Abstract
The HtrA surface protease in gram-positive bacteria is involved in the processing and maturation of extracellular proteins and degradation of abnormal or misfolded proteins. Inactivation of htrA has been shown to affect the tolerance to thermal and environmental stress and to reduce virulence. We found that inactivation of Streptococcus mutans htrA by gene-replacement also resulted in a reduced ability to withstand exposure to low and high temperatures, low pH, and oxidative and DNA damaging agents. The htrA mutation affected surface expression of several extracellular proteins including glucan-binding protein B (GbpB), glucosyltransferases, and fructosyltransferase. In addition, htrA mutation also altered the surface expression of enolase and glyceraldehyde-3-phosphate dehydrogenease, two glycolytic enzymes that are known to be present on the streptococcal cell surface. As expected, microscopic analysis of in vitro grown biofilm structure revealed that the htrA deficient biofilms adopted a much more granular patchy appearance, rather than the relatively smooth confluent layer normally seen in the wild type. These results suggest that HtrA plays an important role in the biogenesis of extracellular proteins including surface associated glycolytic enzymes and in biofilm formation of S. mutans.
Streptococcus mutans has been strongly implicated as the principal etiological agent in human dental caries (44). In addition to dental caries, S. mutans is also an important agent of infective endocarditis (4, 79). More than 20% of cases of viridians streptococcus-induced endocarditis are caused by S. mutans (29, 44). S. mutans expresses plethora surface proteins, and many of them are virulence factors. These include adhesins, specialized transport systems for fermentable sugars, synthesis of soluble and insoluble dextrans and fructans, and factors required for acidogenesis and acidouricity (38). Various cell-surface substances, including serotype-specific polysaccharide antigens, lipoteichoic acid, glucosyltransferases (GTFs), fructosyltransferase (FTF), dextranase, glucan-binding proteins, a 29-kDa protein WapA, and a 190-kDa SpaP (also known as protein P1, PAc, antigen B, and antigen I/II) are thought to play important roles in interaction between the organism and its host (35, 38), and have been considered for vaccine candidates for dental caries (for review, see reference 34).
Biofilms consist of complex mixture of microorganisms that adhere to each other and in most cases to a surface. One of the important virulence properties of mutans streptococci is their ability to form biofilms along with other bacteria (38, 83). This biofilm, known as dental plaque, is one of the best-studied biofilms (20, 36). Biofilm formation is considered to be a two-step sequential process requiring early attachment of the bacterial cells to a surface, followed by growth dependent multilayer accumulation of bacteria involving intra- and intergenic cell-to-cell interactions and adhesions (20). There have been considerable efforts to identify factors of S. mutans and other oral streptococci that are involved in biofilm initiation and development. Surface-associated proteins, such as SpaP and Fap1, function as high-affinity adhesins and are required in the initiation of biofilm (12, 23, 53). Extracellular glucans, synthesized from sucrose by GTFs, are key players in adhesion interactions and accumulation of mutans streptococci on smooth surfaces (38). Glucan-binding proteins (Gbp) such as GbpC are involved in rapid, dextran-dependent aggregation during biofilm formation (70). Interestingly, immunization with GbpB, another glucan-binding protein, induces an immune response in rats that interferes with the accumulation of S. mutans and reduces the level of dental caries (73). There are numerous cell-surface and extracellular proteins that work in concert to successfully establish S. mutans in tooth biofilms (38).
In bacteria, proteolysis plays important roles in many biological processes such as post-translational regulation of gene expression. This includes processing and maturation of various surface associated proteins in the case of gram-positive bacteria (26, 40). In S. mutans, cell surface protein SpaP is found free in culture supernatant, along with a number of lower-molecular-mass forms (65). Another surface associated protein WapA, is also released by proteolytic cleavage (22) and two cell-wall-associated enzymes, dextranase and fructanase, are found predominantly in culture supernatants (14, 31, 32). In addition to the proteolytic events that release these cell-surface-associated proteins and enzymes, extracellular GTFs and FTF are also broken down to lower-molecular-mass-forms (1, 68) and function as glucan-binding proteins (67). Thus, expression of various surface proteins is dependent on proteolysis, which can strongly influence both the level of activity and the cellular localization of these proteins.
There are several different proteases present in various streptococci (for reviews, see references 19 and 62). Among them, the trypsin like serine protease HtrA (also known as DegP) (for reviews, see references 18 and 54) is of scientific interest for several reasons including its role in cellular physiology, in particular in helping bacteria to survive environmental stresses such as elevated temperature, oxidative and osmotic stresses. HtrA homologs have been identified in phylogenetically distinct bacteria including many gram-positive pathogens suggesting a fundamental role of HtrA in response to environmental stress and possibly in pathogenesis. The proteins characteristically possess an amino-terminal hydrophobic region, a trypsin-like catalytic domain with conserved His, Ser and Asp residues and a PDZ domain thought to be involved in protein-protein interactions (69). HtrA proteins have been shown to have a housekeeping function, acting as chaperones and degrading misfolded proteins (74). Interestingly, in Lactococcus lactis, a closely related bacterium to S. mutans, HtrA was shown to be involved in the processing of propeptides and in the maturation of a natural extracellular protein (59). HtrA was thus proposed to be involved not only in degradation of misfolded proteins, but also in the processing/maturation of normal proteins. The Streptococcus pyogenes HtrA was also shown to be involved in production of several virulence factors whose biogenesis requires extensive processing. This includes a cysteine protease SpeB and a hemolysin SLS (46). Recently, the Staphylococcus aureus HtrA was also shown to control expression of several secreted virulence factors, including hemolysins (63). Thus, it appears that HtrA is involved in bacterial pathogenesis or by modulating virulence factor expression.
The regulation and organization of the htrA locus are very different in various gram-positive bacteria. For example, Bacillus subtilis and S. aureus both have more than one functional htrA gene and they are located far from one another in the genome (51, 52, 63). In streptococci, a single copy of htrA is present and it is located near the chromosomal replication origin. The immediate downstream genes are highly conserved and are involved in cell division (24), but the organization of genes upstream of the htrA locus is not well conserved (Fig. 1). Because htrA is located near the replication origin, inactivation by insertional mutagenesis may alter the efficiency of cell division by interfering with the downstream dnaA gene and can cause growth defects as proposed for S. pyogenes (46).
FIG. 1.
Construction of S. mutans htrA-null strain. (A) Organization of open reading frames in the htrA region of various streptococci. The GenBank accession numbers of the sequences are as follows: S. mutans (Smu), NC004350; S. pyogenes (Spy), AE004092; S. pneumoniae (Spn), NC003098; and L. lactis (Lla), NC002662. Except for L. lactis, the region downstream of htrA is conserved and corresponds to chromosome replication origin/function, while the upstream region is not conserved. (B). Schematic diagram of the htrA inactivated S. mutans. The omega-Km cassette (thick black arrow) was used to inactivate the htrA gene by inserting it in the coding region. The chromosomal segments used as the homologous region for gene replacement are indicated by small arrowheads (1 and 2). The putative promoter of htrA gene is indicated by a bent arrow and the putative transcriptional terminator is shown by a lollipop. The region of htrA that was cloned in complementing plasmid pIB108 is indicated by a bar below. (C). PCR verification of htrA inactivation. The primer pairs used for this amplification (arrowheads 1 and 2) generate a diagnostic 1.9-kb fragment from the wild type and 3.9-kb fragment from the htrA mutant strains. The orientation of kamamycin-resistant cassette was verified by PCR using primers 1 and 4 and 3 and 2. The samples are as follows: M, 1-kb NEB ladder; 1, NG8; 2, isogenic htrA mutant (IBS101).
Based on the studies in other bacteria, one would expect S. mutans HtrA to be involved in the processing of extracellular (exported/cell-wall-associated) proteins including adhesins that are involved in biofilm formation and pathogenesis. However, a recent study with a single-crossover htrA mutant of S. mutans showed no significant effect on expression or processing of several extracellular proteins (21). The aim of this present study was to further investigate the role of HtrA on surface protein expression in S. mutans. In particular, the contribution of HtrA to the development of biofilm formation by this organism was examined. As previously observed, a mutant of HtrA showed several stress sensitive phenotypes (21). However, in contrast to the previous study, we found that mutants of HtrA displayed altered expression of several surface proteins including various surface associated glycolytic enzymes as well as formation of a biofilm with altered architecture.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli strain DH5α was grown in Luria-Bertani medium, and, when necessary, ampicillin (100 μg ml−1), kanamycin (100 μg ml−1), and/or spectinomycin (100 μg ml−1) were included. S. mutans strains were routinely grown in Todd-Hewitt medium (BBL; Becton Dickinson) supplemented with 0.2% yeast extract (THY). When necessary, kanamycin (300 μg ml−1) and/or spectinomycin (300 μg ml−1) was included.
For growth experiments involving stress tolerance, THY agar plates (1.5%) containing NaCl (0.5 to 1.5 M), puromycin (0.5 to 2.0 μg/ml; Sigma-Aldrich), H2O2 (1.0 to 2.5 mM; Sigma-Aldrich), mitomycin C (7.5 to 15 ng/ml; Sigma-Aldrich) were poured (we observed that the activity of puromycin and H2O2 varied between batches). Freshly grown overnight cultures were spun down and washed with sterile 0.9% saline. Cultures were resuspended in either the same or 1/10 of the original volume in 0.9% saline and 10-fold serial dilutions were made. A 10-μl aliquot of each dilution was spotted on the THY agar plates containing different stressors and plates were incubated at 37°C aerobically.
For growth experiments involving pH, initial pHs of THY agar were adjusted to pHs 5.5, 6.0, and 7.0 with HCl before sterilization. 50 mM citrate-phosphate buffer of desired pH was added to media after sterilization. Different dilutions of bacterial cultures were spotted as described above and plates were incubated at 37°C anaerobically. Hydrophobicity assay was done as described previously (71).
For some growth experiments and biofilm assay, semidefined biofilm medium (BM) (45) was also used supplemented with 1% glucose or 1% sucrose as the carbon source. Preparation of competent cells and transformation of S. mutans were done as described previously (15, 58).
Construction of strains.
The htrA gene was insertionally inactivated in NG-8 strains by gene-replacement. Based on the UA159 genome sequence information (GenBank accession no. AE015037), a 1.9-kb DNA fragment containing the htrA gene was PCR amplified from NG-8 genomic DNA using primers HtrA-For (5′-GACTAGCATTATTTGGAATTTTCTCATCGG-3′) and HtrA-Rev (5′-GTCAACGAAGTTGTCTTCATATCTCACCTC-3′). The DNA fragment was cloned into the pGEMT-Easy TA cloning vector (Promega), and the resulting construct was confirmed by restriction analysis. A kanamycin-resistant cassette (ΩKm) (57) isolated from plasmid pUC4ΩKm upon digestion by SmaI was then ligated into a unique PacI site (restricted and blunted by T4 polymerase) within the htrA coding sequence (at amino acid position 243), and the resulting construct was named pIB101. Plasmid pIB101 was linearzed by NotI and then used to transform S. mutans NG-8 strains according to a previously published protocol (15). Kanamycin-resistant transformants were selected on THY agar containing appropriate antibiotic, and PCR analysis was done to confirm that htrA inactivation had occurred by double-crossover recombination.
To express the htrA gene in trans, the full-length htrA (with promoter) was amplified by PCR using primers HtrA-For and HtrA-Rev and cloned into pAT28 (78) that had been restricted by SmaI. The resulting plasmid contains the intact hrtA gene under its own promoter.
Preparation of whole-cell extract, cell wall, and supernatant proteins.
Overnight cultures were grown in THY, collected by centrifugation, and washed twice in phosphate-buffered saline (PBS). Cell density was adjusted to 5.0 cell units ml−1 (1 cell unit per ml, equivalent to 1 ml culture at an optical density at 600 nm of 2.0) and total cell extract were prepared by lysing the cell suspension with a glass bead beater as described for S. pyogenes (10). Cell wall preparation was carried out as using mutanolysin and lysozyme as described previously (10). For culture supernatant proteins, cells were removed from fresh overnight culture by centrifugation followed by filtration through a 0.2-μm pore size filter. Proteins were obtained by precipitation with trichloroacetic acid (TCA, 20% [wt/vol] final concentration) on ice for 2 to 4 h, washed with acetone, and resuspended in PBS or 1× gel loading buffer (NEB) to 1/40 volume. Equal amounts of proteins (as measured by Bradford assay) were loaded in each lane for Western analysis.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-4 to 20% polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto a nitrocellulose membrane and blocked with 3% bovine serum albumin (BSA)-0.1% Tween 20 in Tris-buffered saline for 1 h at room temperature. Polyclonal antibodies to S. pyogenes glyceraldehyde-3-phosphate dehydrogenease (GAPDH) or enolase (kindly provided by Vijay Pancholi) and FTF (kindly provided by Gilad Bachrach) were added (1:1,000 dilutions) and incubated for 1 h at room temperature. Membranes were washed thoroughly with 0.1% Tween 20 in Tris-buffered saline, probed with anti-rabbit immunoglobulin G-alkaline phosphatase-conjugated secondary antibodies for 1 h, and washed again. Reactivity was detected using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (BCIP) as substrates.
Detection of proteins on immobilized S. mutans cells.
S. mutans strains grown overnight were collected by centrifugation and washed twice with PBS. Cell density was adjusted to 2.0 cell units ml−1 and serial dilutions were made in PBS. Aliquots of 5 μl were spotted onto nitrocellulose membranes and air dried, and nonspecific sites were blocked with 3% BSA-0.1% Tween 20 in PBS (PBST) at room temperature for 1 h. Binding to digoxigenin (DIG)-labeled proteins was carried out as described previously (10). Binding to anti-GbpB antibody (kindly provided by Daniel Smith) was carried out as described above for Western blot analysis except that anti-rat secondary antibody was used. Binding to antigen P1 was done using a monoclonal antibody to P1 (kindly provided by L. Jeannine Brady). Binding to biotin-dextran was done as described previously (71) with the following modifications. Membranes were blocked in PBS with 3% BSA for one hour and washed three times with PBS plus 0.1% Tween 20 (PBST). Biotin-dextran (200 μg ml−1; Sigma) in PBS with 0.2% BSA were then added and incubated for one hour. Membranes were washed with PBST and then anti-biotin monoclonal antibody (Sigma) was used to detect dextarn binding. All the secondary antibodies used were alkaline phosphatase conjugated. Binding of ligand to the cell wall was detected using nitroblue tetrazolium and BCIP as substrates.
Biofilm formation assay.
Biofilms of S. mutans were grown in two different media. NG-8 and its derivatives were grown overnight in THY medium at 37°C anaerobically. The culture was diluted 1:10 into fresh THY medium and incubated further for 6 h. The culture was then diluted 1:1,000 into either THY or BM (45) media containing 1% sucrose. 0.4 or 0.8 ml of this cell suspension was added to each well of an eight-well or four-well, respectively, glass chamber slide (Lab-Tek; Nalge Nunc International) for biofilm formation on glass. For biofilm formation on polystyrene surface, flat-bottomed 96-well microtiter plates (Corning Inc.) were used. The slides or the microtiter plates were incubated at 37°C for 20 to 24 h as a static culture to allow biofilm formation.
Confocal and scanning election microscopy.
Biofilm formation on glass slides was first examined by confocal laser scanning microscopy (CLSM). Culture media was removed from the slide chamber and the chamber was washed with sterile distilled water. Biofilm cells were stained with labeling solution containing 0.2 μM BacLight Green (Molecular Probes) in 0.9% saline for 1 h to allow live cells to metabolize the dye and develop fluorescence. Slides were analyzed by CLSM.
Biofilms formed on glass slides were also analyzed by scanning electron microscopy (SEM). Slides were washed once with sterile water, fixed with glutaraldehyde, and incubated at room temperature overnight. Following dehydration through a graded series of ethanol washes, slides were air dried, sputter coated with gold and analyzed by SEM (ISI-60A; International Scientific Instruments) at several magnifications at the University of South Dakota core facility.
PM tests.
Phenotype microarray (PM) tests were performed at Biolog, Inc., essentially as described elsewhere. Both the wild type (NG-8) and the htrA mutant (IBS101) were subjected to full 20-panel PM analysis. PM arrays contain different carbon sources, nitrogen sources, phosphorus sources, sulfur sources as well as various inhibitory compounds for stress responses as described previously (11, 84). Readings were recorded for 24 h, and kinetic data were analyzed with OmniLog-PM software. This software generates time course curves for respiration and calculates differences in the areas for mutant and wild-type cells. The differences are averages of values reported for two separate experiments. Detailed information of PM experiments is available at http://www.biolog.com.
RESULTS
Characterization of S. mutans htrA mutants.
The htrA gene of S. mutans strain NG-8 was disrupted by the introduction of a kanamycin-resistant cassette into the coding region of the gene. The inactivation was confirmed by primers flanking htrA to amplify a diagnostic PCR fragment of 4.0 kb from the kanamycin-resistant transformants and a 1.9-kb fragment of the wild-type copy of htrA (Fig. 1).
We first verified the subcellular localization of HtrA protein in S. mutans strains. Different subcellular fractions from NG-8 (wild type) were prepared as described in the Materials and Methods, fractionated by SDS-PAGE, and probed with anti-HtrA antibody generated against the B. subtilis HtrA ((3), kindly provided by Dr. Devine). This antibody reacted with a diagnostic band of ∼43-kDa length from whole-cell lysate, cell wall, and culture supernatant fractions from NG-8 strain. This immunoractive band was absent from any of the fractions from IBS101 (htrA mutant) (data not shown). This suggests that, like B. subtilis HtrA, S. mutans HtrA is also present on the cell surface (extracellular).
Compared to the wild-type strain, the htrA mutant strain (IBS101) was found to grow similarly at 37°C in both rich (THY) and chemically defined (BM) media (data not shown). No differences in colony morphology were noted between the htrA and wild-type strain. However, one striking feature of the htrA mutant was its capability to form longer chains compared to wild-type NG-8 (two to three times longer) (Fig. 2A). Increased chaining was primarily associated with late-exponential- to stationary-phase cultures. In this aspect, the htrA mutant behaved like the biofilm deficient brpA mutant strain described previously (80).
FIG. 2.
Phenotypic characterization of htrA mutant strain. (A) Overnight cultures of the wild type (NG-8) and its isogenic htrA mutant strain (IBS101) were grown in THY medium and the bacteria were visualized by scanning electron microscopy (magnification, ×1,600). (B) Effect of temperature on the growth of S. mutans strains. Different dilutions of fresh overnight cultures were spotted on MS-agar plates and incubated under microaerophilic conditions at the indicated temperature for 40 h before photographed. Samples are NG-8 (wild type), IBS101 (htrA), and IBS101/pIB108 (htrA/htrA+). Experiments were repeated no fewer than three times and relevant areas of representative plates are shown.
As expected, the S. mutans htrA mutant also showed a reduced temperature range for growth. As shown in Fig. 2B, htrA failed to grow on mitis salivarius agar plate at both 30°C and 42°C, whereas the wild-type grew satisfactorily (Fig. 2). To confirm that the lack of HtrA was responsible for the observed phenotype, the intact htrA gene was provided in trans on plasmid pIB108. Introduction of pIB108 in IBS101 restored the growth capacity both at 30°C and 42°C (Fig. 2B).
In S. pneumoniae, disruption of htrA causes a drastic reduction in transformation efficiency (30). However, when we compared the transformation efficiency for replicated plasmid, integrated plasmid and linear chromosomal DNA, we found no differences between wild-type and htrA strains of S. mutans (data not shown).
Stress response phenotypes of htrA mutant.
The HtrA protease in other bacteria has been found to be associated with the ability to survive under different stress conditions. Therefore first we tested the htrA mutant for its ability to withstand treatment with H2O2 for oxidative stresses and treatment with puromycin (which causes premature chain termination during protein synthesis) to mimic thermal stress. In both cases, the htrA mutant was more sensitive to stress conditions compared to the wild-type strain (Fig. 3). Interestingly, there were no significant differences when cells were subjected to superoxide stress generated by paraquate or tert-butyl hydroperoxide (data not shown).
FIG. 3.
HtrA is essential for stress tolerance in S. mutans. (A) Freshly grown overnight cultures of NG-8 (wild type), IBS101 (htrA), and IBS101/pIB108 ((htrA/htrA+) were washed and 10-fold serially diluted in PBS. Ten-microliter samples were spotted on THY agar plates with nothing (37°C), puromycin (0.75 μM, PUR), or H2O2 (1.5 mM). (B) Cultures were spotted on THY agar plates that were buffered to generate pH 7.0, pH 6.0, or pH 5.5. (C) Cultures were spotted on THY agar containing nothing (control), NaCl (1 M), or mitomycin C (12.5 ng/ml, MC). Plates were incubated at 37°C in the presence of ambient air (A and C) or anaerobically (B). Experiments were repeated no fewer than three times and relevant areas of representative plates are shown.
S. mutans is known to adapt to a low pH environment and mount an acid tolerance response (7, 27, 61). Acid tolerance induces several stress-responsive proteins. In order to determine whether htrA is involved in acid tolerance, we measured growth of the htrA mutant under acidic conditions. As shown in Fig. 3B, the growth of the htrA mutant strain was significantly reduced at pH 5.5 as compared to the wild type. However, the htrA mutant grew as the wild type at pH 6.0.
Since many of the proteins involved in acid tolerance are also involved in DNA damage repair (such as uvrA), we tested htrA mutant for its ability to withstand the DNA damaging agent mitomycin C. Mitomycin C alkylates double stranded DNA and thereby blocks DNA replication. As shown in Fig. 3C, survival of S. mutans was greatly reduced when htrA was absent. This suggests that htrA plays an important role in the DNA damage repair pathway. RecA is one of the key proteins involved in DNA repair, however it is also induced under acid stress (42). We therefore tested the strains for UV sensitivity since RecA is involved in UV-irradiated DNA damage repair. We found no differences between wild-type and htrA mutant strains (data not shown) indicating protein other than RecA is directly or indirectly involved in this phenomenon.
In Listeria monocytogenes, htrA was recently shown to be involved in salt tolerance (82). Therefore, we tested whether S. mutans htrA is also involved in salt tolerance. We found that like L. monocytogenes, the S. mutans htrA mutant was sensitive to salt stress. All the stress sensitive phenotypes could be complemented by introducing plasmid pIB108 into the IBS101 strain. Taken together our results show that S. mutans htrA is indeed involved in various stress response pathways including DNA damage repair.
PM analysis of the htrA mutant.
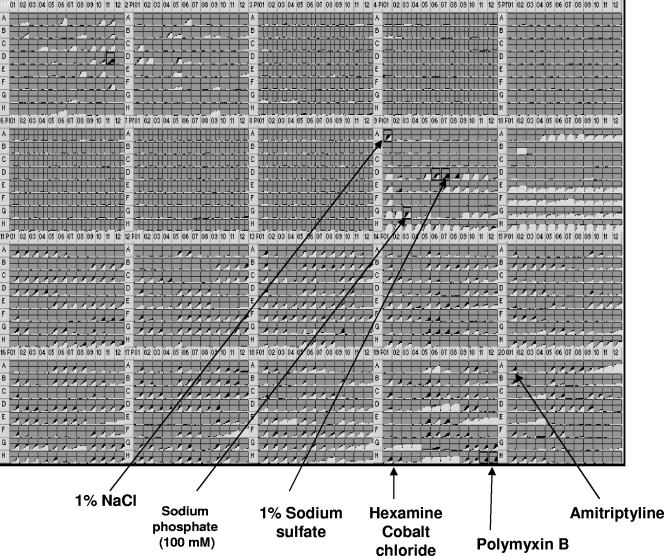
PM assay is a relatively new tool which allows testing of mutants for a large number of phenotypes simultaneously (11, 37, 60, 84). The growth of bacteria in different media is measured with tetrazolium redox dye. Reduction of this dye due to respiration during bacterial growth results in purple color formation and accumulates in the well over the incubation period. Total loss of function will result in no growth and no color formation. Thus, colorimetric detection due to respiration can provide a reporter system for phenomic testing. PM assays were performed on the htrA mutant (IBS101), compared with the wild-type strain (NG-8), in a set of 20 96-well microplates containing various nutrients or inhibitors. This allowed testing of nearly 1,900 cellular phenotypes in a sensitive, highly controlled, and reproducible format. As shown in Fig. 4, the metabolic signal was generally low for nitrogen metabolism (PM3, -6, -7, -8), phosphate and sulfate metabolism (PM4), nutrient stimulation (PM5) with the exception of carbon sources (PM1 and -2). However, the result indicated that the mutant (IBS101) showed slower growth using carbon sources such as sucrose (PM1, D11) and lactulose (PM1, D10) than its parental strain (NG-8), while there was no significant change in utilization of other carbon sources (Fig. 4). There was systematically lower signal for the mutant strain (IBS101) in PM9 through -20, suggesting lower levels of growth and/or metabolism relative to the wild type (NG-8) in these panels. Specifically, there were negative differences in salt and osmotic tolerance for 1% sodium chloride (PM9, A1), 3 to 4% sodium sulfate (PM9, D6 and -7), and phosphate toxicity of 100 mM sodium phosphate (PM9, G3). This observation is consistent with our result with osmotic stress (Fig. 3). Also, there were significant differences for other chemical sensitive panels such as hexamine cobalt chloride (PM19, H1 and -2), polymyxin B (PM19, H11 and -12), and amitriptyline (PM20, A1).
FIG. 4.
PM comparison of NG-8 and htrA mutant. Time course curve for respiration (tetrazolium color formation) was generated with OmniLog-PM software. Grey indicates that growth of the wild-type NG-8 and the mutant IBS101 were similar. Black indicates faster growth of either the NG-8 or the IBS101 strain.
Altered surface protein expression in htrA mutant.
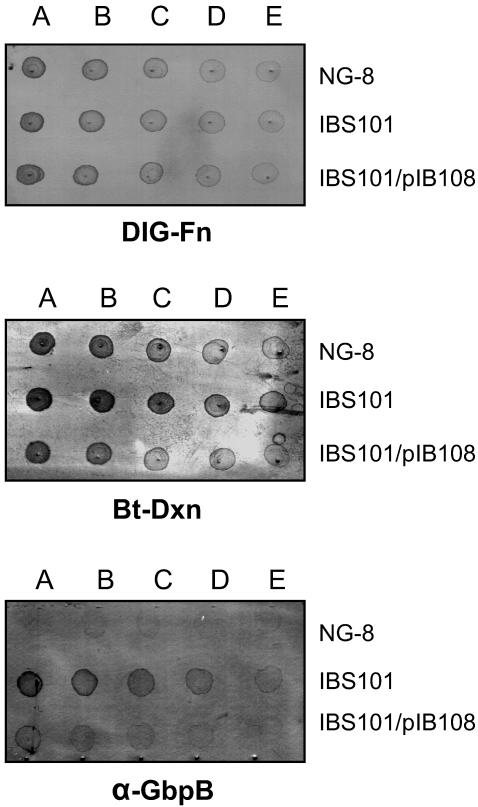
Based on the previous studies in other gram-positive bacteria, it appears that HtrA is involved in the processing and maturation of extracellular proteins. Recently in S. pyogenes it was shown that SpeB protein is processed by HtrA to active form from zymogen (46). In addition, biogenesis of streptolysin S (SLS) activity is altered by htrA mutation in this bacterium (46). We hypothesized that in S. mutans, HtrA is also involved in processing and maturation of various extracellular proteins. If HtrA affects the processing of pre-proteins or cell-associated proteins, one would expect to find a different protein profile in the culture supernatant of htrA mutant, compared to wild-type strain. When we tested for hydrophobicity, it appeared that the htrA mutant showed a drastic reduction, calculated from the proportion of cells associated with the hexadecane phase (29% for wild-type NG-8 compare to 10% for the mutant IBS101). This indicates that surface protein expression is probably altered in the mutant. Therefore, we tested expression of several extracellular proteins in the htrA mutant. A simple test for expression of surface proteins is by spotting whole cells onto nitrocellulose membranes and developing the membranes with various DIG-labeled ligands or antibodies. Since glucans play an important role in S. mutans adhesion and accumulation on tooth surfaces, we assayed for expression of glucan-binding proteins (GBPs) first. At least three GBPs are present in S. mutans and these proteins are very important for bacterial adhesion (5). Most of the GBPs are found both on the cell surface and in the intracellular compartment of the bacterium. GBPs can bind to dextran as ligand. As shown in Fig. 5, binding to biotin-dextran was drastically increased (approximately two- to fourfold) in the htrA mutant IBS101. Since, GbpB was shown to bind glucan (72), we tested for surface expression of GbpB. As expected, GbpB expression was increased (more than eightfold) in IBS101 compared to NG-8. Both of these phenotypes can be complemented with pIB108, a plasmid containing full-length htrA. We also tested surface expression of antigen P1 (data not shown), fibrinogen (data not shown) binding protein, and fibronectin-binding protein (Fig. 5) but did not find any differences.
FIG. 5.
Expression of surface proteins in the htrA mutant. Twofold serial dilutions starting with 2 cell units/ml (A to E) from freshly prepared overnight cultures were spotted onto a nitrocellulose membrane and probed with DIG-labeled fibronectin (DIG-Fn), biotin-dextran (Bt-Dxn), and anti-GbpB. Samples are NG-8 (wild type), IBS101 (htrA), and IBS101/pIB108 (htrA/htrA+).
Expression of several exported virulence factors was recently shown to be affected by htrA mutation in S. aureus (62). Therefore, involvement of HtrA in the expression of exported proteins in S. mutans was examined. Protein extracts were prepared from cells and culture supernatants from stationary phase cultures of the wild-type (NG-8) and htrA mutant (IBS101) cells and analyzed by SDS-PAGE. As shown in Fig. 6A, the expression profile of exoproteins was markedly different. In the IBS101 extracts, we consistently observed increased proteins compared to NG-8. Protein profiles of mid-exponential-phase growing cultures analyzed by SDS-PAGE were essentially the same for wild type and the mutant (data not shown). This was also the case for total cellular fraction profiles from overnight cultures (data not shown), although the resolution in the SDS-PAGE may be likely to mask any differences. Mass spectromectric analysis was performed on several secreted proteins that were present in higher amount in the mutant strain compared to wild-type strain. At least five secreted proteins appear to be accumulated more in the stationary-phase supernatant of IBS101 relative to wild-type strain. Mass spectrometric identification suggests that GtfB, FTF, GbpB, enolase, and GAPDH (Fig. 6A and data not shown) are accumulated more in the IBS101 strain compared to NG-8.
FIG. 6.
Analysis of extracellular proteins from the wild type and the htrA mutant. (A) Supernatant proteins from overnight cultures were precipitated by 20% TCA, washed with acetone and resuspended in PBS. Equal amounts of proteins were loaded in each lane and samples were run on SDS-4 to 20% PAGE gels and stained with Coomassie blue. Bands corresponding to arrowheads were excised from stained gel and identified by mass spectrometry. Lanes: M, NEB prestained marker; 1, NG-8; 2, IBS101. Proteins identified by mass spectrometry are indicated at the right. (B) Western blot analysis of supernatant proteins separated by SDS-4 to 20% PAGE. The antibodies used are indicated below the gels. Lanes contain sample from NG-8 (lanes 1), IBS101 (lanes 2), and IBS101/pIB108 (lanes 3). Open arrows indicate different species of the reacted proteins.
In streptococci, several enzymes involved in glycolysis are known to be present as both extracellular and intracellular forms (9, 55, 56). It was surprising to observe that these enzymes are negatively affected by HtrA. In order confirm our mass-spectrometric identification, Western blot analyses were carried out. Stationary-phase culture supernatants from the wild-type (NG-8), htrA mutant (IBS101), and complemented (IBS101/pIB108) strains were separated by SDS-PAGE and probed with anti-enolase or anti-GAPDH antibodies. As shown in Fig. 6B and C, mutation in htrA caused increased production (approximately two- to threefold) of both enzymes. This increased production is specific to htrA mutation since it can be complemented (Fig. 6B and C, lanes 3). In both cases antibody reacted with two major bands. In both cases the smaller bands probably correspond to the proteolytic products. Taken together, the results described above show that the htrA mutation affects normal exoprotein expression during stationary phase growth.
Effect of HtrA on FTF expression.
S. mutans produces a FTF which synthesizes fructan polymers from sucrose. Enzymatically active FTF can be found in either cell-associated or extracellular form (8). FTF is one of the most abundant proteins in the culture supernatant of NG-8. Since we identified FTF by mass spectrometric analysis as one of the proteins that was accumulated in the culture supernatant of IBS101, we verified our result by Western blot analysis (Fig. 7). Stationary phase culture supernatants from the wild-type (NG-8), htrA mutant (IBS101), and complemented (IBS101/pIB108) strains were separated by SDS-PAGE and probed with anti-FTF antibody. As expected, IBS101 showed increased presence of FTF in the supernatant compared to NG-8. However, we observed only partial complementation when htrA was overexpressed from plasmid pIB108.
FIG. 7.
Western blot analysis of FTF (levansucrase) expression. NG-8 (wild type, lane 1), IBS101 (htrA, lane 2), and IBS101/pIB108 (htrA/htrA+, lane 3) were grown overnight in THY broth and whole-cell extracts were prepared. Equal amounts of cell extracts were separated on SDS-4 to 20% PAGE gels and reacted with anti-FTF antibody. M, prestained molecular mass marker (NEB).
Altered biofilm formation in htrA mutant.
Biofilm formation by S. mutans involves two processes: sucrose independent initial attachment mediated by surface proteins (such as SpaP, GbpC), and sucrose-dependent attachment mediated by glucans synthesized by glucosyltransferases from sucrose. For biofilm formation on in vitro surfaces such as glass and polystyrene, sucrose plays an important role. Since surface protein expression was altered in the htrA mutant strain, we wanted to know whether htrA mutation affected biofilm formation. Biofilms were grown in THY or BM media supplemented with sucrose on glass or polystyrene surfaces. Biofilms formed equally well in both media by the wild-type NG-8 after 20 h of growth. After overnight growth in the chamber slides or the microtiter plates, there was a noticeable difference in biofilm structure formed by IBS101 compared to the parental strain (Fig. 8A). Wild-type biofilm had a very confluent appearance while the htrA mutant showed a patchy appearance. This defect in biofilm formation by htrA mutant was complemented when htrA was provided in trans by plasmid pIB108 (Fig. 8A). The architecture of biofilms formed by the htrA mutant appeared to be altered as compared to the wild type (Fig. 8B). The htrA mutant formed large aggregates heterogenously distributed throughout the biofilm matrix. There is a noticeable large gap in the biofilm matrix and the microcolonies were large dome-like structures. Wild-type biofilms are very uniform with thick layers of cells completely covering the attached surfaces. Thus, HtrA deficiency causes alter biofilm formation in S. mutans.
FIG. 8.
Inactivation of htrA causes altered biofilm formation by S. mutans strains. (A) Crystal-violet-stained 24-h-old biofilm of NG-8 (wild type), IBS101 (htrA), and IBS101/pIB108 (htrA/htrA+). Biofilms were grown in BM media on glass slides (GS) or on polystyrene microtiter wells (PS). (B) Confocal laser scanning micrographs (top panel) and scanning electron micrographs (middle and lower panel) of biofilms accumulated on glass surface. Left panels, wild-type biofilms (NG-8); right panels, htrA mutant biofilms (IBS101). Magnifications, ×40 (top), ×600 (mid), and ×1,600 (bottom).
DISCUSSION
Streptococcal genomes encode only one HtrA protease, unlike other low G+C gram-positive bacteria such as B. subtilis and S. aureus that encodes multiple HtrA paralogues. However, the genomic organization of the htrA locus in streptococci is quite unique (Fig. 1A). In most streptococci, htrA is located near the chromosomal replication origin. While the organization of the downstream region of htrA appears to be highly conserved, the upstream region is not well conserved among streptococci. For example, in S. pneumoniae the com operon is present near the htrA locus (76) whereas in S. mutans the com operon is present elsewhere in the genome (Fig. 1A). Immediately downstream of htrA is a gene with similarity to spo0J. This gene is associated with chromosome partitioning in B. subtilis (33). spo0J is followed by the oriC region and dnaA, whose product is required for initiation of chromosomal replication. In streptococci, the htrA locus also contains several binding sites for DnaA protein (24) and, as suggested by Lyon and Caparon (46), may be involved in chromosome replication and cell division. In other gram-positive bacteria including the closely related L. lactis and Enterococcus faecalis, the htrA locus is not associated with the chromosomal replication origin and therefore may not be involved in cell division.
Like in other bacteria, we have observed stress sensitive phenotypes due to the htrA mutation. S. mutans can grow in temperatures ranging from 28°C to 47°C with optimum growth around 37°C (47). The htrA mutant of S. mutans showed a restricted temperature range and failed to grow at both 30°C and 42°C. In S. pyogenes, a single-crossover insertional mutant showed a temperature sensitive phenotype while a deletion mutant did not (46). Because a large insertion had occurred into the htrA locus during single crossover integration, it was suggested that such a large insertion may have interfered with the normal chromosomal replication. However, our inactivated htrA mutant was made by gene-replacement and we were able to complement the thermo-sensitive phenotype by providing the htrA gene in trans from the pIB108 plasmid. This complementation rules out the possibility of polar effect of mutation on downstream genes. Interestingly, our mutant also failed to grow at low temperature (30°C). Most evidence suggests that htrA acts as a housekeeping protease to degrade unfolded proteins during heat shock (54). While this function is necessary for growth at high temperature, htrA also acts as chaperon at low temperature (74). Possibly due to loss of this latter function, the htrA mutant failed to grow at 30°C.
The S. mutans htrA mutant also showed sensitivity towards osmotic stress generated by NaCl, acid stress, and oxidative stress. These observations are consistent with HtrA being involved in stress response. Although the role of HtrA in osmotic stress was not characterized previously, in L. monocytogenes HtrA was shown to be involved in osmotic stress (82). We also found that in S. mutans HtrA plays a role in osmotic stress. We have observed acid sensitivity of our htrA mutant which is consistent with the earlier observation (21). During acid stress, several proteins including many heat shock proteins and chaperons are overexpressed in S. mutans (42, 81). This general stress induction in S. mutans confers cross-protection to other stresses such thermal and oxidative stress (75). It is possible that htrA expression is also induced during acid stress and plays an important role in acid-tolerance. To understand how HtrA participates in osmotic and acid stress responses requires further investigation.
Interestingly, the htrA mutant showed sensitivity towards the DNA damaging agent mitomycin C. Mitomycin C induces mainly interstrand DNA cross-links (77) and results in induction of SOS response. RecA protein plays a central role in repair of mitomycin C damaged DNA and HtrA could modulate RecA function. However, we tested htrA mutant for UV sensitivity and found no difference compared to the wild type (data not shown). Therefore, a protein other than RecA is probably involved in mitomycin C sensitivity in the htrA mutant. In this context, it important to point out that several DNA repair proteins are also induced during acid stress (42) and thus there may be a direct connection between DNA repair and acid-tolerance mediated by HtrA.
During stationary phase, the expression of surface associated proteins was altered by htrA mutation in S. mutans. Notably, we found that expression of GbpB was greatly enhanced in the htrA mutant. GbpB is a glucan-binding protein which shares extensive homology with peptidoglycan hydrolases from other gram-positive bacteria (50). Interestingly, GbpB is up-regulated during high osmolarity, high temperature, and low pH, and therefore GpbB may act as a general stress protein (17). GbpB is also essential for cell wall integrity and maintenance of cell shape (17). Attempts to knock out the gbpB gene have also supported the notion that expression of GbpB is essential for cell survival (49). In the htrA mutant, we have found increased expression of GbpB not only in the cell surface fraction but also in the culture supernatant and whole-cell lysate fractions (data not shown). Therefore it appears that overall expression of GbpB is increased in the htrA mutant. It is possible that under normal conditions the protease activity of HtrA regulates GbpB expression either directly by degrading the protein or indirectly by degrading a transcriptional regulator necessary for gbpB expression. How HtrA regulates GbpB expression and what is the relationship between GbpB and stress response remains to be explored.
We have tested expression of various other cell surface proteins in the htrA mutant. We found that htrA mutation had no effect on cell surface expression of antigen P1 (data not shown) or binding to fibronectin and fibrinogen (data not shown) consistent with previous results (6, 21). However, in contrast with the previous report, we found that htrA mutation affects GtfB and FTF expression (Fig. 6A and 7). Both GtfB (39) and FTF (64) bind to glucan and play an important role in S. mutans adhesion to the tooth surface. Both proteins are found in the extracellualar fraction in addition to cell-associated forms (5, 38). The predicted molecular mass of S. mutans FTF, after removal of the signal peptide, is ∼83 kDa. However, S. mutans was found to secrete FTFs with various molecular masses ranging from 59 to 106 kDa. Processing of a single ftf gene product at the post-translational level accounts for this diversity (64, 66). The increased production of extracellular FTF in the htrA mutant suggests that HtrA plays a role in FTF secretion. However, unlike GbpB, we did not find increased FTF expression on the cell surface (data not shown), suggesting that association of FTF to the cell surface rather than total production is altered by htrA mutation. Since HtrA is also cell wall associated (3), it is possible that HtrA facilitates proper folding of FTF needed for surface anchoring during or after translocation from cytoplasm to the cell wall.
Previously, Diaz-Torres and Russell characterized an htrA mutant from S. mutans (21). They investigated the effect of htrA mutation on seven extracellular or wall associated proteins including GTF, FTF and WapA, and found no differences between wild-type and htrA mutant strains. However, their htrA mutant was generated by single-crossover insertion using an internal fragment of htrA gene. Single cross over mutant may generate a partially functional truncated HtrA. Indeed, their htrA mutant contained a truncated copy encoding a 392-amino-acid polypeptide, lacking only C-terminal 10 amino acids and therefore might be fully functional. In contrast, our htrA mutant was generated by double crossover by disrupting HtrA at amino acid position 243, thereby deleted the conserved catalytic domain. Moreover, recently in S. pyogenes it was shown that a single-crossover insertional inactivation of htrA resulted in different phenotypes when compared with a clean non-polar deletion mutant (46). Although unlikely, strain differences (LT11 [21] versus NG-8, our strain) might have played a role in observed phenotypic differences. Nevertheless, the single-crossover insertion mutant of S. mutans htrA formed cohesive clumps in liquid media and showed different colony morphology indicating altered surface properties.
Glycolytic enzymes including GAPDH and enolase were also enriched in the stationary phase culture supernatant of the htrA mutant. GAPDH and enolase have been identified on the cell surfaces and culture supernatants of pathogenic streptococci, which raises questions as to their function at these locations (16). None of these proteins contain signal peptide, LPXTG anchoring motif or other known cell-wall-anchoring motifs and thus how these proteins are secreted or anchored to the cell surface remains to be uncovered. Pancholi and Fischetti (56) found that the surface GAPDH of S. pyogenes binds to fibronectin, lysozyme and to the cytoskeletal proteins and contributes to pathogenesis in this organism. In S. oralis, GAPDH binds to Porphyromonas gingivalis fimbriae and may contribute to its colonization in the periodontal pockets (48). In other streptococci, GAPDH also acts as an adhesin and contributes to virulence (13, 43). Surface enolase is also important for pathogenesis. For example, S. pyogenes enolase binds to plasmino(gen) and plays a role in bacterial invasion of the human host and autoimmune sequelae (55). S. mutans enolase binds to salivary mucin and this may help S. mutans to disseminate through oral tissues to enter the bloodstream and cause bacterial endocarditis (25). Thus, these glycolytic enzymes and other surface proteins that bind to components of host extracellular matrices constitute a new class of virulence factors known as anchorless adhesins (16). Since HtrA interferes with the secretion of GAPDH and enolase, it is possible that HtrA also interferes with the other anchorless adhesins.
We found several altered biofilm phenotypes such as increased size of cell aggregates and patchy biofilm structure in the htrA mutant. This suggests that HtrA has a regulatory role for one or more proteins related to biofilm formation. Indeed expression of several proteins involved in biofilm formation was modified by HtrA. For example, both GTF and FTF act as adhesins and their expression was modified in the htrA mutant. Surface expression of glycolytic enzymes is also important for biofilm and their expression was modified by htrA mutation. Since HtrA is a surface protein one could imagine that HtrA itself participates in the adhesions of bacteria in the biofilm. How HtrA influences biofilm is currently being addressed using a proteomics approach.
The main goal of this investigation was to learn whether S. mutans serine protease HtrA influences the expression of extracellular proteins including virulence factors. We found that indeed HtrA alters expression of several extarcellular proteins, some participate in virulence. HtrA also interferes with normal biofilm formation. At this time we can only speculate about the molecular mechanisms by which HtrA regulates the expression of cell surface proteins. In addition to a direct role in surface protein expression, HtrA may have an indirect role in regulating cell surface expression. For example, HtrA may regulate a transcriptional activator that controls surface protein expression. In the absence of a functional HtrA, such transcriptional regulator increases expression of surface proteins. Alternatively, HtrA may degrade some site specific endoproteases, which cleave cell surface proteins. In the absence of HtrA, these proteases can interfere with surface expression of various proteins, for example, the glycolytic enzymes or FTF. Indeed S. mutans expresses several proteases that are known to modulate several surface proteins and release them in the culture supernatant (28, 41). Whether HtrA really regulates other proteases remains to be determined and is the focus of our ongoing research.
After the manuscript had been reviewed, Ahn et al. (2) showed that HtrA in S. mutans (UA159) affects biofilm formation, consistent with our observation.
Acknowledgments
We are grateful to Vijay Pancholi for anti-enolase and anti-GAPDH antibodies, to Gilad Bachrach for anti-FTF antibody, to Daniel Smith for anti-GBP antibody, and to L. Jeannine Brady for anti-P1 monoclonal antibody. We thank Said Suleman for technical help with the SEM analysis and Keith Weaver for critically reviewing the manuscript.
A part of this publication was made possible by NIH grant number 2 P20 RR016479 from the INBRE Program of the National Center for Research Resources.
Editor: A. D. O'Brien
REFERENCES
- 1.Aduse-Opoku, J., M. L. Gilpin, and R. R. Russell. 1989. Genetic and antigenic comparison of Streptococcus mutans fructosyltransferase and glucan-binding protein. FEMS Microbiol. Lett. 50:279-282. [DOI] [PubMed] [Google Scholar]
- 2. Ahn, S.-J., J. A. C. Lemos, and R. A. Burne. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antelmann, H., E. Darmon, D. Noone, J. W. Veening, H. Westers, S. Bron, O. P. Kuipers, K. M. Devine, M. Hecker, and J. M. van Dijl. 2003. The extracellular proteome of Bacillus subtilis under secretion stress conditions. Mol. Microbiol. 49:143-156. [DOI] [PubMed] [Google Scholar]
- 4.Baddour, L. M. 1988. Twelve-year review of recurrent native-valve infective endocarditis: a disease of the modern antibiotic era. Rev. Infect. Dis. 10:1163-1170. [DOI] [PubMed] [Google Scholar]
- 5.Banas, J. A., and M. M. Vickerman. 2003. Glucan-binding proteins of the oral streptococci. Crit. Rev. Oral Biol. Med. 14:89-99. [DOI] [PubMed] [Google Scholar]
- 6.Beg, A. M., M. N. Jones, T. Miller-Torbert, and R. G. Holt. 2002. Binding of Streptococcus mutans to extracellular matrix molecules and fibrinogen. Biochem. Biophys. Res. Commun. 298:75-79. [DOI] [PubMed] [Google Scholar]
- 7.Belli, W. A., and R. E. Marquis. 1991. Adaptation of Streptococcus mutans and Enterococcus hirae to acid stress in continuous culture. Appl. Environ. Microbiol. 57:1134-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergeron, L. J., and R. A. Burne. 2001. Roles of fructosyltransferase and levanase-sucrase of Actinomyces naeslundii in fructan and sucrose metabolism. Infect. Immun. 69:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. Alpha-enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 10.Biswas, I., P. Germon, K. McDade, and J. R. Scott. 2001. Generation and surface localization of intact M protein in Streptococcus pyogenes are dependent on sagA. Infect. Immun. 69:7029-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen, W. H., K. Schilling, E. Giertsen, S. Pearson, S. F. Lee, A. Bleiweis, and D. Beeman. 1991. Role of a cell surface-associated protein in adherence and dental caries. Infect. Immun. 59:4606-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brassard, J., M. Gottschalk, and S. Quessy. 2004. Cloning and purification of the Streptococcus suis serotype 2 glyceraldehyde-3-phosphate dehydrogenase and its involvement as an adhesin. Vet. Microbiol. 102:87-94. [DOI] [PubMed] [Google Scholar]
- 14.Burne, R. A., and J. E. C. Penders. 1992. Characterization of the Streptococcus mutans GS-5 fruA gene encoding exo-β-d-fructosidase. Infect. Immun. 60:4621-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burne, R. A., Z. T. Wen, Y. Y. Chen, and J. E. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chhatwal, G. S. 2002. Anchorless adhesins and invasins of Gram-positive bacteria: a new class of virulence factors. Trends Microbiol. 10:205-208. [DOI] [PubMed] [Google Scholar]
- 17.Chia, J. S., L. Y. Chang, C. T. Shun, Y. Y. Chang, Y. G. Tsay, and J. Y. Chen. 2001. A 60-kilodalton immunodominant glycoprotein is essential for cell wall integrity and the maintenance of cell shape in Streptococcus mutans. Infect. Immun. 69:6987-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 19.Collin, M., and A. Olsen. 2003. Extracellular enzymes with immunomodulating activities: variations on a theme in Streptococcus pyogenes. Infect. Immun. 71:2983-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diaz-Torres, M. L., and R. R. Russell. 2001. HtrA protease and processing of extracellular proteins of Streptococcus mutans. FEMS Microbiol. Lett. 204:23-28. [DOI] [PubMed] [Google Scholar]
- 22.Ferretti, J. J., R. R. Russell, and M. L. Dao. 1989. Sequence analysis of the wall-associated protein precursor of Streptococcus mutans antigen A. Mol. Microbiol. 3:469-478. [DOI] [PubMed] [Google Scholar]
- 23.Froeliger, E. H., and P. Fives-Taylor. 2001. Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gasc, A. M., P. Giammarinaro, S. Richter, and M. Sicard. 1998. Organization around the dnaA gene of Streptococcus pneumoniae. Microbiology 144:433-439. [DOI] [PubMed] [Google Scholar]
- 25.Ge, J., D. M. Catt, and R. L. Gregory. 2004. Streptococcus mutans surface α-enolase binds salivary mucin MG2 and human plasminogen. Infect. Immun. 72:6748-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottesman, S., S. Wickner, and M. R. Maurizi. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815-823. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton, I. R., and N. D. Buckley. 1991. Adaptation by Streptococcus mutans to acid tolerance. Oral Microbiol. Immunol. 6:65-71. [DOI] [PubMed] [Google Scholar]
- 28.Harrington, D. J., and R. R. Russell. 1994. Identification and characterisation of two extracellular proteases of Streptococcus mutans. FEMS Microbiol. Lett. 121:237-241. [DOI] [PubMed] [Google Scholar]
- 29.Horaud, T., and F. Delbos. 1984. Viridans streptococci in infective endocarditis: species distribution and susceptibility to antibiotics. Eur. Heart J. 5(Suppl. C):39-44. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72:3584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Igarashi, T., A. Yamamoto, and N. Goto. 1992. Characterization of an exo-beta-D-fructosidase from Streptococcus mutans Ingbritt. Microbiol. Immunol. 36:643-647. [DOI] [PubMed] [Google Scholar]
- 32.Igarashi, T., A. Yamamoto, and N. Goto. 1995. Sequence analysis of the Streptococcus mutans Ingbritt dexA gene encoding extracellular dextranase. Microbiol. Immunol. 39:853-860. [DOI] [PubMed] [Google Scholar]
- 33.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koga, T., T. Oho, Y. Shimazaki, and Y. Nakano. 2002. Immunization against dental caries. Vaccine 20:2027-2044. [DOI] [PubMed] [Google Scholar]
- 35.Koga, T., Y. Yamashita, Y. Nakano, M. Kawasaki, T. Oho, H. Yu, M. Nakai, and N. Okahashi. 1995. Surface proteins of Streptococcus mutans. Dev. Biol. Stand. 85:363-369. [PubMed] [Google Scholar]
- 36.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 37.Koo, B. M., M. J. Yoon, C. R. Lee, T. W. Nam, Y. J. Choe, H. Jaffe, A. Peterkofsky, and Y. J. Seok. 2004. A novel fermentation/respiration switch protein regulated by enzyme IIAGlc in Escherichia coli. J. Biol. Chem. 279:31613-31621. [DOI] [PubMed] [Google Scholar]
- 38.Kuramitsu, H. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 39.Kuramitsu, H. K., and L. Ingersoll. 1978. Interaction of glucosyltransferase with the cell surface of Streptococcus mutans. Infect. Immun. 20:652-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laskowska, E., D. Kuczynska-Wisnik, J. Skorko-Glonek, and A. Taylor. 1996. Degradation by proteases Lon, Clp and HtrA, of Escherichia coli proteins aggregated in vivo by heat shock; HtrA protease action in vivo and in vitro. Mol. Microbiol. 22:555-571. [DOI] [PubMed] [Google Scholar]
- 41.Lee, S. F., Y. H. Li, and G. H. Bowden. 1996. Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infect. Immun. 64:1035-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Len, A. C., D. W. Harty, and N. A. Jacques. 2004. Stress-responsive proteins are upregulated in Streptococcus mutans during acid tolerance. Microbiology 150:1339-1351. [DOI] [PubMed] [Google Scholar]
- 43.Ling, E., G. Feldman, M. Portnoi, R. Dagan, K. Overweg, F. Mulholland, V. Chalifa-Caspi, J. Wells, and Y. Mizrachi-Nebenzahl. 2004. Glycolytic enzymes associated with the cell surface of Streptococcus pneumoniae are antigenic in humans and elicit protective immune responses in the mouse. Clin. Exp. Immunol. 138:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loesche, W. J. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50:353-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyon, W. R., and M. G. Caparon. 2004. Role for serine protease HtrA (DegP) of Streptococcus pyogenes in the biogenesis of virulence factors SpeB and the hemolysin streptolysin S. Infect. Immun. 72:1618-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma, Y., and R. E. Marquis. 1997. Thermophysiology of Streptococcus mutans and related lactic-acid bacteria. Antonie Leeuwenhoek 72:91-100. [DOI] [PubMed] [Google Scholar]
- 48.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mattos-Graner, R., P. Zucchi, D. J. Smith, and M. J. Duncan. 2002. Mutant analysis of the gene encoding glucan binding protein B indicates an essential role in Streptococcus mutans. J. Dent. Res. 81A:40. [Google Scholar]
- 50.Mattos-Graner, R. O., S. Jin, W. F. King, T. Chen, D. J. Smith, and M. J. Duncan. 2001. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect. Immun. 69:6931-6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Noone, D., A. Howell, R. Collery, and K. M. Devine. 2001. YkdA and YvtA, HtrA-like serine proteases in Bacillus subtilis, engage in negative autoregulation and reciprocal cross-regulation of ykdA and yvtA gene expression. J. Bacteriol. 183:654-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noone, D., A. Howell, and K. M. Devine. 2000. Expression of ykdA, encoding a Bacillus subtilis homologue of HtrA, is heat shock inducible and negatively autoregulated. J. Bacteriol. 182:1592-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oetjen, J., P. Fives-Taylor, and E. Froeliger. 2001. Characterization of a streptococcal endopeptidase with homology to human endothelin-converting enzyme. Infect. Immun. 69:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 55.Pancholi, V., and V. A. Fischetti. 1998. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 56.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate-dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Perry, D., and H. K. Kuramitsu. 1989. Genetic linkage among cloned genes of Streptococcus mutans. Infect. Immun. 57:805-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poquet, I., S. D. Ehrlich, and A. Gruss. 1998. An export-specific reporter designed for gram-positive bacteria: application to Lactococcus lactis. J. Bacteriol. 180:1904-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pruss, B. M., J. W. Campbell, T. K. Van Dyk, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quivey, R. G., Jr., R. C. Faustoferri, K. A. Clancy, and R. E. Marquis. 1995. Acid adaptation in Streptococcus mutans UA159 alleviates sensitization to environmental stress due to RecA deficiency. FEMS Microbiol. Lett. 126:257-261. [DOI] [PubMed] [Google Scholar]
- 62.Rasmussen, M., and L. Bjorck. 2002. Proteolysis and its regulation at the surface of Streptococcus pyogenes. Mol. Microbiol. 43:537-544. [DOI] [PubMed] [Google Scholar]
- 63.Rigoulay, C., J. M. Entenza, D. Halpern, E. Widmer, P. Moreillon, I. Poquet, and A. Gruss. 2005. Comparative analysis of the roles of HtrA-like surface proteases in two virulent Staphylococcus aureus strains. Infect. Immun. 73:563-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rozen, R., D. Steinberg, and G. Bachrach. 2004. Streptococcus mutans fructosyltransferase interactions with glucans. FEMS Microbiol. Lett. 232:39-43. [DOI] [PubMed] [Google Scholar]
- 65.Russell, M. W., L. A. Bergmeier, E. D. Zanders, and T. Lehner. 1980. Protein antigens of Streptococcus mutans: purification and properties of a double antigen and its protease-resistant component. Infect. Immun. 28:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Russell, M. W., and B. Mansson-Rahemtulla. 1989. Interaction between surface protein antigens of Streptococcus mutans and human salivary components. Oral Microbiol. Immunol. 4:106-111. [DOI] [PubMed] [Google Scholar]
- 67.Russell, R. R., A. C. Donald, and C. W. Douglas. 1983. Fructosyltransferase activity of a glucan-binding protein from Streptococcus mutans. J. Gen. Microbiol. 129:3243-3250. [DOI] [PubMed] [Google Scholar]
- 68.Russell, R. R. B., E. Abdulla, M. L. Gilpin, and K. Smith. 1986. Characterization of Streptococcus mutans surface antigens, p. 61-70. In S. Hamada, et al. (ed.), Molecular biology and immunology of Streptococcus mutans. Elsevier, Amsterdam, The Netherlands.
- 69.Sassoon, N., J. P. Arie, and J. M. Betton. 1999. PDZ domains determine the native oligomeric structure of the DegP (HtrA) protease. Mol. Microbiol. 33:583-589. [DOI] [PubMed] [Google Scholar]
- 70.Sato, Y., Y. Yamamoto, and H. Kizaki. 1997. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 65:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shah, D. S., and R. R. Russell. 2004. A novel glucan-binding protein with lipase activity from the oral pathogen Streptococcus mutans. Microbiology 150:1947-1956. [DOI] [PubMed] [Google Scholar]
- 72.Smith, D. J., H. Akita, W. F. King, and M. A. Taubman. 1994. Purification and antigenicity of a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 62:2545-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, D. J., and M. A. Taubman. 1996. Experimental immunization of rats with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries. Infect. Immun. 64:3069-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 75.Svensater, G., B. Sjogreen, and I. R. Hamilton. 2000. Multiple stress responses in Streptococcus mutans and the induction of general and stress-specific proteins. Microbiology 146:107-117. [DOI] [PubMed] [Google Scholar]
- 76.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 77.Tomasz, M. 1995. Mitomycin C: small, fast and deadly (but very selective). Chem. Biol. 2:575-579. [DOI] [PubMed] [Google Scholar]
- 78.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1990. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van de Rijn, I., M. George, A. Bouvet, and R. B. Roberts. 1986. Enzyme-linked immunosorbent assay for the detection of antibodies to nutritionally variant streptococci in patients with endocarditis. J. Infect. Dis. 153:116-121. [DOI] [PubMed] [Google Scholar]
- 80.Wen, Z. T., and R. A. Burne. 2002. Functional genomics approach to identifying genes required for biofilm development by Streptococcus mutans. Appl. Environ. Microbiol. 68:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkins, J. C., K. A. Homer, and D. Beighton. 2002. Analysis of Streptococcus mutans proteins modulated by culture under acidic conditions. Appl. Environ. Microbiol. 68:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wonderling, L. D., B. J. Wilkinson, and D. O. Bayles. 2004. The htrA (degP) gene of Listeria monocytogenes 10403S is essential for optimal growth under stress conditions. Appl. Environ. Microbiol. 70:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou, L., X. H. Lei, B. R. Bochner, and B. L. Wanner. 2003. Phenotype microarray analysis of Escherichia coli K-12 mutants with deletions of all two-component systems. J. Bacteriol. 185:4956-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]