Abstract
To determine whether calcium polyvalent cation-sensing receptors (CaRs) are salinity sensors in fish, we used a homology-based cloning strategy to isolate a 4.1-kb cDNA encoding a 1,027-aa dogfish shark (Squalus acanthias) kidney CaR. Expression studies in human embryonic kidney cells reveal that shark kidney senses combinations of Ca2+, Mg2+, and Na+ ions at concentrations present in seawater and kidney tubules. Shark kidney is expressed in multiple shark osmoregulatory organs, including specific tubules of the kidney, rectal gland, stomach, intestine, olfactory lamellae, gill, and brain. Reverse transcriptase–PCR amplification using specific primers in two teleost fish, winter flounder (Pleuronectes americanus) and Atlantic salmon (Salmo salar), reveals a similar pattern of CaR tissue expression. Exposure of the lumen of winter flounder urinary bladder to the CaR agonists, Gd3+ and neomycin, reversibly inhibit volume transport, which is important for euryhaline teleost survival in seawater. Within 24–72 hr after transfer of freshwater-adapted Atlantic salmon to seawater, there are increases in their plasma Ca2+, Mg2+, and Na+ that likely serve as a signal for internal CaRs, i.e., brain, to sense alterations in salinity in the surrounding water. We conclude that CaRs act as salinity sensors in both teleost and elasmobranch fish. Their tissue expression patterns in fish provide insights into CaR functions in terrestrial animals including humans.
Calcium polyvalent cation-sensing receptor (CaRs) are G protein-coupled receptors that “sense” alterations in divalent and polyvalent cations present in extracellular fluid and are the principal mechanism regulating extracellular Ca2+ in terrestrial organisms including humans (1). Although CaRs' tissue distribution in terrestrial animals highlights their pivotal role in Ca2+ and Mg2+ homeostasis, their presence in mammalian brain and gastrointestinal tract suggest that CaRs might have additional physiological roles that are not currently understood (1–3). Moreover, Ca2+ sensing by human parathyroid CaR is altered by modulating extracellular ionic strength (4), suggesting that CaRs might link divalent and monovalent cation metabolism in terrestrial animals (5). In this article, we provide evidence that CaRs allow fish to sense and respond to alterations in water salinity based on changes in Ca2+, Mg2+, and Na+ concentrations found in freshwater, brackish water, and seawater. Similarly, changes in plasma Ca2+, Mg2+, and Na+ occurring when fish move from freshwater to seawater likely serve as salinity sensing cues for CaRs located within fish internal organs.
Materials and Methods
cDNA Library Manufacture and Isolation of Dogfish Shark Kidney CaR (SKCaR) Clone.
A single SKCaR clone was isolated after screening 1.5 × 106 plaques of an unamplified dogfish shark λZAP kidney cDNA library with a 32P-labeled rat kidney CaR (RaKCaR) cDNA probe under intermediate stringency (0.5 × SSC, 0.1% SDS, 50°C) (6). Nucleotide sequencing of the clone used commercial sequencing services (University of Maine, Orono), and its sequence was compared with that of other CaRs (GenBank).
SKCaR Expression in Human Embryonic Kidney (HEK) Cells.
HEK cells were transiently transfected with SKCaR-pcDNA3 (Invitrogen) (HEK-SKCaR), and responses to alterations in extracellular Ca2+, Mg2+, and NaCl were quantified by using a FURA-2/AM-based assay as described (4, 7) and expressed as % normalized intracellular calcium response to receptor activation.
Polyclonal Antiserum Manufacture, Immunoblotting, and Immunocytochemistry.
A 17-mer peptide (ARSRNSADGRSGDDLP + C for conjugation) corresponding to SKCaR amino acids 965–980 was synthesized and used to raise rabbit antiserum as described (8). Immunoblotting (9) of crude membrane preparations prepared by differential centrifugation of shark kidney, rectal gland, or HEK cell homogenates, as well as immunocytochemistry of frozen (8) and paraffin sections from fixed fish tissues (H.H., unpublished work), was performed by using either anti-SKCaR (1:2–3,000) or another anti-CaR antiserum (A4641) (1:1–2,000) raised to amino acids 214–237 of bovine parathyroid CaR (7–10). Immunolocalization studies in gill chloride cells were conducted on hydrated deparaffinized sections after blocking endogenous peroxidase (10 min of 3% H2O2 in methanol). Sections were exposed to 1% goat serum, then mAb α5 (1:175 of culture supernatant in PBS for 90 min). α5 (anti-Na+K+ ATPase) was developed by D. Fambrough (Developmental Studies Hybridoma Bank, National Institute of Child Health and Human Development, maintained at University of Iowa, Iowa City) and detected with the Vectastain Elite ABC Kit (Vector Laboratories) (8).
Quantitation of Net Water Transport in Isolated Flounder Urinary Bladder Sac Preparations.
Net apical to basolateral water flux (transepithelial water reabsorption, Jv) was measured gravimetrically in 10-min intervals by using individual bladders as described (11, 12).
RNA Blotting Analysis and Reverse Transcriptase–PCR (RT-PCR) of Teleost and Elasmobranch Tissues.
Total RNA was purified with Stat 60 reagent (Tel-Test B, Friendswood, TX) and used for RNA blotting (3, 9) after poly(A)+ purification with the Micro FastTrack Kit or RT-PCR after cDNA production with a cDNA cycle kit (Invitrogen). The cDNA was amplified [30 cycles of 1 min at 94°C, 2 min at 57°C (salmon) or 56°C (flounder), 3 min at 72°C] by using degenerate primers [forward primer dSK-F3 (SKCaR nucleotides 2279–2306) and reverse primer dSK-R4 (SKCaR nucleotides 2904–2934)]. Aliquots of PCRs were subjected to gel electrophoresis and ethidium bromide staining or blotted onto Magnagraph membranes (Osmonics, Westboro, MA) and probed with a 32P-Atlantic salmon genomic PCR product (653-bp sequence corresponding to that shown in Fig. 1), washed (0.1 × SSC, 0.1% SDS at 55°C), and autoradiographed. Amplified PCR products from tissues were sequenced as described above.
Figure 1.
Comparison of SKCaR (S), RaKCaR (R), Fugu CaR (F), and two RT-PCR teleost CaR products from winter flounder (W) or Atlantic salmon (A) tissues. Amino acids identical to SKCaR are shaded, and locations of the seven-transmembrane regions are boxed. Bold underlined amino acids indicate the location of the dSK-F3 and dSK-R4 primers used to amplify 656-nt PCR products.
Measurement of Plasma Ca2+, Mg2+, and Na+ Concentrations.
Plasma Ca2+, Mg2+, and Na+ were quantified (NorDx Diagnostic Laboratories, Scarborough, ME) from blood collected from the caudal artery of Atlantic salmon and anticoagulated with 8% lithium heparin (Sigma).
Fish.
Squalus acanthias, Salmo salar smolt, and Pleuronectes americanus were obtained from the Mount Desert Island Biological Laboratory, Heritage Salmon (New Brunswick, Canada), or T. Linley of Washington County Technical College, Eastport, ME.
Results
Structure of SKCaR.
We obtained a 4,116-nt clone from a dogfish shark kidney cDNA library by using a RaKCaR probe under reduced stringency. The clone contains a 3,081-nt ORF encoding a 1,027-aa protein called SKCaR (GenBank accession AF406649 at http://www.ncbi.nlm.nih.gov). As shown in Fig. 1, SKCaR possesses an overall 74% amino acid identity with RaKCaR (6), including a large extracellular domain (74.8% identity vs. RaKCaR) that binds positively charged ligands including Ca2+, Mg2+, and polyamines (1), a seven-transmembrane G-protein binding region (91.2% identity vs. RaKCaR), and a 156-aa intracellular carboxyl terminus (30.8% identity vs. RaKCaR). SKCaR bears only a 64% homology to the candidate teleost CaR (Fugu rubripes) (13). Major differences between SKCaR and Fugu candidate CaR include a 19-aa deletion in the extracellular domain and a foreshortened carboxyl-terminal domain in Fugu as compared with SKCaR.
SKCaR mRNA and Protein Are Expressed in Multiple Tissues of Dogfish Shark.
Shark kidney contains two major SKCaR poly(A)+ transcripts of ≈7 and 4 kb (Fig. 2A) that are similar to those in mammalian kidney (6, 8). Abundant SKCaR protein is present on the apical (luminal-facing) membranes of kidney proximal tubule segments PIa and PIIb, late distal tubule (LDT) (Fig. 2B), and collecting tubule-collecting duct as well as the basolateral membrane of early distal tubules. A similar pattern of immunostaining was obtained by using anti-CaR antiserum 4641 (raised to 23-aa bovine parathyroid CaR sequence homologous to SKCaR amino acids 247–270) (Fig. 2C). Detailed analyses of the specific anatomical pattern of SKCaR expression in shark kidney suggests that it plays an important role in the sensing and regulation of renal Mg2+ secretion/reabsorption (H. H., unpublished work).
Figure 2.
SKCaR is expressed in a variety of shark tissues. (A) Blot of kidney poly(A)+ RNA showing 7- and 4-kb SKCaR transcripts; gel top (GT) and dye front (DF). Immunocytochemistry with anti-SKCaR (B and D–H), anti-4641 (C), and anti-Na+K+ ATPase (I). Arrows point to specific labeling. (B) Kidney LDT apical surface labeling (red-brown color) (× 1,390). (C) Kidney LDT and collecting duct (CD) (×1,390). (D) Rectal gland central canal (CC) and parenchyma (×265). (E) Intestinal apical (A) surfaces and blood or lymph vessels (v) (×1,390). (F) Stomach epithelial cells (me) and crypts (c) (×1,030). (G) Olfactory epithelial cells (A arrows) (×230). (H and I) Gill tissue (chloride cells show specific labeling) (×180). For various controls see Fig. 6.
SKCaR is present in multiple other osmoregulatory tissues including rectal gland, intestine, stomach, olfactory epithelia, and gill chloride cells as shown by immunocytochemistry (Fig. 2 D–H) and RNA blotting (data not shown). Based on these locations, SKCaR may function to sense changes in the divalent and monovalent cation concentrations in seawater (14) as well as in the lumens of rectal gland, stomach, gut, and associated vessels (15).
SKCaR Protein Is a Salinity Sensor.
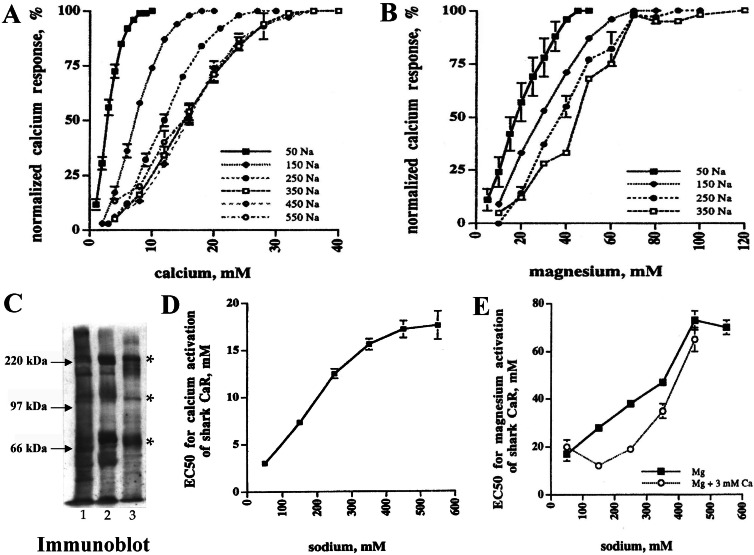
HEK–SKCaR cells respond to alterations in extracellular Ca2+ (Fig. 3A) or Mg2+ (Fig. 3B) as measured by increases in intracellular Ca2+. Comparison of SKCaR's EC50 values for Ca2+ (EC50 ≈7.5 mM, Fig. 3D) or Mg2+ (EC50 ≈30 mM Fig. 3E) shows that it is less sensitive to these divalent cations as compared with mammalian CaRs (EC50 3–5 mM Ca2+, 15–20 mM Mg2+) when bathed in mammalian physiological saline solution (150 mM NaCl) (4, 7). HEK–SKCaR cells contain 200- to 240-kDa, 120- to 140-kDa, and 68-kDa protein bands that comigrate with corresponding SKCaR bands in both kidney and rectal gland by using anti-SKCaR (Fig. 3C). The specificity of anti-SKCaR serum is further illustrated in Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org.
Figure 3.
Ionic pharmacology of SKCaR in HEK cells. (A) SKCaR sensitivity to extracellular Ca2+ concentrations at different NaCl concentrations. (B) SKCaR sensitivity to extracellular Mg2+ concentrations at different NaCl concentrations. (C) HEK–SKCaR cells (lane 3) express SKCaR proteins (*) that comigrate with bands in shark kidney (lane 1) and rectal gland (lane 2). (D) Modulation of Ca2+ EC50 by NaCl. (E) Modulation of Mg2+ EC50 by NaCl that is enhanced by 3 mM Ca2+.
Seawater concentrations of NaCl (450 mM) increase SKCaR's EC50 values for Ca2+ and Mg2+ ≈2.4-fold as compared with values obtained in 150 mM NaCl (Fig. 3). SKCaR's responsiveness under ionic conditions of shark urine (240 mM NaCl, 3 mM Ca2+, 10–100 mM Mg2+) reveals that it is well suited to sense Mg2+ in shark kidney LDT and collecting tubule-collecting duct (Fig. 3E) (16).
In Winter Flounder, CaRs Are Present in Multiple Tissues and Modulate Thiazide-Sensitive NaCl Cotransporter (NCC)-Mediated Water Reabsorption in Urinary Bladder.
Urinary bladder epithelial cells of marine teleosts recover water from ureteral urine via solute-coupled NCC water transport (11, 17). Bladder and stomach contain CaR proteins (Fig. 4 A–C; Fig. 7, which is published as supporting information on the PNAS web site) where immunoblots reveal a CaR band of ≈230 kDa (Fig. 4D). CaR transcripts in flounder tissues were detected by RT-PCR using specific primers to amplify a 653-nt portion of CaR mRNA (Fig. 4E) as well as by RNA blotting (Fig. 4F). The corresponding 198-aa partial sequence of this bladder CaR shares an 85.4% identity with SKCaR (Fig. 1). Omission of reverse transcriptase enzyme from these reactions yields no amplification products (data not shown).
Figure 4.
Winter flounder tissues contain CaRs including a 4.1-kb CaR in urinary bladder that modulates NCC-mediated NaCl-coupled water reabsorption. (A) Immunocytochemistry with anti-SKCaR (A) or 4641 (Fig. 7) antisera reveals CaR in bladder epithelial cells (arrows) (×620). (B) Anti-SKCaR staining of stomach cells (arrows) (×750). (C) Same as B with preimmune antiserum. (D) Immunoblot of stomach homogenate using anti-4641 antiserum in the absence (A) or presence (A+P) of blocking peptide showing specific 230-kDa band. (E) Southern blot of RT-PCR of winter flounder: lane 1, urinary bladder; lane 2, kidney; lane 3, intestine; lane 4, stomach; lane 5, liver; lane 6, brain; lane 7, water blank, and lane 8, positive control. (F) RNA blot of 5 μg poly(A)+ RNA/lane from flounder heart (lane 1), liver (lane 2), spleen (lane 3), and urinary bladder (lane 4) probed first with 32P-RaKCaR and exposed for 10 days (Left), then stripped and reprobed with 32P-NCC cDNA (17) and exposed for 1 hr. Representative of five experiments. (G) Effects on Jv of separate additions of 100 μM hydrochlorothiazide (HCTZ), 100 μM Gd3+, or 200 μM Neo to solution bathing mucosal surface (n = 5 each). (H) Jv inhibition by Gd3+ and Neo. Each data point is mean of at least five independent Jv determinations.
Removal of NaCl and water by bladder epithelia of seawater-adapted flounder increases bladder Mg2+ concentrations to as high as 100 mM (11). We used bladder sac preparations to test whether a resident CaR might sense alterations in ionic content and, in response, reduce bladder water recovery to prevent Mg2+ crystal formation. Bladder Jv was inhibited by CaR agonists Gd3+ and neomycin (Neo) (Fig. 4 G and H) and was fully reversible (Fig. 7). The IC50 for Gd3+ (19 μM) and Neo (84 μM) are 54% and 120%, respectively, of values reported for bovine parathyroid CaR (1). Thus, CaRs modulate transepithelial water transport, preventing excessively high luminal Mg2+ concentrations that might otherwise occur in seawater (15).
CaRs Are Present in Organs of Atlantic Salmon and Permit Sensing of External and Internal Alterations in Ca2+, Mg2+, and Na+.
RT-PCR and immunocytochemistry show that CaRs are present in Atlantic salmon tissues (Fig. 5 A–C; Fig. 8, which is published as supporting information on the PNAS web site). RT-PCR products from salmon urinary bladder, kidney, and olfactory lamellae are 84.9% identical to amino acids of SKCaR (Fig. 1). blast (National Institutes of Health, Bethesda, MD) analysis of flounder and salmon partial CaR sequences confirms they are most similar to other CaR sequences, with significantly lower homology to putative pheromone receptors (1, 13).
Figure 5.
CaRs in Atlantic salmon tissues may sense increases in external and plasma Ca2+, Mg2+, and NaCl that accompany transfer from freshwater to seawater. (A) Ethidium bromide (EtBr)-stained gel and corresponding Southern blot (SB) of RT-PCR products in: lane 1, gill; lane 2, olfactory lamellae; lane 3, urinary bladder; lane 4, kidney; lane 5, intestine; lane 6, stomach; lane 7, liver; lane 8, brain; lane 9, water blank; and lane 10, positive control. Anti-SKCaR immunostaining (arrows) of hindbrain neurons (B) containing CaRs (×840) and gill chloride cells (arrows) and mucous cells (arrowhead) (C) (×175) (also anti-4641 staining of chloride cells; see Fig. 8). (D) Chloride cells stained with anti-Na+K+ATPase antiserum (arrows) (×175). (E) Plasma Ca2+ and Mg2+ and corresponding Na+ concentrations (F) of freshwater-adapted fish (average weight 30 g) transferred to seawater at time 0 and samples obtained at intervals indicated. Each data point is the mean ± SD of at least four independent determinations.
In salmon, CaR is present in brain neurons adjacent to the fourth ventricle within the region of the vagal lobe and commissural nucleus of Cajal (Fig. 5B). This hindbrain region is important in regulation of gustatory and visceral activities including esophageal and intestinal motility (18). CaR is also present in mucous and chloride cells of salmon gill (Fig. 5 C and D).
The presence of CaRs in organs like brain suggests that changes in divalent and monovalent cation concentrations of extracellular fluid in teleost fish might serve as internal signals for sensing alterations in environmental water salinity. As shown in Fig. 5 E and F, transfer of freshwater-adapted Atlantic salmon to seawater increases plasma Ca2+, Mg2+, and Na+. Under these circumstances, the elevations of plasma Na+, Ca2+, and Mg2+ might serve as regulatory signals sensed by internal CaRs during the interval immediately after seawater transfer.
Discussion
Previous efforts to understand the distribution and function of CaRs in fish have focused exclusively on their roles as regulators of divalent mineral ion metabolism (13, 14, 19, 20). The data presented here suggest that CaRs may also act as salinity sensors in multiple organs that are either directly in contact with freshwater, brackish water, or seawater, or tissues that experience alterations in Ca2+, Mg2+, or Na+ concentrations as a consequence of a fish's exposure to varying salinity environments.
In dogfish kidney, SKCaR is present in Mg2+-secreting and reabsorptive segments (H.H., unpublished work) where it may modulate Mg2+ transport and excretion by this organ. Similarly, sensing of luminal Mg2+ concentrations by teleost urinary bladder CaR(s) integrates water recovery and Mg2+ excretion. These functions for CaRs in fish renal tissue are similar to those reported for mammalian thick ascending limb, distal tubule and collecting duct (1, 6, 8). Further studies are needed to determine whether SKCaR homologs in other shark organs function in a manner similar to that reported for amphibian (21) and mammalian CaRs (22).
SKCaR homologs in olfactory and gill epithelia (Figs. 2 and 5) are bathed by a range of salinity gradients that exist between seawater (10 mM Ca2+, 50 mM Mg2+, 450 mM Na+) and freshwater (0.07–2 mM Ca2+, 0.4–3 mM Mg2+, 3–5 mM Na+), which are encountered by teleost (14) and selected elasmobranch fish (23). SKCaR activation by either Ca2+ or Mg2+ is progressively reduced by increasing NaCl concentrations (Fig. 3). Should olfactory and gill SKCaR homologs function similarly, these data will provide insights into cation sensing by teleost olfactory (14, 24) and gill epithelia (25, 26) and suggest that CaRs may be salinity sensors for these organs.
CaRs' sensing of polycationic agonists is modulated by alterations in ionic strength, not extracellular osmolality (4). As reported here, CaRs in fish possess similar structural and functional features as compared with mammalian CaRs and may provide a mechanism to inform internal organs of fish of changes in surrounding water salinity. Additional studies focused on the roles of CaRs in both fish and mammals may provide insights into the possible roles of CaRs as internal salinity sensors in both aquatic and terrestrial animals.
Supplementary Material
Acknowledgments
We acknowledge the expert advice of D. Evans, D. Ward, and D. Russell and the technical assistance of A. Wisinski.
Abbreviations
- CaR
calcium polyvalent cation-sensing receptor
- RaKCaR
rat kidney CaR
- SKCaR
dogfish shark kidney CaR
- HEK
human embryonic kidney
- RT-PCR
reverse transcriptase–PCR
- Jv
transepithelial water reabsorption
- NCC
thiazide-sensitive NaCl cotransporter
- LDT
late distal tubule
- Neo
neomycin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF406649).
References
- 1.Brown E M, Pollack M D, Seidman C E, Seidman J G, Chou Y-H W, Riccardi D, Hebert S C. N Engl J Med. 1995;333:234–240. doi: 10.1056/NEJM199507273330407. [DOI] [PubMed] [Google Scholar]
- 2.Rogers K V, Dunn C K, Hebert S C, Brown E M. Brain Res. 1997;744:47–56. doi: 10.1016/s0006-8993(96)01070-0. [DOI] [PubMed] [Google Scholar]
- 3.Chattopadhyay N, Cheng I, Rogers K, Riccardi D, Hall A, Diaz R, Hebert S C, Soybel D I, Brown E M. Am J Physiol. 1998;274:G122–G130. doi: 10.1152/ajpgi.1998.274.1.G122. [DOI] [PubMed] [Google Scholar]
- 4.Quinn S J, Kifor O, Trivedi S, Diaz R, Vassilev P, Brown E. J Biol Chem. 1998;273:19579–19586. doi: 10.1074/jbc.273.31.19579. [DOI] [PubMed] [Google Scholar]
- 5.Baum M A, Harris H W. Am J Med Sci. 1998;316:321–328. doi: 10.1097/00000441-199811000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Riccardi D, Park J, Lee W-S, Gamba G, Brown E M, Hebert S C. Proc Natl Acad Sci USA. 1995;92:131–135. doi: 10.1073/pnas.92.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bai M, Quinn S, Trivedi S, Kifor O, Pearce S H, Pollak M R, Krapcho K, Hebert S C, Brown E M. J Biol Chem. 1996;271:19537–19545. doi: 10.1074/jbc.271.32.19537. [DOI] [PubMed] [Google Scholar]
- 8.Sands J M, Naruse M, Baum M, Jo I, Hebert S C, Brown E M, Harris H W. J Clin Invest. 1997;99:1399–1405. doi: 10.1172/JCI119299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward D T, Brown E M, Harris H W. J Biol Chem. 1998;273:14476–14483. doi: 10.1074/jbc.273.23.14476. [DOI] [PubMed] [Google Scholar]
- 10.Riccardi D, Hall A E, Chattopadhyay N, Xu J Z, Brown E M, Hebert S C. Am J Physiol. 1998;274:F611–F622. doi: 10.1152/ajprenal.1998.274.3.F611. [DOI] [PubMed] [Google Scholar]
- 11.Renfro J L. Am J Physiol. 1975;228:52–61. doi: 10.1152/ajplegacy.1975.228.1.52. [DOI] [PubMed] [Google Scholar]
- 12.Demarest J R. Am J Physiol. 1984;246:F395–F401. doi: 10.1152/ajprenal.1984.246.4.F395. [DOI] [PubMed] [Google Scholar]
- 13.Naito T, Saito Y, Yamamoto J, Nozaki Y, Tomura K, Hazama M, Nakanishi S, Brenner S. Proc Natl Acad Sci USA. 1998;95:5178–5181. doi: 10.1073/pnas.95.9.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hubbard P C, Barata E N, Canario A V M. J Exp Biol. 2000;203:3821–3829. doi: 10.1242/jeb.203.24.3821. [DOI] [PubMed] [Google Scholar]
- 15.Karnaky K J. In: The Physiology of Fishes. 2nd Ed. Evans D H, editor. Boca Raton, FL: CRC; 1998. pp. 157–176. [Google Scholar]
- 16.Beyenbach K W. In: Cellular and Molecular Approaches to Fish Ionic Regulation. Wood C M, Shuttleworth T J, editors. New York: Academic; 1995. pp. 192–209. [Google Scholar]
- 17.Gamba G, Saltzberg S N, Lombardi M, Miyanoshita A, Lytton J L, Hediger M A, Brenner B M, Hebert S C. Proc Natl Acad Sci USA. 1993;90:2749–2753. doi: 10.1073/pnas.90.7.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wullimann M F. In: The Physiology of Fishes. 2nd Ed. Evans D H, editor. Boca Raton, FL: CRC; 1998. pp. 245–282. [Google Scholar]
- 19.Ingleton P M, Hubbard P A, Danks J A, Elgar G, Sandford R A, Balmeat R J. In: Comparative Endocrinology. Danks J, Dacke C, Flik G, Gay C, editors. Bristol: BioScientifica; 1999. pp. 45–48. [Google Scholar]
- 20.Radman D P, McCudden C, James K, Nemeth E M, Wagner G. Mol Cell Endocrinol. 2002;186:111–119. doi: 10.1016/s0303-7207(01)00643-8. [DOI] [PubMed] [Google Scholar]
- 21.Cima R R, Cheng I, Klingensmith M E, Chattopadhyay N, Kifor O, Hebert S C, Brown E M, Soybel D I. Am J Physiol. 1997;273:G1051–G1060. doi: 10.1152/ajpgi.1997.273.5.G1051. [DOI] [PubMed] [Google Scholar]
- 22.Geibel J P, Wagner C A, Caroppo R, Qureshi I, Gloeckner J, Manuelidis L, Kirchoff P, Radebold K. J Biol Chem. 2001;276:39549–39552. doi: 10.1074/jbc.M107315200. [DOI] [PubMed] [Google Scholar]
- 23.Piermarini P M, Evans D M. J Exp Biol. 2000;203:2957–2966. doi: 10.1242/jeb.203.19.2957. [DOI] [PubMed] [Google Scholar]
- 24.Bodznick D. J Comp Physiol. 1978;127:157–166. [Google Scholar]
- 25.Perry S. Annu Rev Physiol. 1997;59:325–347. doi: 10.1146/annurev.physiol.59.1.325. [DOI] [PubMed] [Google Scholar]
- 26.Seidelin M, Madsen S S, Blenstrup H, Tipsmark C K. Physiol Biochem Zool. 2000;73:446–453. doi: 10.1086/317737. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.