Abstract
Over the past years, family 18 chitinases have been validated as potential targets for the design of drugs against human pathogens that contain or interact with chitin during their normal life cycles. Thus far, only one potent chitinase inhibitor has been described in detail, the pseudotrisaccharide allosamidin. Recently, however, two potent natural-product cyclopentapeptide chitinase inhibitors, argifin and argadin, were reported. Here, we describe high-resolution crystal structures that reveal the details of the interactions of these cyclopeptides with a family 18 chitinase. The structures are examples of complexes of a carbohydrate-processing enzyme with high-affinity peptide-based inhibitors and show in detail how the peptide backbone and side chains mimic the interactions of the enzyme with chitooligosaccharides. Together with enzymological characterization, the structures explain why argadin shows an order of magnitude stronger inhibition than allosamidin, whereas argifin shows weaker inhibition. The peptides bind to the chitinase in remarkably different ways, which may explain the differences in inhibition constants. The two complexes provide a basis for structure-based design of potent chitinase inhibitors, accessible by standard peptide chemistry.
Chitinases, which hydrolyze linear polymers of β-(1,4)-linked N-acetylglucosamine (GlcNAc), have been proposed as attractive targets for the development of drugs against a range of human pathogens. Chitinase inhibitors have shown activity against fungi including Candida albicans (1), insects (2–5) and the human malaria parasite Plasmodium falciparum (6, 7). Furthermore, a knockout experiment confirmed that P. falciparum chitinase is essential for insect invasion (8). Mammalian chitinases also have been identified (9, 10), yet their function is not defined clearly, and they seem not to be essential (11). Thus, the chitinases from human pathogens could be targeted through specific inhibitors without negative side effects on their mammalian hosts.
Although chitinases seem to be an interesting target for the design of pesticides, fungicides, and antimalarials, only a few chitinase inhibitors have been described. The most potent one, the pseudotrisaccharide allosamidin, is a natural product isolated from Streptomyces (3, 12), with Ki values that range from strong (0.48 nM) to relatively weak (3.1 μM) inhibition (13, 14). Allosamidin derivatives were able to kill Lucilia cuprina blowfly larvae in in vitro test systems after contact or feeding (15). Structural analyses of complexes with family 18 chitinases have shown that allosamidin mimics an oxazolinium ion reaction intermediate bound to subsites −3 through −1 (16, 17). During normal catalysis, this oxazolinium ion intermediate is formed by nucleophilic attack of the N-acetyl group of the −1 sugar on the anomeric carbon, which occurs concomitantly with protonation and breakage of the scissile glycosidic bond by the catalytic acid (refs. 17–21; note that this “substrate-assisted” retaining mechanism differs from the “classical” retaining mechanism in glycosyl hydrolases (22–24) in which a carboxylate side chain, usually a glutamic acid, is the nucleophile).
Unfortunately, total synthesis of allosamidin and its derivatives is difficult and expensive (25, 26), making it a less suitable candidate for further optimization as a chemotherapeutical. Other natural products that inhibit family 18 chitinases have been reported such as the styloguanidines isolated from a marine sponge (27), cyclo(Arg-Pro) from marine bacteria (1, 28), and the psammaplins isolated from a marine sponge (29). Unfortunately, all these inhibitors exhibit only weak (>1 μM) inhibition. Ab initio organic synthesis of novel chitinase inhibitors based on oligo-GlcNAcs has been attempted also, yet thus far no potent inhibitors have been identified (ref. 30 and G. Thiele, A. Rottman, A. Germera, E. Kleinpeter, K.-D. Spindler, B.S., V.G.H.E., and M.G.P., unpublished data).
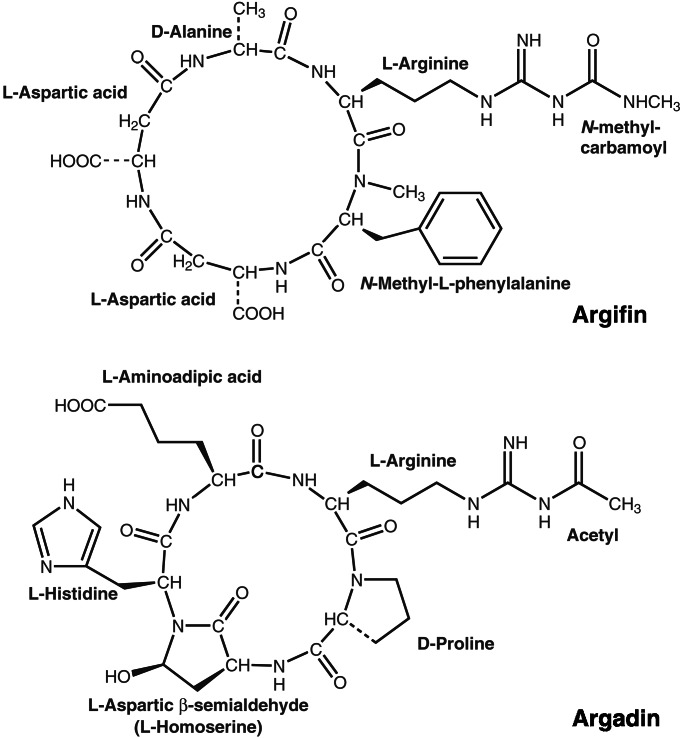
Recently, two previously uncharacterized natural product cyclopentapeptide chitinase inhibitors have been reported, argifin and argadin (Fig. 1), that were isolated from Gliocladium and Clonostachys fungal cultures, respectively (4, 5). When tested against a family 18 chitinase from the blowfly L. cuprina, IC50 values were found to be in the 100 nM range for argifin (4) and in the 1 nM range for argadin (5). These inhibitors thus seem to be as potent as allosamidin yet are more accessible synthetically through standard peptide chemistry. Hence, argifin and argadin could be good candidates for structure-based optimization, possibly leading to subnanomolar inhibition of family 18 chitinases from human pathogens. Here, we describe the structures of argifin and argadin in complex with a family 18 chitinase from Serratia marcescens [chitinase B (ChiB); ref. 31] at 2.0-Å resolution. These structures give an unprecedented view of how high-affinity peptides inactivate carbohydrate-processing enzymes and provide a basis for further structure-based inhibitor design.
Figure 1.
Argifin and argadin two-dimensional structures. The chemical structures of argifin and argadin are shown as published previously (4, 5). The stereochemistry on the histidine Cα carbon and the aspartic β-semialdehyde (ASA) Cα,Cγ carbons could not be determined previously but is defined by the diffraction data presented here.
Methods
Purification and Crystallization.
ChiB from S. marcescens was overexpressed and purified from Escherichia coli as described (32). The protein was crystallized from solutions containing ammonium sulfate and glycerol by vapor diffusion as described in detail elsewhere (17, 31). Crystals then were soaked for 12–24 h in mother liquor containing an ≈125-fold molar excess of argifin or argadin. Crystals then were frozen in a cryostream, and single-crystal diffraction data were collected at beamline ID14-EH4, European Synchrotron Radiation Facility (Table 1). The data were processed with DENZO and reduced with SCALEPACK, both from the HKL suite of programs (33).
Table 1.
Details of data collection and structure refinement
| ChiB–argifin | ChiB–argadin | |
|---|---|---|
| Cell dimensions, Å | a = 55.77 | a = 55.06 |
| b = 103.91 | b = 102.93 | |
| c = 186.61 | c = 185.78 | |
| Resolution range, Å | 35–2.0 (2.07–2.0) | 30–2.0 (2.07–2.0) |
| No. observed reflections | 234,279 (17,389) | 224,646 (18,204) |
| No. unique reflections | 72,253 (6,453) | 69,979 (6,563) |
| Redundancy | 3.2 (2.7) | 3.2 (2.8) |
| I/σI | 6.3 (3.1) | 9.7 (2.6) |
| Completeness, % | 96.9 (88.0) | 97.8 (93.3) |
| Rsym, % | 9.4 (34.1) | 7.6 (43.2) |
| Rcryst, % | 19.2 | 20.4 |
| Rfree, % | 23.1 | 23.1 |
| No. Rfree reflections | 722 | 1,030 |
| No. protein atoms | 7,793 | 7,794 |
| No. water molecules | 1,065 | 590 |
| No. inhibitor atoms | 96 | 96 |
| RMSD from ideal geometry | ||
| Bonds, Å | 0.008 | 0.008 |
| Angles, ° | 1.4 | 1.4 |
| B-factor rmsd, Å2 (bonded, main chain) | 1.4 | 1.5 |
| 〈Bprotein〉, Å2 | 30.2 | 35.3 |
| 〈Binhibitor〉, Å2 | 34.1 | 37.3 |
Values between brackets are for the highest resolution shell. Crystals were of space group P212121 and were cryocooled to 100 K. All measured data were included in structure refinement. rmsd, rms deviation.
Structure Determination.
Refinement was initiated by rigid body fitting using the previously published native ChiB structure (31) followed by iterative model building in O (34) and refinement in CNS (35). Models for argifin and argadin were not included until their conformations were well defined by the unbiased |Fo − Fc|,φcalc electron density maps (Fig. 2). Argifin and argadin starting structures and molecular topologies then were generated by using the PRODRG server (36), built into the density (Fig. 2), and the resulting complexes were refined further against the x-ray data. This refinement resulted in the final models described in Table 1 (see Figs. 2 and 3). For both structures, some small differences were found between the two independent monomers in the asymmetric unit as discussed previously (17, 31), although the inhibitors bound both monomers in identical fashion. In the interest of simplicity, differences between the argifin and argadin complexes are discussed by using the same monomer consistently. Images were generated by using PYMOL (www.pymol.org) and LIGPLOT (37).
Figure 2.
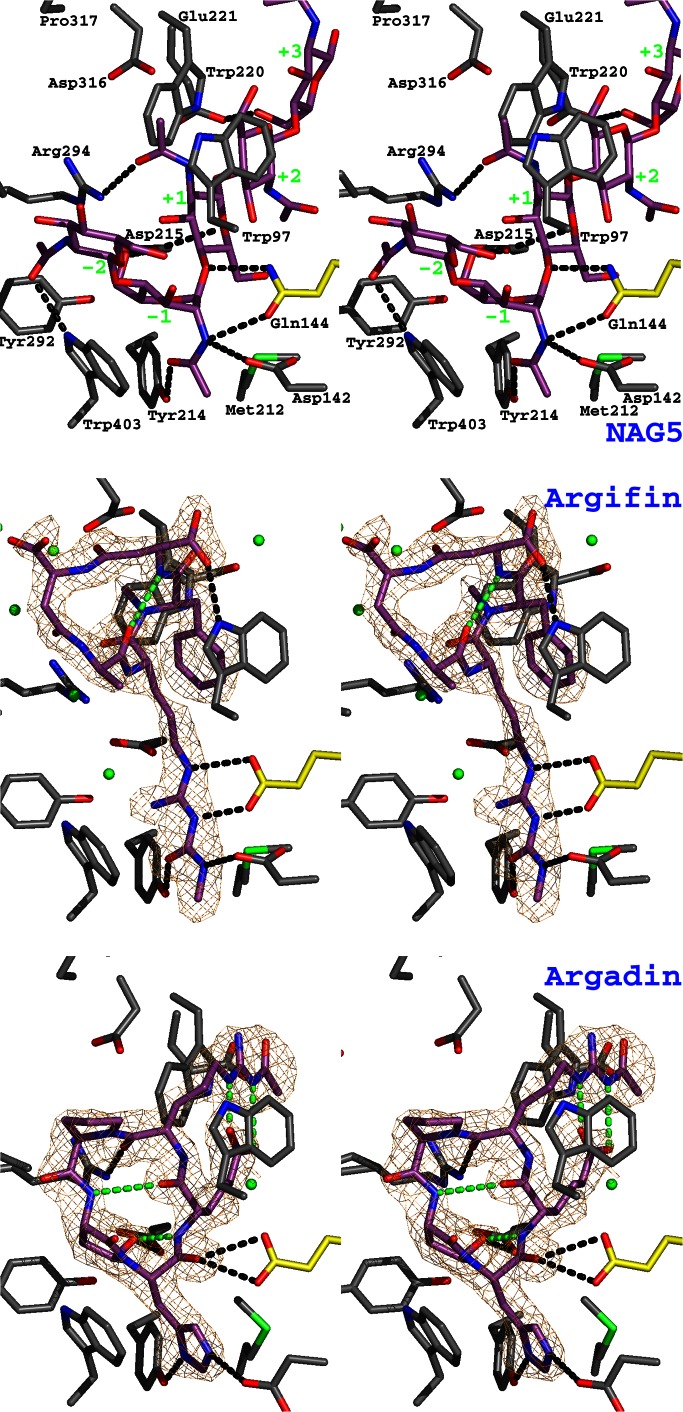
Argifin and argadin complexed to ChiB. The previously published structure of mutationally inactivated ChiB (where the catalytic Glu-144 has been replaced with a glutamine) in complex with GlcNAc5 (NAG5; ref. 17) is shown as a stereo stick model and compared with the ChiB–argifin and ChiB–argadin complexes. Unbiased (i.e., before including any inhibitor model) |Fo − Fc|,φcalc (contoured at 2.5 σ) maps are shown in orange. Ligand carbon atoms are colored purple. Side chains interacting with the cyclopentapeptides are shown in a sticks representation with carbons colored gray except for the catalytic residue 144, for which carbons are shown in yellow. Tyr-145 (which only hydrogen-bonds to GlcNAc5; see also Fig. 3) has been omitted to improve clarity. Water molecules hydrogen-bonding to both protein and inhibitor are shown as green spheres (hydrogen bonds are not shown). Hydrogen bonds between the ligands and the protein side chains are shown as black dotted lines. Argifin/argadin intramolecular hydrogens bonds are shown as green dotted lines. In the complex with GlcNAc5, the sugar subsites are indicated by green labels.
Figure 3.
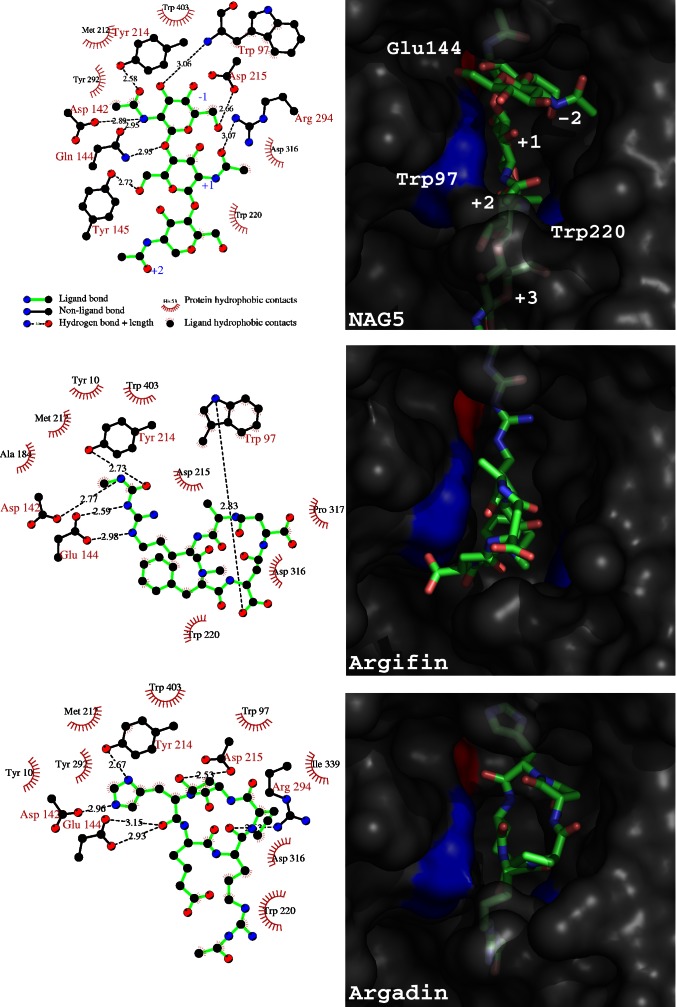
Further details of inhibitor–ChiB interactions. Schematic protein–ligand interactions (Left, calculated with LIGPLOT; ref. 37) and surface plots are shown for three ChiB complexes. The ChiB–GlcNAc5 (NAG5) complex (17) is shown, for comparison purposes, together with the ChiB–argifin and ChiB–argadin complexes described here. In the schematic drawings, only protein–ligand hydrogen bonds are shown (see key). For the complex with GlcNAc5, only the central three sugars are shown. In the surface representations, the protein surface is colored gray with the exception of Trp-97 and Trp-220 (blue) and the catalytic acid Glu-144 (red). The ligands are shown in a sticks representation with carbons colored green. The sugar subsites are labeled in the GlcNAc5 complex.
Enzymology.
Enzyme kinetics were determined by using 4-methylumbelliferyl β-d-N,N-diacetylchitobioside as a substrate in 50 mM citrate-phosphate buffer, pH 6.3. This substrate permits accurate determination of kinetic parameters despite the fact that relatively low substrate concentrations have to be used (<Km; ref. 29). Standard reaction mixtures contained 2.75 nM wild-type ChiB or 21.6 nM Trp-220 mutant, 0.1 mg/ml BSA, and 0–40 or 0–120 μM of the substrate for the wild type and W220A mutant, respectively. In initial inhibition tests, IC50 values at 20 μM substrate concentration were determined (Km values for the uninhibited enzymes were 33 and 97 μM for wild type and W220A, respectively). Reaction mixtures (50 μl) were incubated for 10 min at 37°C, after which the reaction was stopped with 1.95 ml 0.2 M Na2CO3. The amount of 4-methylumbelliferyl released was determined by using a DyNA 200 fluorimeter (Hoefer).
For the determination of kinetic parameters, reaction mixtures were incubated at 37°C, and samples were taken at 0, 2.5, 5.0, and 7.5 min. The production of 4-methylumbelliferyl was linear in all cases, thus permitting straightforward calculation of enzyme velocities. Inhibitors were added at concentrations close to the determined IC50 values. The mode of action for each inhibitor was determined by plotting the data in Lineweaver–Burk plots. Ki values were approximated by using the reciprocal Michaelis–Menten equation for competitive inhibition. The Ki values presented are average values derived from the results of four independent experiments. Because of limited availability of argifin, the (high) Ki value for inhibition of W220A by this compound could not be determined accurately. The value presented is an estimate based on a limited number of experiments.
Results and Discussion
Crystals of wild-type ChiB were soaked in solutions containing the cyclopentapeptides argifin and argadin (Fig. 1) and subsequently used to collect diffraction data to 2.0-Å resolution (Table 1). The structures were solved by molecular replacement and refined to R = 0.192 and Rfree = 0.231 for the ChiB–argifin complex and R = 0.204 and Rfree = 0.231 for the ChiB–argadin complex. Well defined density, completely covering the inhibitors, could be seen in the unbiased |Fo − Fc|,φcalc maps before any inhibitor model was included (Fig. 2) and allowed the building and refinement of the complete argifin and argadin structures. The quality of the maps also enabled us to determine the stereochemistry on the histidine Cα carbon and the ASA Cα,Cγ carbons in argadin, which could not be defined previously [see Fig. 1; we renamed the argadin residue originally identified as homoserine to ASA, because this terminology is more correct chemically (5)]. The stereochemistry of argadin amino acids thus was elucidated as l-arginine, d-proline, l-ASA, l-histidine, and l-aminoadipic acid, and the configuration of ASA–Cγ was R. Both inhibitors have a well defined cyclic backbone, although argifin has a more open and oval-shaped structure than argadin because of its larger number of backbone atoms (argifin has two β-amino acids, Fig. 1). Argifin contains only one intramolecular hydrogen bond (Asp–N to Ala–O), whereas argadin contains four intramolecular hydrogen bonds (ASA–N to adipic acid–O, adipic acid–N to ASA–Oδ, and arginine–Nɛ/η to adipic acid–Oζ1/2) (Fig. 2). Taken together, argadin seems to be more compact and more conformationally restricted than argifin.
The ChiB–argifin and ChiB–argadin complexes are compared with the previously published ChiB–GlcNAc5 complex (17) in Figs. 2 and 3. The ChiB–GlcNAc5 complex [obtained by soaking GlcNAc5 in crystals of the inactive ChiB–E144Q mutant (17)] shows which residues interact with the substrate and how these residues, through a combination of hydrogen bonds and van der Waals interactions, distort the sugar at the −1 position to the boat conformation. Residues Tyr-214 and Asp-142 make an important contribution to the distortion, because they form tight interactions with the N-acetyl group of the −1 sugar. The O7 atom in the N-acetyl group (hydrogen-bonded by Tyr-214; Figs. 2 and 3) is brought close to the anomeric C1 carbon and is optimally aligned for nucleophilic attack (17, 38). In the wild-type enzyme these interactions would lead to the formation of an oxazolinium ion intermediate, in which the C1 and O7 atoms are covalently linked, forming a five-membered ring (17–21). In the other sugar subsites, hydrogen bonding plays only a minor role, because most of the interactions are through stacking with solvent-exposed aromatic residues, most notably Trp-97/Trp-220, which forms a triple sandwich with the +1/+2 sugars (Figs. 2 and 3).
Several of the ChiB–GlcNAc5 interactions are mimicked in the ChiB–argifin complex. Asp-142, the catalytic residue Glu-144, and Tyr-214, which all interact with the distorted −1 sugar in the ChiB–GlcNAc5 complex, form hydrogen bonds with the N-methyl-carbamoylated arginine side chain in argifin (Figs. 2 and 3). Whereas in the unliganded ChiB structure the Asp-142 side chain points toward Asp-140 (and shares a proton with it; refs. 17, 31, and 39), Asp-142 in both the ChiB–GlcNAc5 and ChiB–argifin complexes points toward Glu-144 and the inhibitor (Figs. 2 and 3). Asp-142 is observed in this conformation in all previously published family 18 chitinase inhibitor–substrate complexes (17, 18, 29, 38) and is thought to play a role in distortion of the N-acetyl group and stabilization of the oxazolinium ion intermediate. In the ChiB–argifin complex structure, Asp-142 makes a hydrogen bond with the carbamoyl group (Figs. 2 and 3). Glu-144, the catalytic acid that interacts with the scissile oxygen in the ChiB–GlcNAc5 complex (17), makes hydrogen bonds to the guanidinium group of the arginine side chain in argifin (Figs. 2 and 3). The Tyr-214–N-acetyl interaction in ChiB–GlcNAc5 is mimicked by a hydrogen bond with the carbonyl oxygen of the carbamoyl moiety in argifin. Apart from these tight interactions in the bottom of the active site, there is only one additional hydrogen bond between argifin and the protein (aspartic acid carboxyl to Trp-97 indole nitrogen). The backbone and most of the side chains in argifin occupy a position approximately equivalent to that of the sugars bound to the −2 and −1 subsites in the ChiB–GlcNAc5 complex (Figs. 2 and 3). The argifin phenyl ring occupies a position equivalent to that of the pyranose ring of the sugar bound in the +1 subsite in the ChiB–GlcNAc5 complex, permitting the formation of the triple sandwich with Trp-97/Trp-220 (Figs. 2 and 3). Thus, the cyclopentapeptide argifin approximately occupies space corresponding to GlcNAc3 bound in subsites −2 through +1 while interacting closely with essential catalytic residues.
Although argifin and argadin share a similar structure (both are cyclopentapeptides containing an arginine derivative; Fig. 1), the structure of the ChiB–argadin complex revealed that both the orientation and location of the two peptide backbones is fundamentally different (Figs. 2 and 3). The more compact and restricted argadin backbone occupies a position corresponding to the sugars bound in the −1 and +1 subsites in the ChiB–GlcNAc5 complex. The arginine and adipic acid side chain, which together form a large, flat moiety through two hydrogen bonds (Fig. 2), occupy the +2 position and form a triple sandwich with Trp-97/Trp-220 (Figs. 2 and 3). There are tight interactions in the bottom of the active site, with Asp-142 and Tyr-214 hydrogen-bonding the two nitrogens in the histidine side chain. The backbone oxygen of the histidine occupies almost the same position as the scissile oxygen in the ChiB–GlcNAc5 complex and hydrogen-bonds to the catalytic acid, Glu-144 (Figs. 2 and 3). Argadin further mimics the ChiB–GlcNAc5 interactions through hydrogen bonds of Asp-215 to the ASA side chain (Oδ) and Arg-294 to the backbone oxygen of the argadin arginine.
Further differences between argifin and argadin are observed when the number of water-mediated hydrogen bonds is investigated. In the argifin complex (Fig. 2), six water molecules are observed that hydrogen-bond to the inhibitor and protein side chains. In the argadin complex, only one such water molecule is observed (Fig. 2), which suggests that argifin displaces less water molecules after binding to the ChiB active site than argadin. A similar trend is observed when analyzing the total amount of surface area buried by the ligands (Fig. 3). Binding of GlcNAc5 covers a surface of 200 Å2 (138 Å2, if only the central three sugars are considered) in the ChiB active site, argifin covers 130 Å2, and argadin covers 148 Å2. Overall, argadin is buried deeper into the active site than argifin and makes tighter interactions.
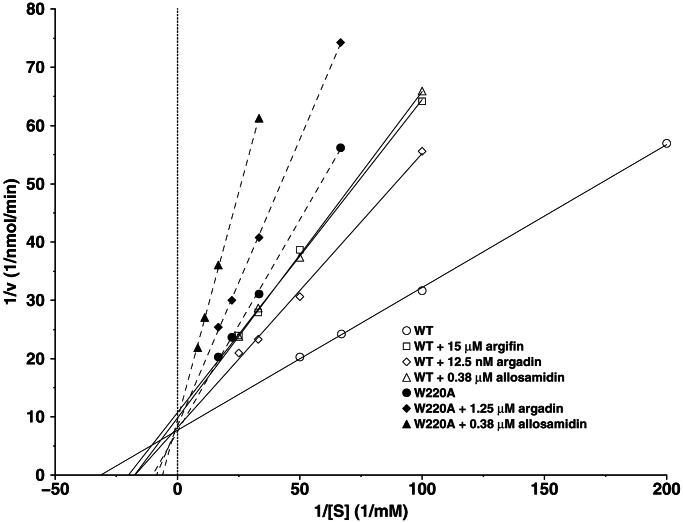
Preliminary analyses of argadin and argifin effects on endochitinase activity from L. cuprina blowfly revealed inhibition in the nanomolar range, with argadin inhibiting at an IC50 of 150 nM (4, 5). We have determined the inhibition constants of argifin and argadin against ChiB, revealing that while allosamidin inhibits ChiB with a Ki of 450 nM, argifin shows weaker inhibition (Ki = 33 μM) and argadin shows ≈20-fold stronger inhibition (Ki = 20 nM) than allosamidin (Fig. 4). Thus, argadin, a synthetically more accessible, peptide-based inhibitor, seems to show stronger inhibition of ChiB than the complex pseudotrisaccharide inhibitor allosamidin. As noted previously, argadin appears to be conformationally more restricted than argifin (Fig. 2), binds deeper in the active site (Fig. 3), and replaces ordered water molecules that were observed in the unliganded ChiB structure (17, 31). Argadin also has more interactions with key residues in the active site (Figs. 2 and 3). It thus is possible that the entropic penalty paid for inhibitor binding is lower for argadin than for argifin, and the enthalpic gain is higher, explaining why inhibition with argadin is 3 orders of magnitude stronger than with argifin. Calorimetric analyses of these effects will be reported in the future after total synthesis of argifin and argadin.
Figure 4.
Enzymology. Lineweaver–Burk plots illustrating inhibition of wild-type (WT) ChiB and the W220A mutant. Km ChiB = 33 μM, kcat ChiB = 15 s−1, Km W220A = 97 μM, and kcat W220A = 1.0 s−1.
Despite their different modes of interaction with the ChiB active site, binding of both argifin and argadin seems to involve interactions with Trp-97 and Trp-220 in the +1/+2 subsites of the enzyme. To estimate the importance of these interactions, we studied the effects on affinity in the ChiB W220A mutant (Fig. 4). The Ki values for inhibition of the W220A mutant were 5 μM and ≈400 μM for argadin and argifin, respectively, representing 250- and 12-fold increases compared with the wild-type enzyme (Fig. 4). In contrast the Ki for allosamidin, which binds to the −3 through −1 subsites and therefore does not interact with the Trp-97/Trp-220 pair, was not affected (530 nM for the W220A mutant and 450 nM for the wild type). These results demonstrate the difference between the well characterized inhibitor allosamidin and the peptide inhibitors argifin and argadin.
It should be noted that the subsites of family 18 chitinases are likely to differ in terms of binding affinity, e.g., depending on whether the enzymes degrade their polymeric substrate from the reducing or nonreducing (e.g., ChiB) end. Argadin and argifin seem optimized for enzymes with high-affinity subsites on the reducing (“+”) side of the catalytic center. Allosamidin, on the other hand, does not interact with this area of the active site as demonstrated by the W220A mutant. Development of peptide inhibitors for enzymes with particularly strong substrate binding on the nonreducing (“−”) side of the catalytic center may require the introduction of side chains that are able to extend into the −2 subsite and beyond.
Concluding Remarks.
The structures presented here give detailed insight into how cyclic peptides can mimic the substrate for a carbohydrate-processing enzyme and inhibit it. Both argifin and argadin interact with side chains (Asp-142, Glu-144, and Tyr-214) in the chitinase active site that are conserved completely and required for catalytic activity in family 18 chitinases (17, 31, 38). Compared with argifin, the tighter interactions of argadin with the chitinase and its more restricted structure (Figs. 2 and 3) could lead to stronger concerted entropic and enthalpic gains and may explain the 1,500-fold stronger inhibition. The mutational data has shown that the interactions of argadin with Trp-220 in the +2 subsite make an important contribution to the binding. The drastic effect of the W220A mutation on Ki values shows that the effectiveness of present inhibitors depends on subsite architecture, which varies significantly from enzyme to enzyme (e.g., Trp-220 is not well conserved in family 18 chitinases). Whereas this variance may cause problems when aiming at the design of “general” chitinase inhibitors, it shows the potential of tailoring inhibitors such that they inhibit only a limited number of specific family 18 chitinases, which is especially important in light of the discovery of human family 18 chitinases (9, 10, 40), which have been implicated in defense against fungal pathogens. The structures of ChiB–argifin and ChiB–argadin complexes provide opportunities for structure-based design and synthesis of derivatives, which may lead to more potent chitinase inhibitors with possible chemotherapeutical potential.
Acknowledgments
We thank the European Synchrotron Radiation Facility (Grenoble, France) for the time at beamline ID14-EH4 and Ian Eggleston for carefully reading the manuscript. We thank Xiaohong Jia for technical assistance. D.M.F.v.A. is supported by a Wellcome Trust Career Development Research Fellowship. B.S. is supported by a grant from the Norwegian Research Council. K.S. is supported partly by the Hoh-Ansha foundation.
Abbreviations
- ASA
aspartic β-semialdehyde
- ChiB
chitinase B
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1h0g (argifin) and 1h0i (argadin)].
References
- 1.Takadera T, Nomoto A, Izumida H, Nishijima M, Sano H. J Antibiot. 1996;49:829–831. doi: 10.7164/antibiotics.49.829. [DOI] [PubMed] [Google Scholar]
- 2.Cohen E. Arch Insect Biochem Physiol. 1993;22:245–261. doi: 10.1002/arch.940220118. [DOI] [PubMed] [Google Scholar]
- 3.Sakuda S, Isogai A, Matsumoto S, Suzuki A. J Antibiot. 1987;40:296–300. doi: 10.7164/antibiotics.40.296. [DOI] [PubMed] [Google Scholar]
- 4.Shiomi K, Arai N, Iwai Y, Turberg A, Koelbl H, Omura S. Tetrahedron Lett. 2000;41:2141–2143. [Google Scholar]
- 5.Arai N, Shiomi K, Yamaguchi Y, Masuma R, Iwai Y, Turberg A, Koelbl H, Omura S. Chem Pharm Bull. 2000;48:1442–1446. doi: 10.1248/cpb.48.1442. [DOI] [PubMed] [Google Scholar]
- 6.Vinetz J M, Dave S K, Specht C A, Brameld K A, Xu B, Hayward R, Fidock D A. Proc Natl Acad Sci USA. 1999;96:14061–14066. doi: 10.1073/pnas.96.24.14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vinetz J M, Valenzuela J G, Specht C A, Aravind L, Langer R C, Ribeiro J M C, Kaslow D C. J Biol Chem. 2000;275:10331–10341. doi: 10.1074/jbc.275.14.10331. [DOI] [PubMed] [Google Scholar]
- 8.Tsai Y-L, Hayward R E, Langer R C, Fidock D A, Vinetz J M. Infect Immun. 2001;69:4048–4054. doi: 10.1128/IAI.69.6.4048-4054.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renkema G H, Boot R G, Muijsers A O, Donker-Koopman W E, Aerts J M F G. J Biol Chem. 1995;270:2198–2202. doi: 10.1074/jbc.270.5.2198. [DOI] [PubMed] [Google Scholar]
- 10.Boot R G, Blommaart E F C, Swart E, Ghauharali van der Vlugt K, Bijl N, Moe C, Place A, Aerts J M F G. J Biol Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 11.Boot R G, Renkema G H, Verhoek M, Strijland A, Bliek J, de Meulemeester T M A M O, Mannens M M A M, Aerts J M F G. J Biol Chem. 1998;273:25680–25685. doi: 10.1074/jbc.273.40.25680. [DOI] [PubMed] [Google Scholar]
- 12.Sakuda S, Isogai A, Matsumoto S, Suzuki A, Koseki K. Tetrahedron Lett. 1986;27:2475–2478. [Google Scholar]
- 13.Spindler-Barth M, Blattner R, Vorgias C E, Spindler K D. Pestic Sci. 1998;52:47–52. [Google Scholar]
- 14.Bokma E, Barends T, Terwisscha van Scheltinga A C, Dijkstra B W, Beintema J J. FEBS Lett. 2000;478:119–122. doi: 10.1016/s0014-5793(00)01833-0. [DOI] [PubMed] [Google Scholar]
- 15.Blattner R, Gerard P J, Spindler-Barth M. Pestic Sci. 1997;50:312–318. [Google Scholar]
- 16.Terwisscha van Scheltinga A C, Kalk K H, Beintema J J, Dijkstra B W. Structure (London) 1994;2:1181–1189. doi: 10.1016/s0969-2126(94)00120-0. [DOI] [PubMed] [Google Scholar]
- 17.van Aalten D M F, Komander D, Synstad B, Gåseidnes S, Peter M G, Eijsink V G H. Proc Natl Acad Sci USA. 2001;98:8979–8984. doi: 10.1073/pnas.151103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terwisscha van Scheltinga A C, Armand S, Kalk K H, Isogai A, Henrissat B, Dijkstra B W. Biochemistry. 1995;34:15619–15623. doi: 10.1021/bi00048a003. [DOI] [PubMed] [Google Scholar]
- 19.Tews I, Terwisscha van Scheltinga A C, Perrakis A, Wilson K S, Dijkstra B W. J Am Chem Soc. 1997;119:7954–7959. [Google Scholar]
- 20.Knapp S, Vocadlo D, Gao Z, Kirk B, Lou J, Withers S G. J Am Chem Soc. 1996;118:6804–6805. [Google Scholar]
- 21.Kobayashi S, Kiyosada T, Shoda S. J Am Chem Soc. 1996;118:13113–13114. [Google Scholar]
- 22.Vocadlo D J, Davies G J, Laine R, Withers S G. Nature (London) 2001;412:835–838. doi: 10.1038/35090602. [DOI] [PubMed] [Google Scholar]
- 23.Uitdehaag J C M, Mosi R, Kalk K H, van der Veen B A, Dijkhuizen L, Withers S G, Dijkstra B W. Nat Struct Biol. 1999;6:432–436. doi: 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 24.Davies G, Henrissat B. Structure (London) 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 25.Griffith D A, Danishefsky S J. J Am Chem Soc. 1991;113:5863–5864. [Google Scholar]
- 26.Blattner R, Furneaux R H, Lynch G P. Carbohydr Res. 1996;296:29–40. doi: 10.1016/s0008-6215(96)90613-8. [DOI] [PubMed] [Google Scholar]
- 27.Kato T, Shizuri Y, Izumida H, Yokoyama A, Endo M. Tetrahedron Lett. 1995;36:2133–2136. [Google Scholar]
- 28.Izumida H, Imamura N, Sano H. J Antibiot. 1996;49:76–80. doi: 10.7164/antibiotics.49.76. [DOI] [PubMed] [Google Scholar]
- 29.Tabudravu J N, Eijsink V G H, Gooday G W, Jaspars M, Komander D, Legg M, Synstad B, van Aalten D M F. Bioorg Med Chem. 2002;10:1123–1128. doi: 10.1016/s0968-0896(01)00372-8. [DOI] [PubMed] [Google Scholar]
- 30. Rottman, A., Synstad, B., Eijsink, V. & Peter, M. G. (1999) Eur. J. Org. Chem. 2293–2297.
- 31.van Aalten D M F, Synstad B, Brurberg M B, Hough E, Riise B W, Eijsink V G H, Wierenga R K. Proc Natl Acad Sci USA. 2000;97:5842–5847. doi: 10.1073/pnas.97.11.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brurberg M B, Nes I F, Eijsink V G H. Microbiology. 1996;142:1581–1589. doi: 10.1099/13500872-142-7-1581. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 35.Brunger A T, Adams P D, Clore G M, Gros P, Grosse-Kunstleve R W, Jiang J-S, Kuszewski J, Nilges M, Pannu N S, Read R J, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.van Aalten D M F, Bywater R, Findlay J B C, Hendlich M, Hooft R W W, Vriend G. J Comput Aided Mol Des. 1996;10:255–262. doi: 10.1007/BF00355047. [DOI] [PubMed] [Google Scholar]
- 37.Wallace A C, Laskowski R A, Thornton J M. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 38.Papanikolau Y, Prag G, Tavlas G, Vorgias C E, Oppenheim A B, Petratos K. Biochemistry. 2001;40:11338–11343. doi: 10.1021/bi010505h. [DOI] [PubMed] [Google Scholar]
- 39.Kolstad G, Synstad B, Eijsink V G H, van Aalten D M F. Acta Crystallogr D. 2002;58:377–379. doi: 10.1107/s0907444901018972. [DOI] [PubMed] [Google Scholar]
- 40. Fusetti, F., von Moeller, H., Houston, D., Rozenboom, H. J., Dijkstra, B. W., Boot, R. G., Aerts, J. M. F. G. & van Aalten, D. M. F. (2002) J. Biol. Chem., in press. [DOI] [PubMed]