convenience and affordability of a small animal model, the use of infant mice to study the H. pylori–host interaction should facilitate screening of mutants for severe colonization defects in the earliest stage of gastric infection. Understanding the earliest events in H. pylori colonization may be critical to the development of effective immunoprophylactic approaches to blocking initial infection. It should be noted that infant mice have been used previously to define protective immune responses by means of passively administered antibodies (50) or with antibody-secreting tumors (51). Although these studies have focused on intestinal pathogens like V. cholerae and Salmonella typhimurium, our results suggest that infant mice also might be explored as a simple model to evaluate the specificity and class of antibodies that are most effective at preventing initial colonization by H. pylori rather than clearance of the organism after chronic infection has been established.
Acknowledgments
We thank Karen Ottemann for strains G27, cheY∷kan G27, Jim Fox for strains AH244, copA∷cat AH244, and frdA∷cat AH244, Jack Pappo for SS1, and Klas Jönsson for pMON/HP0508. We thank Cindy Ku for technical assistance and Su Chiang, Qing Xu, and Jon Blum for critical reading of this manuscript. This work was supported by an American Cancer Society Postdoctoral Fellowship (to B.P.G.) and National Institutes of Health Grant AI26289 (to J.J.M.).
Abbreviations
- cfu
colony-forming units
- C.I.
competitive index
- GI
gastrointestinal
- WT
wild type
- p.i.
postinfection
References
- 1.Marshall B J, Warren J R. Lancet. 1984;1:1311–1315. doi: 10.1016/s0140-6736(84)91816-6. [DOI] [PubMed] [Google Scholar]
- 2.Caruso M L, Fucci L. J Clin Gastroenterol. 1990;12:601–602. [PubMed] [Google Scholar]
- 3.Baskerville A, Newell D G. Gut. 1988;29:465–472. doi: 10.1136/gut.29.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eaton K A, Morgan D R, Krakowka S. Infect Immun. 1989;57:1119–1125. doi: 10.1128/iai.57.4.1119-1125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radin M J, Eaton K A, Krakowka S, Morgan D R, Lee A, Otto G, Fox J. Infect Immun. 1990;58:2606–2612. doi: 10.1128/iai.58.8.2606-2612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yokota K, Kurebayashi Y, Takayama Y, Hayashi S, Isogai H, Isogai E, Imai K, Yabana T, Yachi A, Oguma K. Microbiol Immunol. 1991;35:475–480. doi: 10.1111/j.1348-0421.1991.tb01577.x. [DOI] [PubMed] [Google Scholar]
- 7.Shomer N H, Dangler C A, Whary M T, Fox J G. Infect Immun. 1998;66:2614–2618. doi: 10.1128/iai.66.6.2614-2618.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui H X, del Rosario A, Sonbati H, Lee C Y, George M, Ross J S. Exp Mol Pathol. 1991;55:261–268. doi: 10.1016/0014-4800(91)90006-j. [DOI] [PubMed] [Google Scholar]
- 9.Karita M, Li Q, Cantero D, Okita K. Am J Gastroenterol. 1994;89:208–213. [PubMed] [Google Scholar]
- 10.Lee A, O'Rourke J, De Ungria M C, Robertson B, Daskalopoulos G, Dixon M F. Gastroenterology. 1997;112:1386–1397. doi: 10.1016/s0016-5085(97)70155-0. [DOI] [PubMed] [Google Scholar]
- 11.Marchetti M, Arico B, Burroni D, Figura N, Rappuoli R, Ghiara P. Science. 1995;267:1655–1658. doi: 10.1126/science.7886456. [DOI] [PubMed] [Google Scholar]
- 12.Falk P G, Bry L, Holgersson J, Gordon J I. Proc Natl Acad Sci USA. 1995;92:1515–1519. doi: 10.1073/pnas.92.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomb J F, White O, Kerlavage A R, Clayton R A, Sutton G G, Fleischmann R D, Ketchum K A, Klenk H P, Gill S, Dougherty B A, et al. Nature (London) 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 14.Salama N, Guillemin K, McDaniel T K, Sherlock G, Tompkins L, Falkow S. Proc Natl Acad Sci USA. 2000;97:14668–14673. doi: 10.1073/pnas.97.26.14668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Doorn L J, Figueiredo C, Sanna R, Pena S, Midolo P, Ng E K, Atherton J C, Blaser M J, Quint W G. J Clin Microbiol. 1998;36:2597–2603. doi: 10.1128/jcm.36.9.2597-2603.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blaser M J, Berg D E. J Clin Invest. 2001;107:767–773. doi: 10.1172/JCI12672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ge Z, Feng Y, Dangler C A, Xu S, Taylor N S, Fox J G. Microb Pathog. 2000;29:279–287. doi: 10.1006/mpat.2000.0391. [DOI] [PubMed] [Google Scholar]
- 18.Xiang Z, Censini S, Bayeli P F, Telford J L, Figura N, Rappuoli R, Covacci A. Infect Immun. 1995;63:94–98. doi: 10.1128/iai.63.1.94-98.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo B P, Mekalanos J J. FEMS Immunol Med Microbiol. 2001;30:87–93. doi: 10.1111/j.1574-695X.2001.tb01554.x. [DOI] [PubMed] [Google Scholar]
- 20.Herrington D A, Hall R H, Losonsky G, Mekalanos J J, Taylor R K, Levine M M. J Exp Med. 1988;168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ottemann K M, Lowenthal A C. Infect Immun. 2002;70:1984–1990. doi: 10.1128/IAI.70.4.1984-1990.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dial E J, Hall L R, Serna H, Romero J J, Fox J G, Lichtenberger L M. Dig Dis Sci. 1998;43:2750–2756. doi: 10.1023/a:1026675916421. [DOI] [PubMed] [Google Scholar]
- 24.Wirth H P, Beins M H, Yang M, Tham K T, Blaser M J. Infect Immun. 1998;66:4856–4866. doi: 10.1128/iai.66.10.4856-4866.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salama N R, Otto G, Tompkins L, Falkow S. Infect Immun. 2001;69:730–736. doi: 10.1128/IAI.69.2.730-736.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eaton K A, Cover T L, Tummuru M K, Blaser M J, Krakowka S. Infect Immun. 1997;65:3462–3464. doi: 10.1128/iai.65.8.3462-3464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chevalier C, Thiberge J M, Ferrero R L, Labigne A. Mol Microbiol. 1999;31:1359–1372. doi: 10.1046/j.1365-2958.1999.01271.x. [DOI] [PubMed] [Google Scholar]
- 28.McGovern K J, Blanchard T G, Gutierrez J A, Czinn S J, Krakowka S, Youngman P. Infect Immun. 2001;69:4168–4173. doi: 10.1128/IAI.69.6.4168-4173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim J S, Chang J H, Chung S I, Yum J S. J Bacteriol. 1999;181:6969–6976. doi: 10.1128/jb.181.22.6969-6976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwao E, Hirayama F, Takagi S, Yokoyama Y, Ikeda Y. J Gastroenterol. 1999;34:47–54. [PubMed] [Google Scholar]
- 31.Eaton K A, Morgan D R, Krakowka S. J Med Microbiol. 1992;37:123–127. doi: 10.1099/00222615-37-2-123. [DOI] [PubMed] [Google Scholar]
- 32.Foynes S, Dorrell N, Ward S J, Stabler R A, McColm A A, Rycroft A N, Wren B W. Infect Immun. 2000;68:2016–2023. doi: 10.1128/iai.68.4.2016-2023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsuda M, Karita M, Morshed M G, Okita K, Nakazawa T. Infect Immun. 1994;62:3586–3589. doi: 10.1128/iai.62.8.3586-3589.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eaton K A, Brooks C L, Morgan D R, Krakowka S. Infect Immun. 1991;59:2470–2475. doi: 10.1128/iai.59.7.2470-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foynes S, Dorrell N, Ward S J, Zhang Z W, McColm A A, Farthing M J, Wren B W. FEMS Microbiol Lett. 1999;174:33–39. doi: 10.1111/j.1574-6968.1999.tb13546.x. [DOI] [PubMed] [Google Scholar]
- 36.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.McLennan A G. Int J Mol Med. 1999;4:79–89. doi: 10.3892/ijmm.4.1.79. [DOI] [PubMed] [Google Scholar]
- 38.Bessman M J, Frick D N, O'Handley S F. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 39.Yang H, Slupska M M, Wei Y F, Tai J H, Luther W M, Xia Y R, Shih D M, Chiang J H, Baikalov C, Fitz-Gibbon S, et al. J Biol Chem. 2000;275:8844–8853. doi: 10.1074/jbc.275.12.8844. [DOI] [PubMed] [Google Scholar]
- 40.Baselski V, Briggs R, Parker C. Infect Immun. 1977;15:704–712. doi: 10.1128/iai.15.3.704-712.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastoris M C, Proietti E, Mauri C, Chiani P, Cassone A. J Med Microbiol. 1997;46:647–655. doi: 10.1099/00222615-46-8-647. [DOI] [PubMed] [Google Scholar]
- 42.Tang H, Kays M, Prince A. Infect Immun. 1995;63:1278–1285. doi: 10.1128/iai.63.4.1278-1285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mackinnon F G, Gorringe A R, Funnell S G, Robinson A. Microb Pathog. 1992;12:415–420. doi: 10.1016/0882-4010(92)90004-8. [DOI] [PubMed] [Google Scholar]
- 44.Bertin A. Ann Rech Vet. 1983;14:169–182. [PubMed] [Google Scholar]
- 45.Field L H, Pope L M, Cole G T, Guentzel M N, Berry L J. Infect Immun. 1981;31:783–791. doi: 10.1128/iai.31.2.783-791.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thelin K H, Taylor R K. Infect Immun. 1996;64:2853–2856. doi: 10.1128/iai.64.7.2853-2856.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Attridge S R, Manning P A, Holmgren J, Jonson G. Infect Immun. 1996;64:3369–3373. doi: 10.1128/iai.64.8.3369-3373.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tacket C O, Taylor R K, Losonsky G, Lim Y, Nataro J P, Kaper J B, Levine M M. Infect Immun. 1998;66:692–695. doi: 10.1128/iai.66.2.692-695.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Logan S M, Conlan J W, Monteiro M A, Wakarchuk W W, Altman E. Mol Microbiol. 2000;35:1156–1167. doi: 10.1046/j.1365-2958.2000.01784.x. [DOI] [PubMed] [Google Scholar]
- 50.Sun D X, Mekalanos J J, Taylor R K. J Infect Dis. 1990;161:1231–1236. doi: 10.1093/infdis/161.6.1231. [DOI] [PubMed] [Google Scholar]
- 51.Winner L, 3rd, Mack J, Weltzin R, Mekalanos J J, Kraehenbuhl J P, Neutra M R. Infect Immun. 1991;59:977–982. doi: 10.1128/iai.59.3.977-982.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]