Abstract
Borrelia burgdorferi, the etiologic agent of Lyme disease, is genetically heterogeneous. Previous studies have shown a significant association between the frequency of hematogenous dissemination in Lyme disease patients and the genotype of the infecting B. burgdorferi strain. Comparative transcriptional profiling of two representative clinical isolates with distinct genotypes (BL206 and B356) was undertaken. A total of 78 open reading frames (ORFs) had expression levels that differed significantly between the two isolates. A number of genes with potential involvement in nutrient uptake (BB0603, BBA74, BB0329, BB0330, and BBB29) have significantly higher expression levels in isolate B356. Moreover, nearly 25% of the differentially expressed genes are predicted to be localized on the cell surface, implying that these two isolates have cell surface properties that differ considerably. One of these genes, BBA74, encodes a protein of 257 amino acid residues that has been shown to possess porin activity. BBA74 transcript level was >20-fold higher in B356 than in BL206, and strain B356 contained three- to fivefold more BBA74 protein. BBA74 was disrupted by the insertion of a kanamycin resistance cassette into the coding region. The growth rates of both wild-type and mutant strains were essentially identical, and cultures reached the same final cell densities. However, the mutant strains consistently showed prolonged lags of 2 to 5 days prior to the induction of log-phase growth compared to wild-type strains. It is tempting to speculate that the absence of BBA74 interferes with the enhanced nutrient uptake that may be required for the entry of cells into log-phase growth. These studies demonstrate the value of comparative transcriptional profiling for identifying differences in the transcriptomes of B. burgdorferi clinical isolates that may provide clues to pathogenesis. The 78 ORFs identified here are a good starting point for the investigation of factors involved in the hematogenous dissemination of B. burgdorferi.
Lyme disease, a multisystem disorder with possible neurologic, cardiac, and arthritic manifestations, is caused by an infection with the spirochete Borrelia burgdorferi (5, 71). The spirochete is transmitted to the host during the feeding of an infected Ixodes tick (69, 70). In unfed ticks, spirochetes are present in the midgut and migrate during a blood meal to the salivary glands, from which they are transmitted to the host via saliva. Numerous studies have demonstrated that the spirochete alters the expression of certain genes as it migrates from the tick midgut to the salivary glands (4, 63, 72). Likewise, genes that are expressed uniquely in the mammalian host have been identified (2, 4, 22, 73). The strategies that enable B. burgdorferi to evade the immune response have not been fully defined, although several mechanisms to explain their ability to persist in the host and disseminate to various organs, causing serious clinical manifestations, have been proposed (34, 82). Thus, the spirochete must adjust its gene expression program so as to adapt to a variety of different environments.
Molecular analysis of B. burgdorferi isolates has resulted in the differentiation of B. burgdorferi sensu lato into 10 distinct species (77). In North America, all known cases of Lyme disease are caused by infection with B. burgdorferi sensu stricto (77). Previous studies have shown that B. burgdorferi sensu stricto isolates obtained from patients with early Lyme disease can be distinguished on the basis of several genotypic parameters (77), and isolates have been classified into several rRNA gene spacer types (RST) (29, 38, 39). We have previously shown that the distributions of these genotypes differ significantly between isolates cultured from the skin of patients with early Lyme disease and those cultured from the blood of such patients (80) and that there is a highly significant association between the frequency of hematogenous dissemination in patients with early Lyme disease and the genotype of the infecting B. burgdorferi strain. These findings were confirmed in a mouse model demonstrating that B. burgdorferi RST1 clinical isolates were more invasive and produced more-severe pathology in experimentally infected animals than certain RST3 isolates did (75, 76). This suggests that the genotypic heterogeneity of B. burgdorferi clinical isolates results in differing potentials for bloodstream dissemination. This variability in virulence properties could be the result of differences in genetic content and/or alterations in gene expression between isolates. It is therefore of interest to identify genes which may be responsible for the observed differences in pathogenic potentials.
Genome array technology has proven to be a powerful tool for the assessment of bacterial gene expression under differing physiological conditions, and comparative transcriptional profiling has been employed to identify virulence genes (17, 18, 61). We recently reported the preparation and use of B. burgdorferi genome arrays to assess gene expression in B. burgdorferi B31 grown at 23 and 35°C (46, 47), and these arrays have been used for several other global transcriptional analyses of B. burgdorferi (3, 10, 44, 74, 83). In the present study, these genome arrays were employed to assess differences in gene expression between two B. burgdorferi clinical isolates with differing capacities for hematogenous dissemination.
MATERIALS AND METHODS
Strains and growth conditions.
B. burgdorferi strain B31 was from our laboratory collection. This is a high-passage-number isolate that lacked plasmids lp25, cp32-6, and cp32-8 and had a high-transformation-level phenotype relative to low-passage-number B31. In addition, two clinical isolates of B. burgdorferi were used in this study: BL206 (RST1), which was cultured from the blood of a patient with erythema migrans, and B356 (RST3), which was cultivated from a skin biopsy of an erythema migrans lesion of a different patient. BL206 was used at passage 6, and B356 was used at passage 5. Initially, isolates were grown in Barbour-Stoenner-Kelly (BSK)-H medium at 34°C until reaching mid-log phase (2.5 × 107 bacteria/ml), diluted to densities of 2.5 × 105 bacteria/ml, and further incubated at 34°C until reaching mid-exponential phase and the same densities (5 × 107 bacteria/ml). Spirochetes were enumerated by dark-field microscopy as described previously (64).
To determine B. burgdorferi growth characteristics, wild-type and mutant strains were grown in modified BSK medium (79) without antibiotics at 34°C to densities of 1 × 108 spirochetes/ml. Dilutions (1:1,000) were inoculated into triplicate 10-ml cultures and cultivated at either 25o or 34°C. Spirochetes were enumerated daily by dark-field microscopy. These studies were repeated a minimum of two times, yielding at least five values for each time point.
RNA and DNA isolation.
RNA was prepared from four separate 50-ml mid-exponential-phase cultures by using a Purescript RNA isolation kit according to the manufacturer's instructions (Gentra Systems). Genomic DNA was removed by DNase I treatment (DNA-free kit; Ambion). The final RNA was resuspended in RNase-free water, and the RNAs from the four separate cultures were pooled and quantitated. DNA was prepared from 10-ml cultures of each isolate by use of a commercial nucleic acid extraction kit (Isoquick; Orca Research).
cDNA probe synthesis.
Hybridization probes for array experiments were generated from each RNA sample by standard cDNA synthesis as described previously (46, 47). For each isolate, two cDNA synthesis reactions were performed, with the first using a mixture of all 3′ B. burgdorferi open reading frame (ORF)-specific primers (Sigma-Genosys Biotechnologies, Inc.) and the second using random hexamer oligonucleotides (Promega). In each case, 5 μg of RNA was mixed with 0.4 mM each of dTTP, dGTP, and dCTP, 200 units of Superscript II reverse transcriptase (Invitrogen, Life Technologies), 10 U of RNase inhibitor, and 20 μCi of [α-33P]dATP (Amersham) in a final volume of 20 μl of 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2 (first-strand buffer). The cDNA synthesis reaction mixtures were incubated at 42°C for 2 h. Trichloroacetic acid precipitation was used to determine the labeling efficiency of each reaction, which was typically 2 × 106 to 5 × 106 cpm/μg. Unincorporated nucleotides were removed from the labeling reaction by passage through a Sephadex G-25 spin column. After purification, both labeling reactions for each isolate were pooled and used as a probe.
Genomic DNA labeling.
Five μg of sheared genomic DNA isolated from strain BL206 or strain B356 was labeled radioactively by random priming using the Klenow fragment of DNA polymerase (21).
Hybridization and array data acquisition.
The B. burgdorferi genome arrays have been described previously (46, 47). Prior to hybridization, a pair of the membrane arrays was placed in a prehybridization solution (ExpressHyb; Clontech) and incubated at 55°C for 1 hour. The labeled cDNA probes were denatured at 95°C for 5 minutes, added to 10 ml of ExpressHyb solution, and incubated with the membrane for 14 to 16 h at 55°C. After hybridization, the membrane was washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.05% sodium dodecyl sulfate (SDS) for 30 min at room temperature and then with 0.1× SSC, 0.1% SDS at 65°C for 30 min. The arrays were exposed to a phosphorimager screen (Molecular Dynamics, California) for 48 h. Array hybridizations for each isolate were performed four times, and the same membranes were probed reciprocally (i.e., by switching the membranes) after stripping in 0.4 M NaOH for 20 min at 42°C, a procedure followed by two 10-min incubations in 200 mM Tris-HCl (pH 7.0), 0.1× SSC, 0.2% SDS at room temperature.
The exposed phosphorimager screen was scanned with a pixel size of 100 μm on a Storm 840 PhosphorImager (Molecular Dynamics). The image files were analyzed using ArrayVision software (Imaging Research, Canada), and the intensities for each spot were exported into ArrayStat (Imaging Research, Canada) for statistical analysis as described in detail elsewhere (47). Briefly, the background-corrected pixel intensity values for each spot were log transformed, common error was determined, and outliers were removed. The values were then normalized by the mean across conditions by an iterative process, and a false-positive error correction was achieved by an application of the false-discovery rate method (6). The z test for four independent conditions (eight independent spots for each ORF) using the false-discovery rate method with a nominal alpha value for P being ≤0.01 was used as the basis for computing confidence intervals.
An alternate method for identifying differentially expressed ORFs was employed (3). ORFs for which the raw pixel values were not >2 standard deviations (SD) above the pixel values for blank spots were eliminated from further analysis. Individual pixel values for the remaining ORFs were divided by the total pixel values for all ORFs, yielding the fractional intensity for each ORF. The ratios of fractional intensity values for each ORF for the two isolates were calculated and converted to log10. ORFs with log ratio values that were greater or smaller than the mean log ratio for all ORFs by >2 SD were considered to be differentially expressed.
Standard and real-time quantitative RT-PCR.
The presence or the absence of BBA74 transcript in wild-type and mutant B. burgdorferi was determined by standard reverse transcription (RT)-PCR. RNA was isolated from spirochetes grown to late log phase in 10 ml of modified BSK (wild-type) or modified BSK containing 200 μg/ml kanamycin (mutant) as described above, and total RNA was treated with DNase (AMBION DNase kit). cDNA was generated by reverse transcription. Each reaction contained 0.4 mM of each dNTP, 10 units of RNase inhibitor, 500 ng of random hexamer primers, 500 ng of total RNA, and 10 units of Superscript II reverse transcriptase (Invitrogen) in first-strand buffer. Following a 1-h incubation at 42°C, 1 μl of cDNA was used as a template to perform PCR using BBA74-specific primers (Table 1). Each RT-PCR run included controls containing no reverse transcriptase to rule out DNA contamination.
TABLE 1.
Oligonucleotide primers used in this study
| Name | Sequence (5′→3′)
|
|
|---|---|---|
| Forward | Reverse | |
| BB0329 | GCAGGATGGATAGGCGATTATGCTG | CTGTTCCCATATATGTATATTGGTGC |
| BB0330 | CACACACATACTTTACTATATTCACAAG | GCCCAGAATATATGTATATTGGTGCAGC |
| BB0365 | GTTTATGCAGGAGAGGCTCCAATTAGTATC | TCGTTAACATAGGTGAAATTTTTTCAACGCCGTCG |
| BB0603 | GCCTTATATCTTGATTATGCAATTCC | GCTTCCGCTGTAGGCTATTTTGAATTGC |
| BB0767 | CGCTAATGCTTTAAACAACCTTGC | CTCCTCTAGAGCCTTTTTTAAATGG |
| BB0768 | AGCGACGGTAGTTGTTACAAGCG | AGAAATTTTCAATTCGGACCCC |
| BB0769 | CTGGTATGATTTTTCATACTCTGGAC | TACATATTAAATCCAAGTCCCGCAATCATTGCTCC |
| BBA03 | TTGCAAAAACAAAAGTAATGATGCCGAGCC | TAAGTGTGGTTTTGGAATCTGC |
| BBA74 | GGAATGGTTGCTGAGGGTGCAAACAAGG | AGAAATAGCCATTACTTTTGTAAGATCTCTTGCGCCTTGAGC |
| BBB29 | GTATTAGAGGGTCTTGGAGG | CATAAGCTTGTCGATTTCCA |
| BBL22 | ATGGAAGACAATGATGATGTAACTC | CAGTTTTAAAAGATCTGGGACTGG |
| recA | GTGGATCTATTGTATTAGATGAGGCTCTCG | GCCAAAGTTCTGCAACATTAACACCTAAAG |
| flaB | AACAGCTGAAGAGCTTGGAATG | CTTTGATCACTTATCATTCTAATAGC |
| FL-571F | GCAGCTAATGTTGCAAATCTTTTC | |
| FL-677R | GCAGGTGCTGGCTGTTGA | |
| A74F344 | AAGAAGCAACTATTGTGGCAAAGTG | |
| A74R425 | ACAGCCTTTTTAGACATTTCAACAAC | |
| pA73F | GAGCTTGATGTTTGCGCGAGT | |
| pA76R | CTTGCCAAGCTGTAGCATCC | |
| pKNF | ATAGCTTGTAAATTCTATCATAATTGTGGT | |
| pKNR | ATCCATGATTTTGTTAAGTAGGTCATT | |
Microarray data were validated using real-time quantitative RT-PCR on a LightCycler PCR system with the SYBR green master mix following the manufacturer's instructions (Roche Diagnostics, Mannheim, Germany). Oligonucleotide primers for the tested ORFs are listed in Table 1. All primer pairs produced only one amplicon of the expected size when B. burgdorferi genomic DNA was used as template for PCR, indicating that all primers were specific for their respective targets. cDNA for RT-PCR was generated using 500 ng of total RNA in a 20-μl reaction mixture as described above. The real-time PCR step was performed in triplicate using 5 ng of cDNA and ORF-specific primers. For the experiments shown in Fig. 1, flaB and BBA74 transcripts were quantitated by SYBR green-based assay on an ABI 7900HT system using primers FL-571F/FL-677R and A74F344/A74R425 (Table 1) for amplification of flaB and BBA74, respectively. Each reaction mixture contained 20 ng of cDNA (prepared as described above) and 20 pmol of each primer in SYBR green PCR master mix (Applied Biosystems, Foster City, CA). Reactions containing cDNA generated without reverse transcriptase or containing primers alone were also included as controls. A relative quantitation method (ΔΔCt) was used to determine the relative expression of each gene between isolates BL206 and B356 (36).
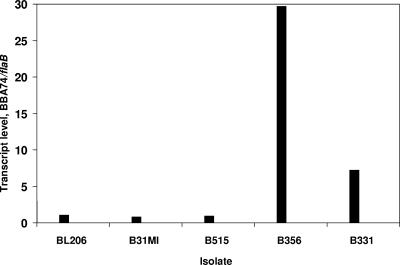
FIG. 1.
BBA74 transcript levels are higher in attenuated B. burgdorferi isolates. BBA74 mRNA and flaB mRNA were quantitated for five B. burgdorferi isolates by real-time RT-PCR. Data for each isolate are the average for two separate cultures, and for each culture the RT-PCR was performed in triplicate.
SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblot analysis.
B. burgdorferi cells (2× 108 to 5 × 108 cells) grown at 34°C were collected by centrifugation at 10,000 × g and washed three times with phosphate-buffered saline (pH 7.4) at 4°C. Each pellet was resuspended in 50 μl of 50 mM Tris-HCl (pH 8.0) containing 0.3% SDS, 10 mM dithiothreitol and stored at −20° or −80°C until use. One hundred μl of phosphate-buffered saline was added, and the cells were lysed by sonication with three 30-second pulses at 1-minute intervals (setting 8) at 4°C using a Branson model 250 sonifier (Branson Ultrasonics, Danbury, CT). The protein concentration was determined using a Coomassie Plus protein assay kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's instructions.
Protein lysates (0.5 to 5 μg) were separated on 10- to 15%-gradient SDS-PAGE gels. Proteins were transferred to polyvinylidene difluoride membranes using a semidry blotting apparatus and probed with either preimmune rabbit serum (1:1,000 dilution), polyclonal anti-BBA74 rabbit serum (1:1,000 dilution), polyclonal anti-OspA/B rabbit serum (1:10,000 dilution), or anti-OspA mouse monoclonal antibody (1:2,000 dilution), each diluted in Tris-buffered saline containing 0.05% Tween 20 (TBS-T), 5% nonfat dry milk. Alkaline phosphatase-conjugated goat anti-rabbit or anti-mouse antibodies (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD) were used as the secondary antibodies at dilutions of 1:1,000. The blots were washed three times with TBS-T to remove the unbound conjugated antibody. Visualization was achieved by treating the blots with the substrate BCIP/NBT 1 (5-bromo-4-chloro-3-indoyl-phosphate/nitroblue tetrazolium) solution (Kirkegaard & Perry Laboratories, Inc.) for 3 min at room temperature. The color reaction was stopped by washing the blots thoroughly with deionized water. Blots were dried overnight at room temperature and scanned on an Alpha Imager 2200 instrument (Alpha Innotech Corporation, San Leandro, CA), and band densitometry was performed with the provided imaging software (AlphaEase FC).
Mutagenesis of BBA74.
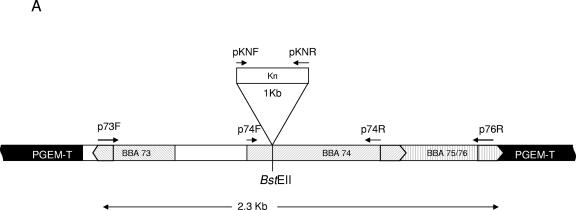
The mutation of BBA74 was accomplished by allelic exchange of the wild-type gene with an inactivated copy. A schematic diagram showing the relevant regions of plasmid lp54 is presented in Fig. 3A. A 2,327-bp DNA fragment spanning BBA73 to BBA76 (nucleotides 50601 to 52908 on plasmid lp54; GenBank accession number, AE000790) was amplified by PCR using primers pA73F and pA76R (Table 1) as forward and reverse primers, respectively, and cloned into the plasmid vector pGEM-T (Promega). A kanamycin resistance cassette from Streptococcus faecalis (65) was PCR amplified using primers pKNF and pKNR (Table 1) as forward and reverse primers, respectively. The recombinant pGEM vector was linearized by cleavage at a unique BstEII site (nucleotide 195 of BBA74) and treated with T4 DNA polymerase to produce blunt ends, and the kanamycin cassette was inserted. The pGEM-T ampicillin resistance locus was inactivated by the insertion of an irrelevant 248-bp DNA fragment into the unique ScaI site in the β-lactamase gene so as to avoid the introduction of ampicillin resistance into B. burgdorferi, and the plasmid phenotype (Knr and Amps) was confirmed in Escherichia coli DH5α. Plasmid DNA was purified from host E. coli by standard methods.
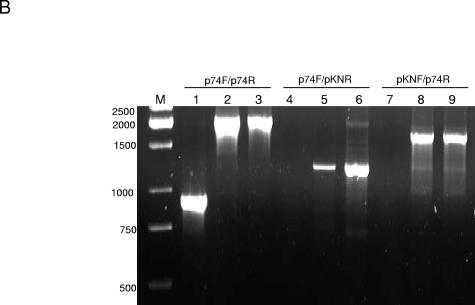
FIG. 3.
Disruption of BBA74 by insertion of kanamycin resistance cassette. (A) Schematic diagram of mutant construct. The region of lp54 between BBA73 and BBA76 was amplified by PCR using primers p73F/p76R and cloned into pGEM-T. The insertion of the kanamycin cassette (Kn) in the unique BstEII site in BBA74 is shown. Locations of forward and reverse primers used for confirmation of the disruption are indicated. (B) Confirmation of BBA74 gene disruption. Wild-type BL206 (lanes 1, 4, and 7), mutant BBA74 (lanes 2, 5, and 8), and pGEM-T vector construct containing disrupted BBA74 (lanes 3, 6 and 9) were subjected to PCR with primers p74F/p74R (lanes 1 to 3), p74F/pKNR (lanes 4 to 6), and pKNF/p74R (lanes 7 to 9). Products were separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. Lane M, DNA molecular size markers.
A high-passage-number B. burgdorferi B31 isolate and the clinical isolates BL206 and B356 were grown in modified BSK medium (79) at 34°C until mid-log phase (5 × 107 cells/ml) was achieved, and the isolates were made competent for DNA uptake by the method of Samuels (59). Competent cells (8.5 × 108), in a 50-μl volume, were mixed with 25 μg of plasmid DNA on ice for 1 minute and were electroporated as described by Hubner et al. (27). Transformed cells were allowed to recover by overnight incubation at 34°C in the absence of kanamycin. Subsequently, kanamycin was added to a final concentration of 200 μg/ml in 20 ml of modified BSK medium, and 200-μl aliquots were plated in 96-well microtiter plates for the selection of positive transformants. Viable B. burgdorferi transformants were selected from wells which produced a color change in the growth medium after 1 to 5 weeks of incubation at 34°C in 5% CO2. These transformants were further purified by limiting dilution and cultivation in modified BSK medium containing 200 μg/ml kanamycin.
Plasmid profiles.
The plasmid content of B. burgdorferi mutants and transformants was determined by plasmid-specific PCR as previously described (28).
RESULTS
Strain B31 sequence-based arrays can be employed for analysis of other B. burgdorferi isolates.
The B. burgdorferi membrane arrays used here contain PCR-amplified ORFs from B. burgdorferi strain B31 MI, the strain whose genome sequence has been previously elucidated (13, 24). Array description, including ORF selection and PCR primer design, has been described previously (46). In this study, the differences in global gene expression between two B. burgdorferi clinical isolates designated BL206 and B356 were assessed. BL206 has been shown to readily disseminate and cause severe disease in experimentally infected mice, whereas B356 does not disseminate (75, 76). All B31-like linear plasmids were present in both isolates, and both lacked circular plasmid cp32-8. In addition, cp32-1 was not present in B356. To ensure that B31 MI sequence-based arrays could be employed for measuring gene expression in other B. burgdorferi isolates, the arrays were hybridized in triplicate with 33P-labeled DNA derived from randomly sheared BL206 and B356 genomic DNA and compared to results obtained with B31 MI DNA. There was a high degree of correlation between the hybridization signals of BL206 and B356 and that of strain B31 to each ORF, with r values of 0.96 and 0.77 for the comparison of B31 to BL206 and to B356, respectively. The better hybridization observed for isolate BL206 DNA was expected, since BL206 is genotypically more similar to B31 than isolate B356 is (37, 76). This verifies that the arrays may be employed for measuring transcript levels in other B. burgdorferi isolates.
Transcript analysis and comparison between B. burgdorferi clinical isolates BL206 and B356.
An underlying hypothesis of this study is that the differences in the pathogenic potentials of the two B. burgdorferi isolates are to some measure the result of differences between them in terms of gene expression. By characterizing isolate-specific patterns of gene expression, it should be possible in principle to identify genes and proteins responsible for the differential pathogenesis among isolates. As a first step in this process, the gene expression profiles of a virulent isolate (BL206) and of an attenuated strain (B356) cultivated at a temperature of 34°C were determined using B. burgdorferi genome arrays. Probe synthesis and array hybridization were performed in quadruplicate for each isolate. Since each ORF is spotted in duplicate on a given array, eight data points were analyzed for each ORF. The intensities for each spot were analyzed using ArrayVision and statistically analyzed with ArrayStat software. In a second approach, the expression of each ORF was determined as the percentage of the total expression for all ORFs, and the expression ratios for each ORF between the isolates were calculated. Expression ratios were converted to log10 values, the mean log ratio for all ORFs was determined, and those ORFs with logged expression ratios of >2 SD above or below the mean were identified. Only those ORFs with significant differences in expression ratios between isolates by both analytical methods were considered to be differentially expressed. These ORFs are listed in Table 2.
TABLE 2.
Genes with significantly differing expression in isolates B356 and BL206
| Relative expression level | Gene | Description | Replicon | Expression ratio by:
|
|||
|---|---|---|---|---|---|---|---|
| Gene array
|
Real-time RT-PCR
|
||||||
| B356/BL206 | BL206/B356 | B356/BL206 | BL206/B356 | ||||
| B356 > BL206 | BB0032 | Hypothetical protein | Chromosome | 3.0 | |||
| BB0036 | DNA topoisomerase IV (ParE) | Chromosome | 3.1 | ||||
| BB0084 | NifS | Chromosome | 5.8 | ||||
| BB0247 | Conserved hypothetical protein | Chromosome | 3.4 | ||||
| BB0282 | Flagellar protein (FlbD) | Chromosome | 3.3 | ||||
| BB0329 | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (OppA-2) | Chromosome | 4.0 | 18.6 | |||
| BB0330 | Oligopeptide ABC transporter, periplasmic oligopeptide-binding protein (OppA-3) | Chromosome | 7.1 | 28.0 | |||
| BB0365 | Lipoprotein LA7 | Chromosome | 12.3 | 18.0 | |||
| BB0519 | grpE protein (GrpE) | Chromosome | 3.7 | ||||
| BB0548 | DNA polymerase I (PolA) | Chromosome | 3.7 | ||||
| BB0603 | Membrane-associated protein P66 | Chromosome | 4.3 | 10.0 | |||
| BB0767 | UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase (MurG) | Chromosome | 3.9 | 30.0 | |||
| BB0768 | Pyridoxal kinase (PdxK) | Chromosome | 5.7 | 3.0 | |||
| BB0769 | Sialoglycoprotease (gcp) | Chromosome | 4.5 | 6.2 | |||
| BBA03 | Outer membrane protein | lp54 | 11.6 | 8.5 | |||
| BBA47 | Hypothetical protein | lp54 | 10.5 | ||||
| BBA52 | Outer membrane protein | lp54 | 8.0 | ||||
| BBA53 | Hypothetical protein | lp54 | 4.6 | ||||
| BBA58 | Hypothetical protein | lp54 | 4.6 | ||||
| BBA59 | Lipoprotein | lp54 | 3.7 | ||||
| BBA67 | Hypothetical protein | lp54 | 17.9 | ||||
| BBA68 | Complement regulatory protein, CRASP-1 | lp54 | 3.4 | ||||
| BBA69 | Hypothetical protein | lp54 | 6.0 | ||||
| BBA74 | Outer membrane porin (Oms28) | lp54 | 27.9 | 20.2 | |||
| BBA76 | Thymidylate synthase-complementing protein (Thy1) | lp54 | 4.4 | ||||
| BBB19 | Outer surface protein C (OspC) | cp26 | 6.4 | ||||
| BBB28 | Hypothetical protein | cp26 | 5.9 | ||||
| BBB29 | PTS system, maltose- and glucose-specific IIABC component (MalX) | cp26 | 2.3 | 2.0 | |||
| BBD18 | Hypothetical protein | lp17 | 6.9 | ||||
| BBJ09 | Outer surface protein D (OspD) | lp38 | 5.2 | ||||
| BBJ23 | Hypothetical protein | lp38 | 7.4 | ||||
| BBJ31 | Hypothetical protein | lp38 | 2.9 | ||||
| BBJ39.1 | Multidrug efflux transporter, possible pseudogene | lp38 | 3.7 | ||||
| BBO32 | Plasmid partition protein, putative | cp32-7 | 2.8 | ||||
| BBQ39 | Conserved hypothetical protein | lp56 | 4.6 | ||||
| BL206 > B356 | BB0199 | Hypothetical protein | Chromosome | 3.6 | |||
| BB0236 | Hypothetical protein | Chromosome | 3.4 | ||||
| BB0300 | Cell division protein (FtsA) | Chromosome | 2.8 | ||||
| BB0372 | Glutamyl-tRNA synthetase (GltX) | Chromosome | 2.1 | ||||
| BB0423 | Hypothetical protein | Chromosome | 5.3 | ||||
| BB0425 | Hypothetical protein | Chromosome | 6.8 | ||||
| BB0802 | Ribosome-binding factor A (RbfA) | Chromosome | 4.7 | ||||
| BBD13 | Hypothetical protein | lp17 | 4.3 | ||||
| BBE09 | Protein P23 | lp25 | 3.7 | ||||
| BBE11 | Hypothetical protein | lp25 | 6.6 | ||||
| BBE19 | Plasmid partition protein, putative | lp25 | 7.5 | ||||
| BBF06 | Conserved hypothetical protein | lp28-1 | 60.7 | ||||
| BBG12 | Hypothetical protein | lp28-2 | 3.0 | ||||
| BBG30 | Hypothetical protein | lp28-2 | 6.6 | ||||
| BBI03 | Hypothetical protein | lp28-4 | 16.3 | ||||
| BBI18 | Hypothetical protein | lp28-4 | 4.7 | ||||
| BBI19 | Conserved hypothetical protein | lp28-4 | 7.9 | ||||
| BBI29 | Hypothetical protein | lp28-4 | 8.2 | ||||
| BBJ08 | Hypothetical protein | lp38 | 4.6 | ||||
| BBJ19 | Conserved hypothetical protein | lp38 | 8.6 | ||||
| BBK01 | Hypothetical protein | lp36 | 3.6 | ||||
| BBK13 | Conserved hypothetical protein | lp36 | 4.3 | ||||
| BBK18 | Conserved hypothetical protein | lp36 | 10.7 | ||||
| BBK21 | Plasmid partition protein, putative | lp36 | 4.2 | ||||
| BBK34 | Hypothetical protein | lp36 | 9.7 | ||||
| BBK40 | Conserved hypothetical protein | lp36 | 38.0 | ||||
| BBK45 | Immunogenic protein P37, putative | lp36 | 13.5 | ||||
| BBK46 | Immunogenic protein P37, authentic frameshift | lp36 | 6.0 | ||||
| BBK47 | Hypothetical protein | lp36 | 5.9 | ||||
| BBK51 | Hypothetical protein | lp36 | 6.0 | ||||
| BBK52 | Protein P23 | lp36 | 6.1 | ||||
| BBL22 | Conserved hypothetical protein | cp32-8 | 3.7 | 28.0 | |||
| BBL38 | Conserved hypothetical protein | cp32-8 | 2.5 | ||||
| BBM40 | Hypothetical protein | cp32-6 | 8.5 | ||||
| BBO20 | Conserved hypothetical protein | cp32-7 | 2.9 | ||||
| BBP28 | Lipoprotein, Mlp | cp32-1 | 15.3 | ||||
| BBQ06 | Conserved hypothetical protein | lp56 | 7.1 | ||||
| BBQ07 | Conserved hypothetical protein | lp56 | 3.4 | ||||
| BBQ35 | Lipoprotein, Mlp | lp56 | 5.1 | ||||
| BBQ84 | Hypothetical protein | lp56 | 9.1 | ||||
| BBR04 | Hypothetical protein | cp32-4 | 2.6 | ||||
| BBS22 | Conserved hypothetical protein | cp32-3 | 3.1 | ||||
| BBS30 | Lipoprotein, Mlp | cp32-3 | 3.5 | ||||
| Other | BB0131 | RecA | Chromosome | 1.5 | 2 | ||
| BB0147 | FlaB | Chromosome | 1.1 | 1.1 | |||
A total of 78 ORFs had significantly different expression levels in the two isolates. Thirty-five ORFs were expressed at significantly higher levels in isolate B356 than in BL206, whereas 43 genes had significantly greater expression in BL206 than in B356. Twenty-one (27%) of the differentially expressed genes are chromosomally encoded, with the remainder located on various plasmids. More than half of these genes (45/78; 58%) have no database match or have no assigned known function. Among those ORFs with assigned functions, the largest single group consists of putative cell envelope components (16/78).
To independently validate the microarray data, the relative expressions of selected ORFs were determined by real-time RT-PCR. As shown in Table 2, the array results were well supported by the real-time RT-PCR findings.
Comparative expression of selected genes. (i) Linear plasmids lp54 and lp36.
A striking observation is the distinctive natures of gene expression for the two isolates with respect to two sets of plasmid-encoded genes. Eleven genes, each on both plasmids lp54 and lp36, were differentially expressed. Surprisingly, all lp54-encoded genes had significantly greater expression in isolate B356. By contrast, the lp36-encoded ORFs all had higher expression levels in isolate BL206. Whereas the observed differences in transcript levels between the isolates for lp54-encoded ORFs appear to reflect true differential expression (i.e., substantial transcript is produced in both isolates), this may not be the case for lp36-encoded genes, since none of the latter genes appear to have substantial expression in isolate B356.
(ii) Possible operons.
Examination of the differentially expressed ORFs revealed two regions that may be expressed as operons. The chromosomal region between BB0328 and BB0335 contains genes encoding components of an oligopeptide permease (Opp). These ABC transporter systems are composed of transmembrane components (OppB and OppC), ATP-binding proteins (OppD and OppF), and peptide-binding proteins (OppA). BB0328 to BB0330 encode binding proteins that have been designated OppA-1, OppA-2, and OppA-3, respectively; BB0332 to BB0335 encode the transmembrane and ATP-binding components of the transporter. BBB16 (on plasmid cp26) and BBA34 (on lp54) encode two additional OppA proteins. BB0329 and BB0330 showed significantly greater expression in B356 than in BL206 (expression ratios of 4.1 and 7.1, respectively). BB0328, BB0333 to BB0335, and BBB16 also showed elevated expression in B356 by both array and RT-PCR analysis, although the differences measured by array were significant by only one of the two statistical methods (data not shown). The data are consistent with earlier studies suggesting that BB0328 to BB0330 and BB0332 to BB0335 may be cotranscribed (7). Alternatively, the three OppA genes may be individually transcribed but coordinately upregulated in B356.
BB0767 to BB0769 were expressed at significantly higher levels in isolate B356 than in BL206 (expression ratios of 3.9, 5.7, and 4.5, respectively). These genes code for three seemingly unrelated proteins: MurG [UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase], pyridoxal kinase, and sialoglycoprotease. They are oriented in the same direction (opposite to the gene numbering), and the coding regions are tightly packed; the end of BB0769 overlaps BB0768 by 25 nucleotides, and BB0768 and BB0767 are separated by 149 nucleotides. Interestingly, the start codon of BB0769 overlaps the termination codon of the upstream BB0770, as do the initiation and termination codons of BB0766 and BB0767, but neither BB0766 nor BB0770 is expressed at significantly different levels in the two isolates. The data suggest that BB0767 to BB0769 may be cotranscribed.
(iii) Outer surface protein genes.
Several well-studied genes encoding outer surface proteins showed significantly higher transcript levels in isolate B356. These include ospC, ospD, BB0603 (encoding P66), and BBA74 (expression ratios of 6.4, 5.2, 4.3, and 27.9, respectively).
Differential expression of BBA74.
The transcript levels of BBA74 were >20-fold higher in B356 than in BL206 (Table 2), and this gene was chosen for more-careful study. To explore whether the differences in expression were due to sequence variation in either upstream or coding regions of BBA74, the region between BBA73 and BBA75 from B. burgdorferi BL206 and B356 was amplified by PCR, and the products were sequenced. The BL206 sequence was identical to that of B31 MI (GenBank accession number, AE000790). The B356 sequence showed only two nucleotide changes: A→G at position −83 and T→A at position −375 relative to the BBA74 initiation codon. Whether either of these changes contributes to the differential expression of BBA74 remains to be determined.
BBA74 transcript levels were measured by real-time RT-PCR in two additional clinical isolates with differing capacities for dissemination. Results are presented in Fig. 1. In strain B515, which disseminates in mice (76), the ratio of mRNA levels of BBA74 and flaB was 0.94; this is comparable to that observed for two other disseminating isolates, BL206 and B31 MI (ratios of 1.06 and 0.81, respectively). In contrast, the BBA74 transcript level was substantially higher (BBA74-to-flaB ratio of 7.22) in strain B331, which does not disseminate (76).
BBA74 protein levels are higher in B. burgdorferi B356.
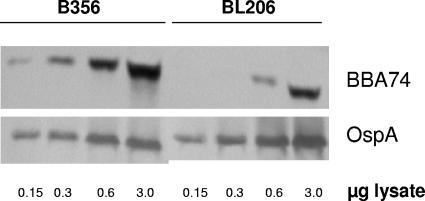
Given the >20-fold increase in the level of BBA74 transcript in isolate B356, it was of interest to determine whether the level of BBA74 protein was also elevated in this isolate relative to BL206. This determination was accomplished by semiquantitative immunoblotting using anti-BBA74 polyclonal antibodies. The densitometry of the blots shown in Fig. 2 revealed that the amount of BBA74 protein is three- to fivefold higher in B356 than in BL206. Similarly, the densitometry of BBA74 spots on two-dimensional gels also yielded a four- to fivefold-higher level for the protein in isolate B356 (data not shown). These results correlate with the increased mRNA levels in B356 as determined by array and RT-PCR analysis.
FIG. 2.
BBA74 protein levels are higher in B. burgdorferi B356. Increasing amounts of whole-cell lysates of strains B356 and BL206 were subjected to SDS-PAGE followed by immunoblotting with either BBA74-specific antibodies (top panel) or OspA-specific antibodies (bottom panel). Lanes 1 to 4, strain B356; lanes 5 to 8, strain BL206.
Mutagenesis of BBA74.
The disruption of BBA74 was accomplished in a strain B31 derivative and the two clinical isolates, BL206 and B356, by allelic exchange of the wild-type gene with a copy of the gene disrupted by the insertion of a kanamycin resistance cassette (Fig. 3A). Mutant clones were assayed by PCR using BBA74-specific primers (p74F and p74R; Table 1) in combination with kanamycin cassette-specific primers (pKNF and pKNR). Results for strain BL206 are shown in Fig. 3B; identical results were obtained with mutants selected in strains B31 and B356 (data not shown). Amplification with primers p74F/p74R yielded a single product in the mutant that was 1 kb larger than in the wild type (compare lanes 1 and 2). PCR using kanamycin cassette primers in combination with BBA74-specific primers resulted in a product only in the mutant isolates (compare lanes 5 and 8 to lanes 4 and 7). The sizes of these products confirm that the kanamycin resistance gene is inserted in an orientation the same as that of BBA74. Furthermore, the results are consistent with the replacement of the resident copy of BBA74 with the disrupted copy, since only a single band was obtained by PCR with primers p74F/p74R (Fig. 3B, lane 2). Plasmid content analysis revealed that the mutant clones lacked plasmids lp25 and lp28-1.
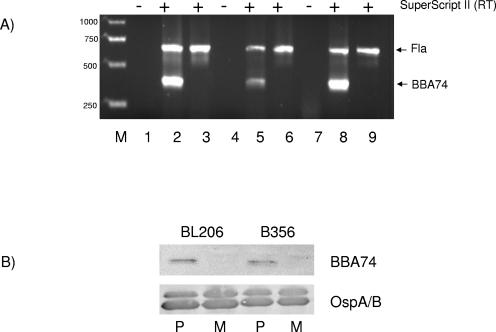
The inactivation of BBA74 in the mutants was confirmed by RT-PCR and Western blotting. As shown in Fig. 4, the mutants did not produce BBA74 transcript (Fig. 4A) or BBA74 protein (Fig. 4B) in any of the B. burgdorferi strain backgrounds.
FIG. 4.
Transcription and translation of BBA74 in mutant strains. (A) RT-PCR. Lanes 1 and 2, wild-type B31; lane 3, mutant B31; lanes 4 and 5, wild-type BL206; lane 6, mutant BL206; lanes 7 and 8, wild-type B356; lane 9, mutant B356. Reactions depicted in lanes 1, 4, and 7 lacked reverse transcriptase. Amplified products were separated by electrophoresis on a 1.5% agarose gel and stained with ethidium bromide. Lane M, DNA molecular size markers. (B) Immunoblotting. Total cell lysate (3.5 μg) was separated by 12.5% SDS-PAGE and transferred to polyvinylidene difluoride membranes, and blots were developed with either OspA/B-specific or BBA74-specific antibodies. P, wild type; M, mutant.
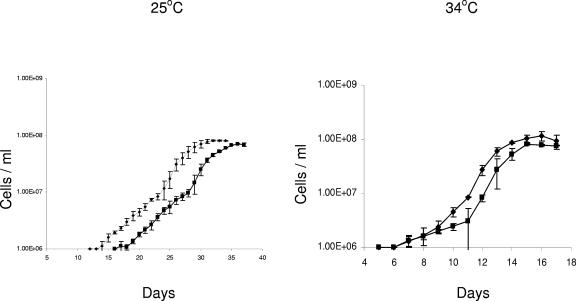
The effect of BBA74 mutagenesis on spirochete growth in vitro at 25 ° and 34°C was determined for BL206 wild-type and mutant isolates (Fig. 5). The growth rates for both the wild type and the mutant were essentially identical (22.4 and 22 h at 34°C for the wild type and the mutant, respectively; 57.5 and 61.9 h at 25°C for the wild type and the mutant, respectively). Wild-type and mutant spirochetes achieved the same final cell densities at both temperatures. Interestingly, there was a consistent lag in the initiation of the growth of the BBA74 mutants at either temperature (2 to 3 days at 34°C and 5 days at 25°C). Furthermore, although there was no significant difference in the calculated growth rates between wild-type and mutant isolates, the mutant required 2 to 4 additional days of growth to reach the maximal cell density, consistent with the observed lag in the initiation of growth. Essentially identical results were obtained by comparing wild-type and mutant growth characteristics for strains B31 and B356.
FIG. 5.
Growth characteristics of wild-type (•) and BBA74 mutant (▪) BL206 at 25 ° and 34°C. Aliquots of starter cultures were inoculated into fresh media at densities of 1 × 105 cells/ml, and growth was monitored daily by dark-field microscopy.
DISCUSSION
This study was undertaken to identify genes that may contribute to the hematogenous dissemination of B. burgdorferi by comparing the global transcription of two clinical isolates differing in this property. In principle, several types of microarray approaches can yield information with regard to potential virulence genes. Comparative genome hybridization analysis scores for differences in the genomic contents of isolates. This approach has been employed to compare the genomes of pathogenic and nonpathogenic isolates of several bacterial strains (14, 31, 58). An alternative strategy is to explore differences in global gene expression between a pathogen cultivated in vitro and that in an infected host (17, 60). Such studies are particularly difficult because the paucity of organisms in infected tissues precludes global microarray-based expression studies without preamplification (60). Comparative transcriptional profiling of virulent and avirulent strains provides a third methodology. Genes with significantly altered expression in such an analysis could be the result of differences in genomic content (presence or absence of gene) or differences in the expression program in one of the isolates. In the present study, the latter approach was employed.
The majority of the genes with expressions that differ significantly between the two isolates studied here are plasmid encoded. Interestingly, genes on two linear plasmids, lp36 and lp54, accounted for 22 of these 57 genes. Eleven lp36-encoded genes had substantially greater expression in BL206 than in B356. Little is known regarding the 54 putative ORFs encoded on this plasmid. BBK32 was identified as a fibronectin-binding protein (53), and BBK17 is annotated as adenine deaminase (24). BBK32 and BBK50 mRNA and protein were detected in engorged ticks and infected mouse tissue, indicating that the genes are expressed in vivo (22). Both of these genes have measurable and equivalent expression levels in both isolates studied here. In contrast, BBK18, BBK34, BBK40, BBK45 to -47, BBK51, and BBK52 had at least fivefold-greater expression levels in BL206. The expression of BBK45 and BBK47 had been shown previously to be downregulated by between 2.8- and 5-fold in B. burgdorferi B31 growing in dialysis membrane chambers (DMC) that mimic the host-adapted state (10, 56). The observed differences in expression of these lp36-encoded genes in the two B. burgdorferi isolates may be the result of true differences in transcript levels. Alternatively, the differences may result from sequence variation in these genes or from their complete absence in the B356 genome. Since the array is composed of ORFs amplified by PCR from strain B31 DNA, substantial genome sequence differences between B31 and B356 would be expected to result in a variation in hybridization efficiency. Although there was only a minor difference in the global hybridization efficiencies of BL206 and B356 genomic DNAs to the array, genomic sequences located on lp36 hybridized almost sevenfold more intensely with BL206 than with B356 genomic DNA. It is, therefore, likely that the differences observed for lp36-encoded genes result from substantial genomic differences between the two isolates relative to this plasmid. The absence of one or more of these genes may contribute to the attenuated phenotype of B356.
In contrast, 11 lp54-encoded genes had significantly greater expression in isolate B356. Numerous studies have shown that the expression of lp54-encoded genes is regulated in response to physiological and environmental conditions (10, 25, 47, 56, 62). These include genes encoding the major outer surface proteins OspA and OspB and decorin-binding proteins DbpA and DbpB. Eight of the 11 genes that exhibited differential expression in this study have been shown previously to have regulated expression. Interestingly, six of these (BBA52, BBA59, BBA67 to -69, and BBA74) were shown to have reduced expression in DMC (10, 56). lp54 DNA from both BL206 and B356 hybridized equally well to the array, so the observed differences in RNA hybridization reflect a true alteration in transcript level.
Based on B31 genome annotation (24), nearly one-fourth (19/78) of the differentially expressed genes are classified as encoding cell envelope or transport proteins. This is consistent with other global analyses of B. burgdorferi transcription (10, 47, 56, 74) and with the notion that the spirochete remodels its surface in response to changes in environmental conditions (35, 52, 81). Of particular interest is the fact that the expression levels in B356 of BB0328 and BB0329, which encode OppA-2 and OppA-3, were increased by 4.1-fold and 7.1-fold, respectively. As noted above, these genes are part of an opp operon spanning from BB0328 to BB0335, and other genes in this region also displayed expression levels elevated by approximately two- to sevenfold, although this increase was significant by only one of the two statistical tests. Previous studies by Bono et al. (7) and Wang et al. (78) demonstrated that (i) BB0328 to BB0330 can be transcribed as individual mRNAs or as a polycistronic transcript; (i) only BBA34 (encoding OppA-5) is subject to temperature regulation; (iii) BB0328, BB0330, and BBA34 are relatively well expressed in vitro at 37°C, but the expression of BB0329 and BBB16 is weak; (iv) BB0329 is not appreciably expressed in ticks, and expression is moderately enhanced in mice; and (v) BB0330 transcription is increased in both ticks and mice relative to in vitro-cultivated spirochetes. Taken together, these studies suggested that different OppA alleles are subject to alternative environmental signals. In the present study, only spirochetes grown at 34°C were analyzed, and while it is clear that oligopeptide permease-related genes have significantly altered expression in isolate B356 relative to that in BL206, whether the regulation of expression under other conditions is also changed remains to be elucidated.
In bacteria, oligopeptide permeases are membrane-associated complexes belonging to the ABC transporter family. Although the major role for most bacterial oligopeptide permeases is in nutrient uptake, they may also be involved in sensing environmental signals in the form of peptides, culminating in diverse responses, such as the expression of virulence determinants, quorum sensing, chemotaxis, sporulation, conjugation, and competence (33, 51, 57). Peptide transport systems have been shown to be associated with the virulences of Salmonella enterica serotype Typhimurium (50), Streptococcus pneumoniae (19), and Enterococcus faecalis (20). Based on these observations, it has been proposed that the OppA proteins may also have a role in the adaptation of B. burgdorferi to different environmental niches, such as the tick or the mammalian host (7, 78). This family of peptide-binding proteins also may be important in regulating spirochetal chemotaxis and/or gene expression for the targeting and penetration of specific tissues in invertebrate and vertebrate hosts.
Outer membrane proteins can serve a role in solute and protein transport, and substances can be taken up by bacteria by diffusion through porin proteins embedded in the outer membrane (1). Three porin activities have been identified for B. burgdorferi. These are encoded by BB0034 (P13), BB0603 (P66 or Oms66), and BBA74 (Oms28) (49, 66, 67). The latter two genes were transcribed in isolate B356 at levels significantly greater than in BL206. P66 localizes to the outer membrane (11, 12) and has been shown to bind β3-chain integrins (15).
BBA74 encodes a product of 257 amino acids with a putative peptidase I signal sequence (66). Skare et al. demonstrated that native BBA74 protein possesses porin activity and designated it Oms28. The protein is associated with B. burgdorferi outer membrane vesicles, although only a fraction of recombinant BBA74 expressed in Escherichia coli is associated with the outer membrane (66). BBA74 expression is regulated by temperature (47, 74), the host environment (56), and the redox state (J. Skare, personal communication). In the present study, BBA74 expressions were found to be 7- to 28-fold higher in two nondisseminating strains (B356 and B331) than in disseminating strains (BL206 and B515). Furthermore, BBA74 protein levels were also higher in B356. The differences in transcript levels are unlikely to be the result of variation in a cis-acting sequence element, since the intergenic region between BBA73 and BBA74 contained only two nucleotide differences in B356 (at positions −83 and −375). Although a role for either of these nucleotides in transcriptional regulation cannot be completely ruled out, it is more likely that differences in the functions or levels of a trans-acting factor are responsible for the observed variation in BBA74 transcription. Interestingly, the mutagenesis of the C-terminal protease, CtpA, results in a dramatic increase in BBA74 protein in strain B31 cultivated at 35°C (48). In the present study, a 3.5- to 5-fold difference in BBA74 protein levels between B356 and BL206 grown at 34°C was noted. This suggests that the levels of BBA74 protein are subject to both transcriptional and posttranslational regulation.
Mutagenesis of BBA74 did not impair the growth of B. burgdorferi in vitro at either 25° or 34°C, and there were no significant differences in growth rates or final cell densities between the mutant and the wild type. However, the BBA74 mutant displayed a 2- to 5-day growth lag at both temperatures. Given the demonstrated porin activity of BBA74 (66), it is tempting to speculate that inactivation of the protein interferes with the nutrient uptake that may be required for the entry of cells into log-phase growth. Similar transitions may be required for the adjustment to growth in different environments during the natural tick-mammal cycle. It is of interest that the mutagenesis of P66, which has also been shown to possess porin activity (67), did not result in any growth impairment in vitro (16). In the present study, all mutants lacked plasmids lp25 and lp28-1 and are, therefore, noninfectious (26, 32, 54, 55). Thus, a definitive role for this protein in vivo has yet to be established.
The present study has several potential limitations. One is the nature of the B. burgdorferi genome itself. Nearly two-thirds of the annotated B31 ORFs have no known function (24), and more than half of the differentially expressed genes identified in this investigation are in this category. To the extent that some of these ORFs may play roles in dissemination, the lack of information regarding their functions is a drawback. A further possible limitation is the use of gene arrays that are based on the genome sequence of strain B31 for analysis of other isolates whose genomes have not been sequenced. Thus, genes with possible relevance to virulence that may be present in the other isolates (e.g., BL206 and B356 in the present investigation) but not in B31 would not be subject to analysis. In principle, substantial sequence differences between B31 and the tested strains could result in reduced signals for certain ORFs. Indeed, signals were somewhat lower when arrays were hybridized with B356 genomic DNA. This problem was addressed in our experimental design by scoring the expression of each ORF as a percentage of the total expression for all ORFs.
The present study was carried out with in vitro-cultivated organisms. It may be argued that certain virulence genes may be expressed only in the infected host. As noted earlier, however, this ideal situation has rarely been achieved in microarray studies of bacterial virulence, due to the low numbers of bacteria typically present in infected tissues (60). As a result, most studies of this kind have relied on the use of in vitro cultivation conditions, e.g., low iron or pH, designed to mimic the in vivo state (9, 17, 60). Two B. burgdorferi global expression studies have been performed with organisms cultivated in dialysis membrane chambers implanted in rats (10, 56). Unfortunately, a number of methodological differences between these two investigations resulted in a low concordance between the two data sets. Interestingly, nearly 30% of the ORFs listed in Table 2 (including BB0603, BBA74, ospC, and ospD) were identified as having significantly altered expression in DMC in at least one of these studies (10, 56). Narasimhan et al. have used a preamplification approach to study B. burgdorferi gene expression in feeding ticks (45) and in heart or central nervous system of nonhuman primates (44). Methodological differences between those reports and the present study make comparisons difficult. One of these studies utilized membrane arrays the same as those employed here. It is worthy of note that expression was detected for only 470 of the ORFs spotted on the membrane (44), suggesting that this preamplification method may alter the mRNA distribution or underestimate the levels of mRNAs with low expression.
Only two clinical isolates, a virulent strain (BL206) and an attenuated strain (B356), were analyzed by transcriptional profiling in this study. Numerous studies have demonstrated substantial genotypic variability among B. burgdorferi isolates (23, 39, 40, 77). Thus, the extent to which these two isolates are representative of all virulent and attenuated strains is unclear, although increased expression of BBA74 was observed in the two attenuated strains studied thus far (B356 and B331). BL206 belongs to a monophyletic clade based on rRNA gene and ospC sequence analysis, and all isolates of this genotype that have been tested in vivo produce severe disseminated infections in mice (76). Thus, it is reasonable to expect that BL206 is representative of other RST1 isolates. The question of whether the observed differences in global gene expressions between BL206 and B356 are reflective of all attenuated isolates characterized to date, which are more genotypically diverse (76, 80), warrants further study. Regardless, this study was designed to explore differences in gene expression for isolates that differ in their potentials for hematogenous dissemination, as is the case for BL206 and B356.
Despite these limitations, several of the present findings are of interest and may provide insights into B. burgdorferi pathogenesis. Evolution of a pathogenic lineage from a mildly pathogenic or commensal ancestor most frequently results from the acquisition of a virulence gene or genes. However, examples of enhanced pathogenicity arising from the loss of a gene or a change in its function have been described for Pseudomonas aeruginosa, E. coli, Shigella spp., and other bacterial pathogens (8, 41, 43, 68). Such alterations have been called pathoadaptive mutations (68). This may be the case for the disseminating lineage of B. burgdorferi. Differences in expression levels of lp54- and lp36-encoded genes in disseminating and nondisseminating strains resulting from either differential transcription or genomic variation were noted. Either mechanism would produce spirochetes with different protein contents. Additionally, a number of genes with potential involvement in nutrient uptake (BB0603, BBA74, BB0329, BB0330, and BBB29) have significantly higher expression in isolate B356. Moreover, nearly 25% of the differentially expressed genes are predicted to be localized on the cell surface. This latter observation implies that these two isolates have considerably different cell surface properties. Mouse infection with isolate BL206 resulted in dissemination to all peripheral tissues tested and a relatively high pathogen burden. In contrast, infection with B356 yielded no disseminated infection, and spirochetes could not be recovered from the inoculation site within 24 h after infection. Similar results were obtained with immunodeficient SCID mice (75, 76). This suggests that isolate B356 is rapidly cleared by components of the innate immune system. The recognition of pathogen-associated molecular patterns plays a central role in the innate immune response to infection (30, 42). Thus, the distinctive natures of the surfaces of BL206 and B356 may result in differential recognition by the innate immune system, leading to the observed differences in spirochete dissemination.
The ORFs listed in Table 2 were scored as having significantly different expression levels in the two tested isolates. These differences can be the result of either alterations in the transcriptional regulation of these genes or differences between the two isolates in terms of genetic content. In either case, these 78 ORFs are a good starting point for the investigation of factors involved in the hematogenous dissemination of B. burgdorferi. Ultimately, the possible roles of these putative virulence genes must be confirmed by mutagenesis and in vivo studies in the rodent-tick infectious cycle. The inactivation of BBA74 is a first step in this process, and more extensive studies with this and other genes are currently under way.
Acknowledgments
We thank Nikhat Parveen for providing one of the B31 strains used in this study; Jon Skare for BBA74-specific antibodies; Doris Bucher for OspA-specific antibodies; Morayo Fakiya, Sabina Sandigursky, and Manjira Ghosh for technical assistance; and Gary Wormser for advice and valuable critical comments.
This work was supported by NIH grant AI45801 and the G. Harold and Leila Y. Mathers Charitable Foundation.
Editor: J. B. Bliska
REFERENCES
- 1.Achouak, W., T. Heulin, and J. M. Pages. 2001. Multiple facets of bacterial porins. FEMS Microbiol. Lett. 199:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homologue. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 3.Anderton, J. M., R. Tokarz, C. D. Thill, C. J. Kuhlow, C. S. Brooks, D. R. Akins, L. I. Katona, and J. L. Benach. 2004. Whole-genome DNA array analysis of the response of Borrelia burgdorferi to a bactericidal monoclonal antibody. Infect. Immun. 72:2035-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anguita, J., M. N. Hedrick, and E. Fikrig. 2003. Adaptation of Borrelia burgdorferi in the tick and the mammalian host. FEMS Microbiol. Rev. 27:493-504. [DOI] [PubMed] [Google Scholar]
- 5.Benach, J. L., E. M. Bosler, J. P. Hanrahan, J. L. Coleman, G. S. Habicht, T. F. Bast, D. J. Cameron, J. L. Ziegler, A. G. Barbour, W. Burgdorfer, R. Edelman, and R. A. Kaslow. 1983. Spirochetes isolated from the blood of two patients with Lyme disease. N. Engl. J. Med. 308:740-742. [DOI] [PubMed] [Google Scholar]
- 6.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57:289-300. [Google Scholar]
- 7.Bono, J. L., K. Tilly, B. Stevenson, D. Hogan, and P. Rosa. 1998. Oligopeptide permease in Borrelia burgdorferi: putative peptide-binding components encoded by both chromosomal and plasmid loci. Microbiology 144:1033-1044. [DOI] [PubMed] [Google Scholar]
- 8.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyce, J. D., P. A. Cullen, and B. Adler. 2004. Genomic-scale analysis of bacterial gene and protein expression in the host. Emerg. Infect. Dis. 10:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brooks, C. S., P. S. Hefty, S. E. Jolliff, and D. R. Akins. 2003. Global analysis of Borrelia burgdorferi genes regulated by mammalian host-specific signals. Infect. Immun. 71:3371-3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bunikis, J., L. Noppa, and S. Bergstrom. 1995. Molecular analysis of a 66-kDa protein associated with the outer membrane of Lyme disease Borrelia. FEMS Microbiol. Lett. 131:139-145. [DOI] [PubMed] [Google Scholar]
- 12.Bunikis, J., L. Noppa, Y. Ostberg, A. G. Barbour, and S. Bergstrom. 1996. Surface exposure and species specificity of an immunoreactive domain of a 66-kilodalton outer membrane protein (P66) of the Borrelia spp. that cause Lyme disease. Infect. Immun. 64:5111-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 14.Chen, T., Y. Hosogi, K. Nishikawa, K. Abbey, R. D. Fleischmann, J. Walling, and M. J. Duncan. 2004. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis. J. Bacteriol. 186:5473-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coburn, J., W. Chege, L. Magoun, S. C. Bodary, and J. M. Leong. 1999. Characterization of a candidate Borrelia burgdorferi beta3-chain integrin ligand identified using a phage display library. Mol. Microbiol. 34:926-940. [DOI] [PubMed] [Google Scholar]
- 16.Coburn, J., and C. Cugini. 2003. Targeted mutation of the outer membrane protein P66 disrupts attachment of the Lyme disease agent, Borrelia burgdorferi, to integrin alphavbeta3. Proc. Natl. Acad. Sci. USA 100:7301-7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conway, T., and G. K. Schoolnik. 2003. Microarray expression profiling: capturing a genome-wide portrait of the transcriptome. Mol. Microbiol. 47:879-889. [DOI] [PubMed] [Google Scholar]
- 18.Cummings, C. A., and D. A. Relman. 2000. Using DNA microarrays to study host-microbe interactions. Emerg. Infect. Dis. 6:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cundell, D. R., B. J. Pearce, J. Sandros, A. M. Naughton, and H. R. Masure. 1995. Peptide permeases from Streptococcus pneumoniae affect adherence to eucaryotic cells. Infect. Immun. 63:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunny, G. M., B. A. Leonard, and P. J. Hedberg. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J. Bacteriol. 177:871-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg, A. P., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed] [Google Scholar]
- 22.Fikrig, E., S. W. Barthold, W. Sun, W. Feng, S. R. Telford III, and R. A. Flavell. 1997. Borrelia burgdorferi P35 and P37 proteins, expressed in vivo, elicit protective immunity. Immunity 6:531-539. [DOI] [PubMed] [Google Scholar]
- 23.Foretz, M., D. Postic, and G. Baranton. 1997. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 47:11-18. [DOI] [PubMed] [Google Scholar]
- 24.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J. F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fuji, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 25.Gilmore, R. D., Jr., M. L. Mbow, and B. Stevenson. 2001. Analysis of Borrelia burgdorferi gene expression during life cycle phases of the tick vector Ixodes scapularis. Microbes Infect. 3:799-808. [DOI] [PubMed] [Google Scholar]
- 26.Grimm, D., C. H. Eggers, M. J. Caimano, K. Tilly, P. E. Stewart, A. F. Elias, J. D. Radolf, and P. A. Rosa. 2004. Experimental assessment of the roles of linear plasmids lp25 and lp28-1 of Borrelia burgdorferi throughout the infectious cycle. Infect. Immun. 72:5938-5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubner, A., A. T. Revel, D. M. Nolen, K. E. Hagman, and M. V. Norgard. 2003. Expression of a luxS gene is not required for Borrelia burgdorferi infection of mice via needle inoculation. Infect. Immun. 71:2892-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer, R., O. Kalu, J. Purser, S. Norris, B. Stevenson, and I. Schwartz. 2003. Linear and circular plasmid content in Borrelia burgdorferi clinical isolates. Infect. Immun. 71:3699-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer, R., D. Liveris, A. Adams, J. Nowakowski, D. McKenna, S. Bittker, D. Cooper, G. P. Wormser, and I. Schwartz. 2001. Characterization of Borrelia burgdorferi isolated from erythema migrans lesions: interrelationship of three molecular typing methods. J. Clin. Microbiol. 39:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janeway, C. A., Jr., and R. Medzhitov. 2002. Innate immune recognition. Annu. Rev. Immunol. 20:197-216. [DOI] [PubMed] [Google Scholar]
- 31.Koide, T., P. A. Zaini, L. M. Moreira, R. Z. Vencio, A. Y. Matsukuma, A. M. Durham, D. C. Teixeira, H. El Dorry, P. B. Monteiro, A. C. da Silva, S. Verjovski-Almeida, A. M. da Silva, and S. L. Gomes. 2004. DNA microarray-based genome comparison of a pathogenic and a nonpathogenic strain of Xylella fastidiosa delineates genes important for bacterial virulence. J. Bacteriol. 186:5442-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labandeira-Rey, M., and J. T. Skare. 2001. Decreased infectivity in Borrelia burgdorferi strain B31 is associated with loss of linear plasmid 25 or 28-1. Infect. Immun. 69:446-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leonard, B. A., A. Podbielski, P. J. Hedberg, and G. M. Dunny. 1996. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc. Natl. Acad. Sci. USA 93:260-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, F. T., F. K. Nelson, and E. Fikrig. 2002. Molecular adaptation of Borrelia burgdorferi in the murine host. J. Exp. Med. 196:275-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(− delta delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 37.Liveris, D., A. Gazumyan, and I. Schwartz. 1995. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 33:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liveris, D., S. Varde, R. Iyer, S. Koenig, S. Bittker, D. Cooper, D. McKenna, J. Nowakowski, R. B. Nadelman, G. P. Wormser, and I. Schwartz. 1999. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J. Clin. Microbiol. 37:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liveris, D., G. P. Wormser, J. Nowakowski, R. Nadelman, S. Bittker, D. Cooper, S. Varde, F. H. Moy, G. Forseter, C. S. Pavia, and I. Schwartz. 1996. Molecular typing of Borrelia burgdorferi from Lyme disease patients by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:1306-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolbert, E. D. Tullson, B. J. Johnson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 41.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medzhitov, R. and C. Janeway, Jr. 2000. Innate immunity. N. Engl. J. Med. 343:338-344. [DOI] [PubMed] [Google Scholar]
- 43.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Narasimhan, S., M. J. Caimano, F. T. Liang, F. Santiago, M. Laskowski, M. T. Philipp, A. R. Pachner, J. D. Radolf, E. Fikrig, and M. J. Camaino. 2003. Borrelia burgdorferi transcriptome in the central nervous system of non-human primates. Proc. Natl. Acad. Sci. USA 100:15953-15958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Narasimhan, S., F. Santiago, R. A. Koski, B. Brei, J. F. Anderson, D. Fish, and E. Fikrig. 2002. Examination of the Borrelia burgdorferi transcriptome in Ixodes scapularis during feeding. J. Bacteriol. 184:3122-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ojaimi, C., C. Brooks, D. Akins, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katonah, J. Radolf, M. Caimano, J. Skare, K. Swingle, S. Sims, and I. Schwartz. 2002. Borrelia burgdorferi gene expression profiling with membrane-based arrays. Methods Enzymol. 358:165-177. [DOI] [PubMed] [Google Scholar]
- 47.Ojaimi, C., C. Brooks, S. Casjens, P. Rosa, A. Elias, A. Barbour, A. Jasinskas, J. Benach, L. Katona, J. Radolf, M. Caimano, J. Skare, K. Swingle, D. Akins, and I. Schwartz. 2003. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun. 71:1689-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ostberg, Y., J. A. Carroll, M. Pinne, J. G. Krum, P. Rosa, and S. Bergstrom. 2004. Pleiotropic effects of inactivating a carboxyl-terminal protease, CtpA, in Borrelia burgdorferi. J. Bacteriol. 186:2074-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ostberg, Y., M. Pinne, R. Benz, P. Rosa, and S. Bergstrom. 2002. Elimination of channel-forming activity by insertional inactivation of the p13 gene in Borrelia burgdorferi. J. Bacteriol. 184:6811-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parra-Lopez, C., M. T. Baer, and E. A. Groisman. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12:4053-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173-185. [DOI] [PubMed] [Google Scholar]
- 52.Porcella, S. F., and T. G. Schwan. 2001. Borrelia burgdorferi and Treponema pallidum: a comparison of functional genomics, environmental adaptations, and pathogenic mechanisms. J. Clin. Investig. 107:651-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Probert, W. S., and B. J. Johnson. 1998. Identification of a 47 kDa fibronectin-binding protein expressed by Borrelia burgdorferi isolate B31. Mol. Microbiol. 30:1003-1015. [DOI] [PubMed] [Google Scholar]
- 54.Purser, J. E., M. B. Lawrenz, M. J. Caimano, J. K. Howell, J. D. Radolf, and S. J. Norris. 2003. A plasmid-encoded nicotinamidase (PncA) is essential for infectivity of Borrelia burgdorferi in a mammalian host. Mol. Microbiol. 48:753-764. [DOI] [PubMed] [Google Scholar]
- 55.Purser, J. E., and S. J. Norris. 2000. Correlation between plasmid content and infectivity in Borrelia burgdorferi. Proc. Natl. Acad. Sci. USA 97:13865-13870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Revel, A. T., A. M. Talaat, and M. V. Norgard. 2002. DNA microarray analysis of differential gene expression in Borrelia burgdorferi, the Lyme disease spirochete. Proc. Natl. Acad. Sci. USA 99:1562-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rudner, D. Z., J. R. LeDeaux, K. Ireton, and A. D. Grossman. 1991. The spo0K locus of Bacillus subtilis is homologous to the oligopeptide permease locus and is required for sporulation and competence. J. Bacteriol. 173:1388-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. USA 97:14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samuels, D. S. 1995. Electrotransformation of the spirochete Borrelia burgdorferi. Methods Mol. Biol. 47:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schoolnik, G. K. 2002. Functional and comparative genomics of pathogenic bacteria. Curr. Opin. Microbiol. 5:20-26. [DOI] [PubMed] [Google Scholar]
- 61.Schoolnik, G. K. 2002. Microarray analysis of bacterial pathogenicity. Adv. Microb. Physiol. 46:1-45. [DOI] [PubMed] [Google Scholar]
- 62.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schwartz, I., G. P. Wormser, J. J. Schwartz, D. Cooper, P. Weissensee, A. Gazumyan, E. Zimmermann, N. S. Goldberg, S. Bittker, and G. L. Campbell. 1992. Diagnosis of early Lyme disease by polymerase chain reaction amplification and culture of skin biopsies from erythema migrans lesions. J. Clin. Microbiol. 30:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Skare, J. T., C. I. Champion, T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1996. Porin activity of the native and recombinant outer membrane protein Oms28 of Borrelia burgdorferi. J. Bacteriol. 178:4909-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Skare, J. T., T. A. Mirzabekov, E. S. Shang, D. R. Blanco, H. Erdjument-Bromage, J. Bunikis, S. Bergstrom, P. Tempst, B. L. Kagan, J. N. Miller, and M. A. Lovett. 1997. The Oms66 (p66) protein is a Borrelia burgdorferi porin. Infect. Immun. 65:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sokurenko, E. V., D. L. Hasty, and D. E. Dykhuizen. 1999. Pathoadaptive mutations: gene loss and variation in bacterial pathogens. Trends Microbiol. 7:191-195. [DOI] [PubMed] [Google Scholar]
- 69.Spach, D. H., W. C. Liles, G. L. Campbell, R. E. Quick, D. E. Anderson, Jr., and T. R. Fritsche. 1993. Tick-borne diseases in the United States. N. Engl. J. Med. 329:936-947. [DOI] [PubMed] [Google Scholar]
- 70.Steere, A. C. 2001. Lyme disease. N. Engl. J. Med. 345:115-125. [DOI] [PubMed] [Google Scholar]
- 71.Steere, A. C., R. L. Grodzicki, A. N. Kornblatt, J. E. Craft, A. G. Barbour, W. Burgdorfer, G. P. Schmid, E. Johnson, and S. E. Malawista. 1983. The spirochetal etiology of Lyme disease. N. Engl. J. Med. 308:733-740. [DOI] [PubMed] [Google Scholar]
- 72.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tokarz, R., J. M. Anderton, L. I. Katona, and J. L. Benach. 2004. Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun. 72:5419-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, G., C. Ojaimi, R. Iyer, V. Saksenberg, S. A. McClain, G. P. Wormser, and I. Schwartz. 2001. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun. 69:4303-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang, G., C. Ojaimi, H. Wu, V. Saksenberg, R. Iyer, D. Liveris, S. A. McClain, G. P. Wormser, and I. Schwartz. 2002. Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J. Infect. Dis. 186:782-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang, G., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang, X. G., B. Lin, J. M. Kidder, S. Telford, and L. T. Hu. 2002. Effects of environmental changes on expression of the oligopeptide permease (opp) genes of Borrelia burgdorferi. J. Bacteriol. 184:6198-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wormser, G. P., S. Bittker, D. Cooper, J. Nowakowski, R. B. Nadelman, and C. Pavia. 2000. Comparison of the yields of blood cultures using serum or plasma from patients with early Lyme disease. J. Clin. Microbiol. 38:1648-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wormser, G. P., D. Liveris, J. Nowakowski, R. B. Nadelman, L. F. Cavaliere, D. McKenna, D. Holmgren, and I. Schwartz. 1999. Association of specific subtypes of Borrelia burgdorferi with hematogenous dissemination in early Lyme disease. J. Infect. Dis. 180:720-725. [DOI] [PubMed] [Google Scholar]
- 81.Yang, X., M. S. Goldberg, T. G. Popova, G. B. Schoeler, S. K. Wikel, K. E. Hagman, and M. V. Norgard. 2000. Interdependence of environmental factors influencing reciprocal patterns of gene expression in virulent Borrelia burgdorferi. Mol. Microbiol. 37:1470-1479. [DOI] [PubMed] [Google Scholar]
- 82.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of VMP-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]
- 83.Zhong, J., and A. G. Barbour. 2004. Cross-species hybridization of a Borrelia burgdorferi DNA array reveals infection- and culture-associated genes of the unsequenced genome of the relapsing fever agent Borrelia hermsii. Mol. Microbiol. 51:729-748. [DOI] [PubMed] [Google Scholar]