Abstract
The wealth of comparative genomics data from yeast species allows the molecular evolution of these eukaryotes to be studied in great detail. We used “proximity plots” to visually compare chromosomal gene order information from 14 hemiascomycetes, including the recent Génolevures survey, to Saccharomyces cerevisiae. Contrary to the original reports, we find that the Génolevures data strongly support the hypothesis that S. cerevisiae is a degenerate polyploid. Using gene order information alone, 70% of the S. cerevisiae genome can be mapped into “sister” regions that tile together with almost no overlap. This map confirms and extends the map of sister regions that we constructed previously by using duplicated genes, an independent source of information. Combining gene order and gene duplication data assigns essentially the whole genome into sister regions, the largest gap being only 36 genes long. The 16 centromere regions of S. cerevisiae form eight pairs, indicating that an ancestor with eight chromosomes underwent complete doubling; alternatives such as segmental duplications can be ruled out. Gene arrangements in Kluyveromyces lactis and four other species agree quantitatively with what would be expected if they diverged from S. cerevisiae before its polyploidization. In contrast, Saccharomyces exiguus, Saccharomyces servazzii, and Candida glabrata show higher levels of gene adjacency conservation, and more cases of imperfect conservation, suggesting that they split from the S. cerevisiae lineage after polyploidization. This finding is confirmed by sequences around the C. glabrata TRP1 and IPP1 loci, which show that it contains sister regions derived from the same duplication event as that of S. cerevisiae.
The same features—small genome size, high gene density, and a near-absence of introns—that made Saccharomyces cerevisiae an ideal subject for the first eukaryotic genome project also make the hemiascomycete fungi attractive for comparative genomics studies. After some initial studies that demonstrated that gene order can be conserved between S. cerevisiae and hemiascomycetes from genera such as Ashbya, Kluyveromyces, and Candida (1–4), and that the extent of gene order conservation decreases with increasing evolutionary distance (4, 5), the Génolevures project (6) systematically surveyed the genomes of 13 hemiascomycete species. These genomes were sequenced to between 0.2× and 0.4× coverage, by using paired sequence reads from both ends of plasmid library clones, thereby gathering extensive information on their gene content and order.
By analysis of the locations of duplicated genes, we proposed in 1997 that S. cerevisiae is a paleopolyploid—i.e., that the entire genome became duplicated at some point in its evolutionary past and subsequently sustained rearrangements and gene loss (7). Under this hypothesis, every part of the S. cerevisiae genome should be paired with another part, but in our original study we were able to map only about 50% of the genome into sister regions because these could be recognized only if they contained several duplicated genes. We predicted that the remainder of the genome could be paired up if the sequences of more genomes became available (8). The Génolevures project, as well as other sequencing surveys (5, 9) and the Candida albicans genome project, now provides the necessary information. We show here that almost the entire genome of S. cerevisiae lies in sister regions derived by descent with modification from single ancestral regions. The Génolevures data strongly confirm the polyploidy model, contrary to the claims made in papers reporting these data (6, 10–12).
Methods
The Génolevures data, comprising 2,500–6,000 plasmid end-sequences for each of 13 hemiascomycete species (6), were downloaded from www.genoscope.fr/externe/sequences/banque_Projet_AR. The contig assemblies described in the Génolevures publications (6) were not available, so we assembled contigs for each species by using phrap (www.phrap.org), after augmenting the datasets for some species by including sequences from GenBank and other projects (5, 9). Sequence assembly 6 for C. albicans was obtained from the Stanford Genome Technology Center website at www-sequence.stanford.edu/group/candida.
We used the S. cerevisiae genome annotation of Wood et al. (13), downloaded from ftp.sanger.ac.uk/pub/yeast/SCreannotation. All assembled contigs were used as queries in blastx searches against the 5,583 annotated proteins (excluding “very hypothetical” proteins and pseudogenes), with the seg filter (14). For any gene-sized region in a contig, we considered the S. cerevisiae protein with the strongest blastx hit to be the ortholog, provided that the Expect value (E-value) for this hit (i) was <1e−10, and (ii) was more than 1e10 times lower than the E-value for the second-best hit to the same region of the contig. Any hits failing the second criterion were flagged as ambiguous.
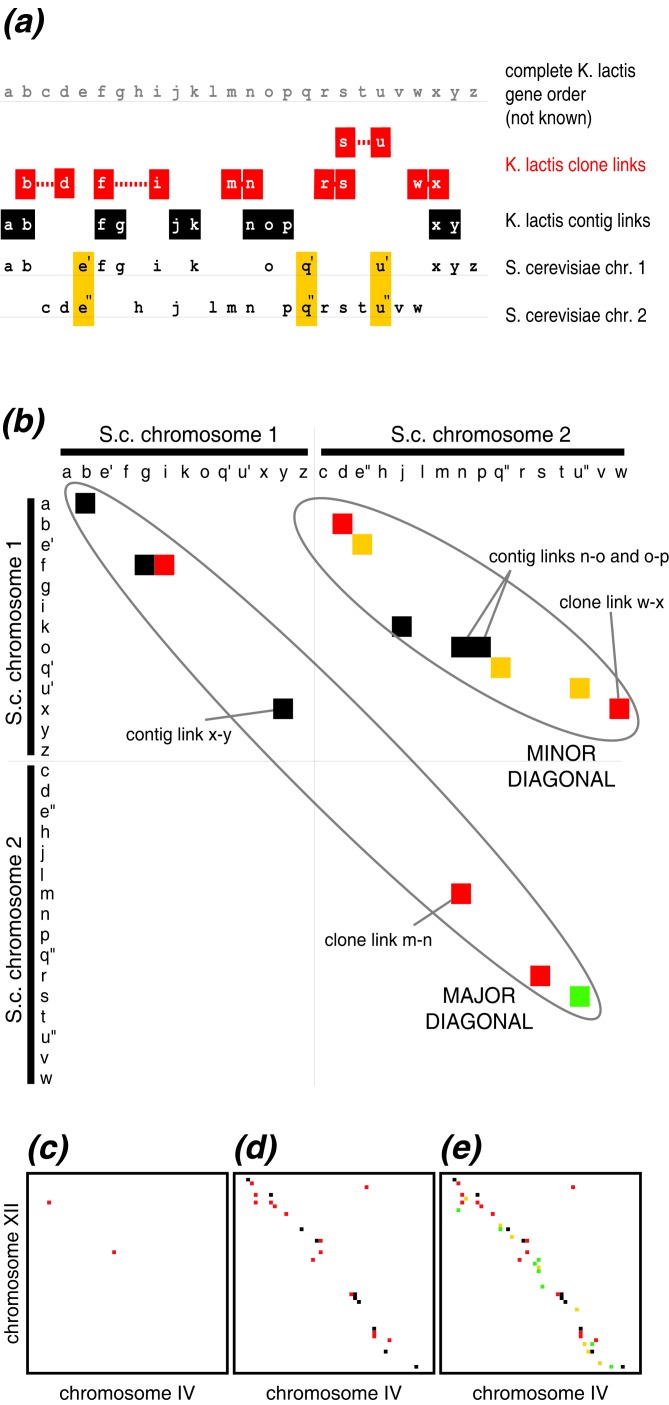
For a particular species (say, Kluyveromyces lactis), the available gene order information is of two types (Fig. 1a). First, a single sequence read, or an assembled sequence contig, may contain two adjacent K. lactis genes. We refer to these genes as contig-linked neighbors. Second, because the Génolevures project sequenced both ends of small plasmid clones, in some cases we know that two genes are a short distance apart on a K. lactis chromosome even where the sequence between them remains unknown and might contain some other genes. We refer to these gene pairs as clone-linked neighbors.
Figure 1.
The proximity plot method. (a) Hypothetical example of fragmentary gene order information from one ascomycete species (K. lactis) and its relationship to the S. cerevisiae genome. Letters a-z represent genes. Gene order information from K. lactis consists of a mixture of contig links (genes that are adjacent in the assembled sequence contigs; black background) and clone links (genes found on opposite ends of a plasmid clone, separated by unsequenced DNA that might contain other genes; red background). Yellow shading indicates duplicated genes in S. cerevisiae. (b) Proximity plot of the clone link (red dots) and contig link (black dots) information from the example in a. The major and minor diagonals are marked by ovals. Yellow dots show duplicated gene pairs in S. cerevisiae, and the green dot shows an ambiguous clone link (the link s–u in K. lactis, where the blast hit between u and u′′ is only marginally stronger than the hit between u and u′). (c) Real example of a minor diagonal between S. cerevisiae chromosomes IV and XII, showing the two pieces of K. lactis clone-link information from Génolevures for a 50-gene section of chromosomes IV and XII. The K. lactis clone links are orthologs of YDR110W-YLR090W and YDR129C-YLR104W. (d) The same minor diagonal, showing gene order information from all 14 species. Red dots are clone links and black dots are contig links. (e) The same minor diagonal, superimposing S. cerevisiae duplicate gene information (yellow dots) and ambiguous clone or contig links (green dots) onto the proximity plot.
The “proximity plots” method was developed to visualize fragmentary gene order data (Fig. 1b). First, we use a matrix that can display any possible gene order arrangement in a yeast species, using the S. cerevisiae genome as a reference. The matrix is depicted as a 5,583 × 5,583 pixel plot, in which the x and y axes both represent gene positions in the S. cerevisiae genome. The position of a gene in S. cerevisiae is simply a number between 1 and 5,583, obtained by numbering the genes sequentially from the left arm of chromosome I thru the right arm of chromosome XVI. On the matrix, dots are plotted to represent gene order information from species other than S. cerevisiae.
For example, if we know that in K. lactis gene A is beside gene B (either as contig-linked neighbors or clone-linked neighbors), a dot is drawn on the plot at the coordinate (A,B). We use different colors to distinguish contig links (black dots) from clone links (red dots). In addition, links involving ambiguous hits appear as green dots (Figs. 1 and 2). To make a proximity plot by using data from K. lactis, all available information about neighboring genes in K. lactis is plotted. It is possible to combine gene order data from multiple source species into a single plot. It is also possible to superimpose the locations of duplicated S. cerevisiae gene pairs onto the proximity plot, even though this is a different type of information (sequence similarity instead of gene order). To do this, duplicated genes within the S. cerevisiae genome were identified by using ssearch (15), and pairs of proteins that are mutual best hits, with E < 1e−5, were plotted as yellow dots (Figs. 1 and 2). The complete proximity plot is too large to reproduce here, but parts of it are shown in Figs. 1–3, and the whole plot can be browsed interactively at wolfe.gen.tcd.ie/prox.
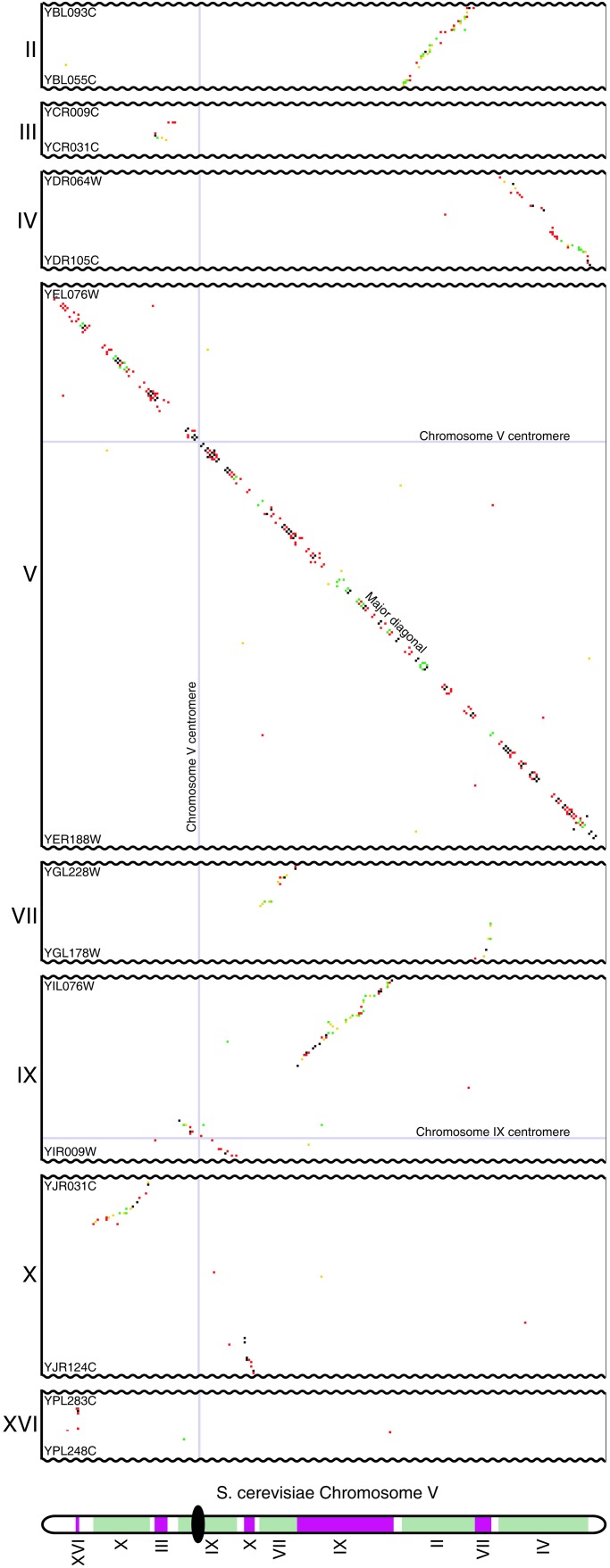
Figure 2.
Sections of the proximity plot involving chromosome V (x axis) compared with other chromosomes (y axis). Gene order data from 14 species, and gene duplication data from S. cerevisiae, are plotted by using the same color scheme as in Fig. 1. Thin gray lines show the locations of centromeres. Parts of chromosomes without significant diagonals are not shown. Genes named on the left indicate the extent of the regions plotted. The bottom diagram shows how the sister regions tile together on chromosome V (colors are arbitrary).
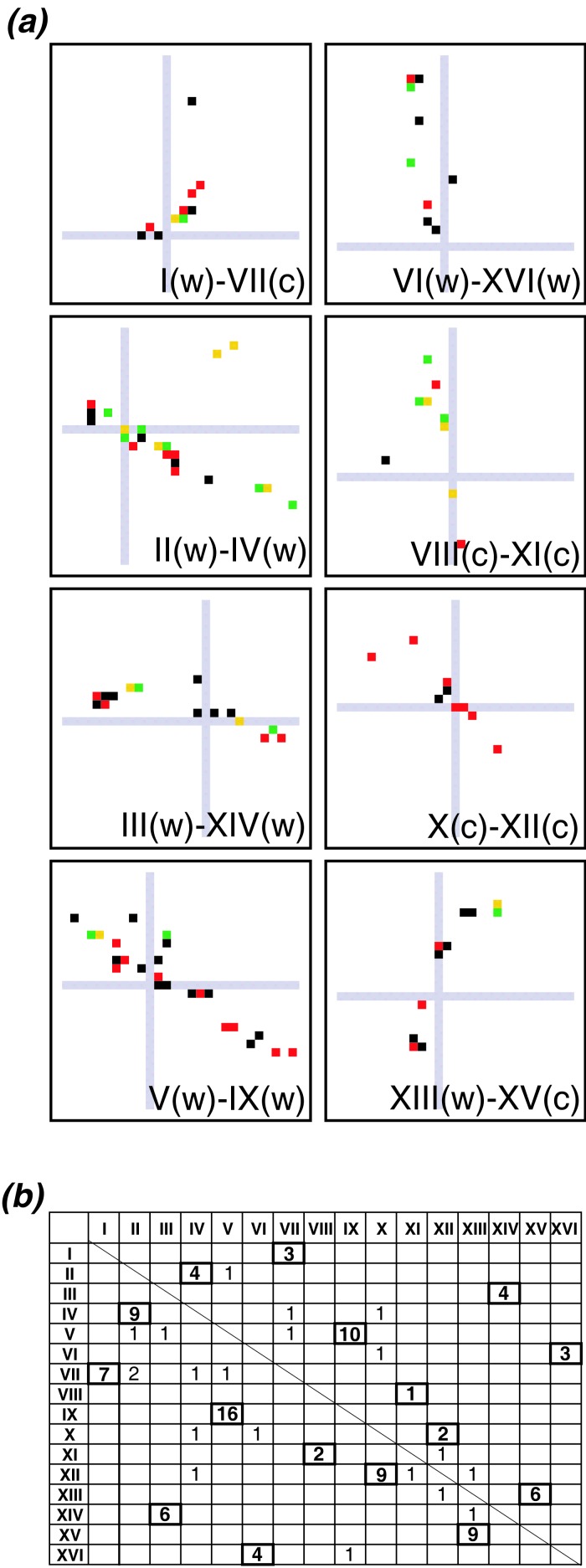
Figure 3.
(a) Proximity plots showing minor diagonals near centromeres. Gray lines indicate the position of centromeres (actually, the first gene on the right arm of each chromosome). The lower-numbered chromosomes are on the x axes. The strand polarity (CDE I → CDE III) of centromeres is indicated by (w) or (c). The field of view is 40 × 40 genes. (b) Numbers of contig links (above diagonal), and total of contig links plus clone links (below diagonal), linking all pairs of centromeres. The 8 proposed centromere pairs are highlighted. Only the first 10 genes left and right of each centromere were considered. Data were pooled from 14 hemiascomycetes, but each link was counted only once, even if it was supported by evidence from more than 1 species.
For phylogenetic analyses, contigs containing the complete ribosomal DNA repeating unit from each species were identified. The contiguous sequence between the 5′ end of the SSU gene and the 3′ end of the LSU gene (5,873 aligned sites) was aligned by using t_coffee (16) and analyzed by the NJ method as implemented in clustalw (17), and by trexml (18).
Candida glabrata TRP1 clones (pT2, pT16) were gifts from Dr. K. Kitada (Nippon Roche Research Center, Kanagawa, Japan; ref. 19). The C. glabrata region between IPP1 and SOK1 was amplified in two segments (IPP1 to HHT2, and HHT2 to SOK1) by long-range PCR. Primers for IPP1 and SOK1 were designed by using codehop (20) from multispecies alignments. Primers for HHT2 were based on the sequence of HHT1 from the TRP1 region. DNA was sequenced commercially by Medigenomix (Martinsreid, Germany) and MWG-Biotech (Ebersberg, Germany) (GenBank accession nos. AF520060 and AF520061).
Results and Discussion
Proximity Plots.
We compared fragmentary information about gene order in 14 hemiascomycete genomes to the S. cerevisiae genome sequence. The “proximity plots” method was devised to visualize the results (see Methods). These plots resemble dot matrix plots, but they display information about gene order instead of sequence similarity. Gene order conservation between S. cerevisiae and another species appear as dots on the major (central) diagonal (Fig. 1 a and b). Dots anywhere else in the plot indicate rearrangements. Several previous studies have reported examples where K. lactis (or Kluyveromyces marxianus) has an “ancestral” gene order consisting of intermingled genes from parts of two different S. cerevisiae chromosomes (2, 4, 11, 21). In a proximity plot, this intermingling creates “minor” diagonals of dots between the two S. cerevisiae chromosomes involved (Fig. 1b). The minor diagonals identify paired regions of the S. cerevisiae genome that are sisters produced by duplication of an original genomic region, which had a gene order similar to what is currently found in K. lactis, followed by extensive gene deletion.
An example of a minor diagonal is shown in Fig. 1 c–e, representing sister regions between parts of chromosomes IV and XII. The proximity plot method allows gene order information from multiple species to be superimposed. When gene order data from only K. lactis is plotted, the minor diagonal is scarcely visible (Fig. 1c). It becomes much clearer when data from all 14 species is plotted simultaneously because many of the species have fragments of ancestral gene order (Fig. 1d). In Fig. 1e, S. cerevisiae duplicate gene pairs (yellow dots) and gene order information involving ambiguous assignments (green dots) have been added. It is noteworthy that this method of comparison is useful only if the genome forming the axes (S. cerevisiae) contains duplicated regions. If the reference genome is not duplicated relative to at least some of the other species, no minor diagonals will be present.
Coverage.
The minor diagonals indicate which parts of the S. cerevisiae genome are sister regions produced by duplication. Many of these correspond to the duplicated chromosomal regions identified in our earlier study (7), but it is important to emphasize that the earlier study used only information about the location of duplicated genes in S. cerevisiae, whereas the proximity plots use only information about gene order in other hemiascomycetes. The two data sources are independent ways of identifying regions of the S. cerevisiae genome that have been produced by duplication. The advantage of the gene order method is that sister regions can be identified even if they do not retain any duplicated genes.
Using proximity plots made from gene order information alone (i.e., red and black dots in Fig. 1; duplicated genes were not used) from the 14 hemiascomycetes, we identified 88 minor diagonals, thereby assigning 176 sections of S. cerevisiae chromosome into sister pairs. These regions cover 3,900 of the 5,583 genes in the genome (70%). There is almost no overlap between the regions, with only 23 genes located within more than one sister. Lack of overlap between sister regions is a key prediction of the polyploidy hypothesis (7).
To maximize the map of sister regions, we superimposed information about duplicated genes onto the proximity plots. Pairs of S. cerevisiae genes that are mutual best hits in Smith-Waterman protein searches are plotted as yellow points (Fig. 1 b and e). We also found that gene order data could be included even for genes whose orthology relationships are uncertain; these are shown as green dots in Fig. 1e. Using these combined plots, almost the entire S. cerevisiae genome could be assigned to 112 pairs of sister regions. Sister regions cover 4,561 genes (82% of the genome), with many of the remaining gaps lying in subtelomeric regions. The largest unpaired section of the genome is only 36 genes long. Only 35 genes are assigned to overlapping regions, and 11 of these are not problematic because they are single genes located at boundaries between one minor diagonal and the next.
The major and minor diagonals involving chromosome V are shown in Fig. 2 as an example of how the sister regions abut one another. In addition to the previously described sister region between the right arm of chromosome V and the left arm of chromosome IX (7), a second large sister region is present at the centromeres of these chromosomes (Fig. 2). This region was not detected previously because it contains only one duplicated gene.
The fact that the sister regions tile together to cover almost the whole genome, and almost without overlap, provides clear support for the hypothesis of paleopolyploidy in S. cerevisiae (7). Each part of the genome has exactly one sister, because the whole genome became duplicated in a single event and subsequently became rearranged into the smaller fragments that we now recognize as sister regions. The model of successive independent duplications of chromosomal regions proposed by Llorente et al. (11) can be largely ruled out because it predicts that significant numbers of overlapping sister regions should be present.
Pairs of Centromeres.
Several of the sister regions span centromeres (Fig. 3a), allowing the following centromere pairs to be identified: I/VII, II/IV, III/XIV, V/IX, X/XII, and XIII/XV. Centromere pairs can also be made between chromosomes VI/XVI and VIII/XI, although in these cases the sister regions end very close to the centromere rather than running across it. In all eight cases, the relative orientation of the two centromeres matches the overall orientation of the paired regions. Thus, the slopes of the minor diagonals for the I/VII and XIII/XV pairs (where one centromere's CDE I—CDE III elements are on the w strand and the other's are on the c strand) are opposite to the slopes for the other six centromere pairs, which are all w/w or c/c (Fig. 3a). The assignment of the centromeres to eight pairs can be verified quantitatively by counting the numbers of dots in proximity plots that are close to pairs of centromeres, for all possible combinations of centromeres (Fig. 3b).
This result confirms and extends previous proposals (5, 22–24) and indicates that the 16 centromeres of S. cerevisiae were produced by duplicating a set of eight centromeres. Because the number of centromeres must equal the number of chromosomes, the S. cerevisiae genome must have evolved from an ancestor with eight chromosomes by duplicating each centromere once. This result could have occurred only through polyploidization.
Genome Status of Each Species.
As well as generating a map of sister regions in the S. cerevisiae genome, the proximity plots of contig links allow the relationship of gene orders between each other species and S. cerevisiae to be quantified. If duplicated genes are ignored, and if gene deletion after polyploidization proceeds randomly on each of the daughter chromosomes, the proximity plot for a species that diverged from S. cerevisiae before the polyploidization is expected to have 50% of its points on the major diagonal and 50% on minor diagonals (Fig. 1b). This result is essentially what we find for K. lactis: after eliminating 26 stray points (points that are on neither diagonal and possibly correspond to gene transpositions, orthology misassignment, or cloning artifacts; ref. 25), 44% of the 161 remaining contig-link points are on the major diagonal, 54% are on the minor diagonal, and the other 2% are “off-major” (Fig. 4a). This third category represents cases where, if the S. cerevisiae gene order is A-B-C, the K. lactis order is A-C, so that gene order is almost conserved and the corresponding points appear slightly off the major diagonal in proximity plots. Clone link information cannot be used to detect off-major points reliably, so only contig links were used for this analysis. Off-major points may be the result of gene transpositions in either species, or complete gene loss from the K. lactis genome.
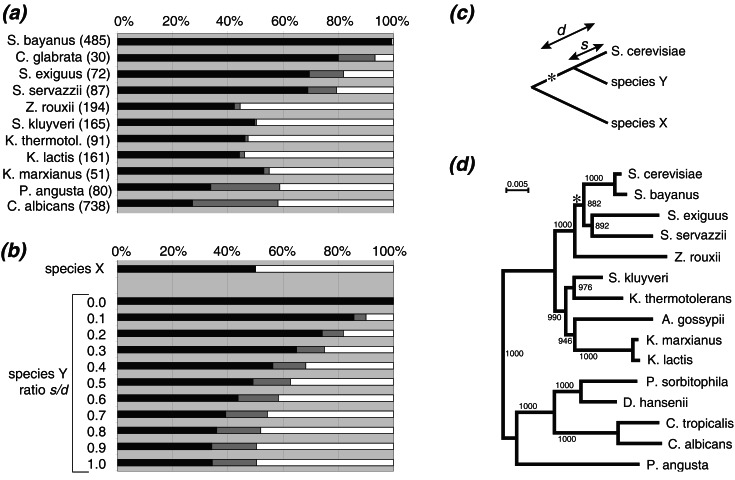
Figure 4.
(a) Distribution of contig-linked points between the major diagonal (black), minor diagonals (white), and in the off-major category (gray). The total number of contig links for each species is shown in parentheses, after excluding stray points that were not near any diagonal. Species that are not shown yielded fewer than 50 non-stray points. Only the 82% of the genome for which minor diagonals have been identified was analyzed, and only unambiguous contig links were used. (b) Theoretical expectations of the distribution of major, minor, and off-major points in species that diverged from S. cerevisiae before the genome duplication (species X) or after it (species Y), as shown in c. The asterisk represents genome duplication. For species Y, “d” represents the duplication date and “s” the speciation date, and the data were produced by computer simulation assuming that genes are deleted at a constant rate. (d) Phylogenetic tree drawn from rDNA sequences (17). The asterisk shows the proposed point of genome duplication. The same topology was obtained for a tree that omitted the bottom five species, and by trexml (18). A. gossypii data are from ref. 27.
The distribution of points between major and minor diagonals is not significantly different from 50:50 in Zygosaccharomyces rouxii, Saccharomyces kluyveri, Kluyveromyces thermotolerans, K. lactis, and K. marxianus (Fig. 4a; P > 0.05 by χ2 test), indicating that these five species diverged from the S. cerevisiae lineage before the polyploidization event. For the more distantly related species C. albicans and Pichia angusta, the number of points on the minor diagonal remains close to 50%, but the number of off-major points increases significantly at the expense of the major diagonal itself. This result is probably due to the accumulation of large numbers of local gene inversions at large evolutionary distances (11, 26). For Saccharomyces bayanus, almost all of the points are on the major diagonal because it diverged from S. cerevisiae long after the polyploidization. The small number of gene order differences between S. bayanus and S. cerevisiae has been analyzed in detail by Fischer et al. (25).
Saccharomyces exiguus, Saccharomyces servazzii, and C. glabrata.
The distributions of points for S. exiguus and S. servazzii are noticeably different from those of the K. lactis group (Fig. 4a), with about 69% of points lying on the major diagonal and a larger number of off-major points (10–13%) than seen in either S. bayanus or the K. lactis group. The increased gene order conservation in these species was also noted by Llorente et al. (11). By computer simulation, we find that their genome arrangements could be explained if those species diverged from the S. cerevisiae lineage after polyploidization, but before the process of gene deletion from sister regions was complete (Fig. 4 b and c). Phylogenetic analysis of ribosomal RNA genes (Fig. 4d) gives a well-resolved tree where S. servazzii and S. exiguus form a monophyletic group more closely related to the S. cerevisiae/S. bayanus pair than to any other species in Génolevures. S. exiguus and S. servazzii are estimated to have 16 and 12 chromosomes, respectively (28), as compared with the basal number of 6–8 chromosomes in K. lactis, Z. rouxii, and S. kluyveri (4, 6). It is therefore possible that the genome duplication occurred at the point marked with an asterisk in Fig. 4d, with a secondary reduction (or possible underestimate; ref. 28) of chromosome numbers in S. servazzii.
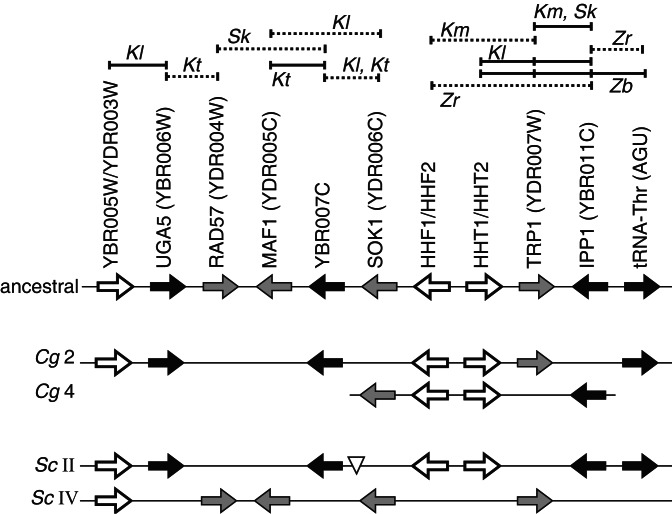
We have previously proposed that the pathogenic yeast C. glabrata may also have diverged from the S. cerevisiae lineage after polyploidization (29). C. glabrata was not included in the Génolevures study, but the small amount of gene order data available for it shows a distribution of points similar to those from S. exiguus and S. servazzii (Fig. 4a). To examine the gene order relationship between C. glabrata and S. cerevisiae in more detail, we completely sequenced two regions of the C. glabrata genome around the TRP1 and IPP1 loci. These loci were chosen because extensive data are already available for them from other species (Fig. 5). In S. cerevisiae, TRP1 and IPP1 are on chromosomes IV and II, respectively, within a pair of sister regions. In species such as K. lactis, genes from these regions are intermingled at a single locus, and a putative ancestral gene order can be inferred (Fig. 5). In C. glabrata, there are two separate genomic regions with gene orders similar to S. cerevisiae chromosomes II and IV. For these loci, and probably for the whole genome, the duplication of an ancestral region and much of the sorting-out of genes onto the two daughter regions clearly occurred before the speciation of C. glabrata and S. cerevisiae.
Figure 5.
Gene organization around the C. glabrata TRP1 and IPP1 genes, in relation to other species. Cg2 and Cg4 are two regions of the C. glabrata genome with similarity to parts of S. cerevisiae (Sc) chromosomes II and IV, which are sisters (block 3 in ref. 7). The extent of sequence determined from C. glabrata is shown by the thin horizontal lines. Shading indicates duplicated genes (white), or homologs of unique genes on S. cerevisiae chromosome II (black) or IV (gray). The ancestral gene order was reconstructed as shown at the top by using contig (solid lines) or clone link (dashed lines) data from Génolevures and GenBank, for K. lactis (Kl), K. marxianus (Km), K. thermotolerans (Kt), S. kluyveri (Sk), Z. rouxii (Zr), and Z. bailii (Zb). The triangle on S. cerevisiae chromosome II shows the position of FLR1, which may have moved onto this chromosome recently (25).
The phylogenetic position of C. glabrata is uncertain; different analyses have placed it at different positions relative to Saccharomyces and Kluyveromyces species, never with strong bootstrap support (4, 30–32). It could not be included in Fig. 4d because its large subunit rRNA gene has not been sequenced. The analysis by Belloch et al. of mitochondrial COX2 sequences placed C. glabrata and S. exiguus closer to the Saccharomyces sensu stricto clade than to the K. lactis/S. kluyveri/K. thermotolerans clade, albeit with low bootstrap support (32). We therefore suggest that C. glabrata, as well as S. exiguus and S. servazzii, is related to S. cerevisiae as shown by the hypothetical species Y in Fig. 4c, whereas K. lactis, K. marxianus, Ashbya gossypii, K. thermotolerans, S. kluyveri, and Z. rouxii hold positions similar to species X. This proposal is largely compatible with the numbers of chromosomes in each species (4, 28). Further comparative genomics studies on C. glabrata, S. exiguus, or S. servazzii will be informative concerning the process of diploidization (33).
Acknowledgments
We thank Dr. K. Kitada for C. glabrata plasmids and C. Kenny for help. Candida albicans sequencing at the Stanford Genome Technology Center was supported by the National Institute of Dental and Craniofacial Research and the Burroughs Wellcome Fund. This study was supported by Science Foundation Ireland.
Footnotes
References
- 1.Altmann-Jöhl R, Philippsen P. Mol Gen Genet. 1996;250:69–80. doi: 10.1007/BF02191826. [DOI] [PubMed] [Google Scholar]
- 2.Ozier-Kalogeropoulos O, Malpertuy A, Boyer J, Tekaia F, Dujon B. Nucleic Acids Res. 1998;26:5511–5524. doi: 10.1093/nar/26.23.5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartung K, Frishman D, Hinnen A, Wolfl S. Yeast. 1998;14:1327–1332. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1327::AID-YEA321>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Keogh R S, Seoighe C, Wolfe K H. Yeast. 1998;14:443–457. doi: 10.1002/(SICI)1097-0061(19980330)14:5<443::AID-YEA243>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.Langkjaer R B, Nielsen M L, Daugaard P R, Liu W, Piskur J. J Mol Biol. 2000;304:271–288. doi: 10.1006/jmbi.2000.4209. [DOI] [PubMed] [Google Scholar]
- 6.Souciet J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, de Montigny J, Dujon B, et al. FEBS Lett. 2000;487:3–12. doi: 10.1016/s0014-5793(00)02272-9. [DOI] [PubMed] [Google Scholar]
- 7.Wolfe K H, Shields D C. Nature (London) 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 8.Seoighe C, Wolfe K H. Proc Natl Acad Sci USA. 1998;95:4447–4452. doi: 10.1073/pnas.95.8.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cliften P F, Hillier L W, Fulton L, Graves T, Miner T, Gish W R, Waterston R H, Johnston M. Genome Res. 2001;11:1175–1186. doi: 10.1101/gr.182901. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann H. FEBS Lett. 2000;487:1–2. doi: 10.1016/s0014-5793(00)02304-8. [DOI] [PubMed] [Google Scholar]
- 11.Llorente B, Malpertuy A, Neuveglise C, de Montigny J, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, et al. FEBS Lett. 2000;487:101–112. doi: 10.1016/s0014-5793(00)02289-4. [DOI] [PubMed] [Google Scholar]
- 12.Llorente B, Durrens P, Malpertuy A, Aigle M, Artiguenave F, Blandin G, Bolotin-Fukuhara M, Bon E, Brottier P, Casaregola S, et al. FEBS Lett. 2000;487:122–133. doi: 10.1016/s0014-5793(00)02291-2. [DOI] [PubMed] [Google Scholar]
- 13.Wood V, Rutherford K M, Ivens A, Rajandream M-A, Barrell B. Comp Funct Genomics. 2001;2:143–154. doi: 10.1002/cfg.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pearson W R, Lipman D J. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Notredame C, Higgins D G, Heringa J. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf M J, Easteal S, Kahn M, McKay B D, Jermiin L S. Bioinformatics. 2000;16:383–394. doi: 10.1093/bioinformatics/16.4.383. [DOI] [PubMed] [Google Scholar]
- 19.Kitada K, Yamaguchi E, Arisawa M. Gene. 1995;165:203–206. doi: 10.1016/0378-1119(95)00552-h. [DOI] [PubMed] [Google Scholar]
- 20.Rose T M, Schultz E R, Henikoff J G, Pietrokovski S, McCallum C M, Henikoff S. Nucleic Acids Res. 1998;26:1628–1635. doi: 10.1093/nar/26.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladrière J M, Georis I, Guerineau M, Vandenhaute J. Gene. 2000;255:83–91. doi: 10.1016/s0378-1119(00)00310-3. [DOI] [PubMed] [Google Scholar]
- 22.Wolfe K H, Lohan A J. Yeast. 1994;10:S41–S46. doi: 10.1002/yea.320100006. [DOI] [PubMed] [Google Scholar]
- 23.Lalo D, Stettler S, Mariotte S, Slonimski P P, Thuriaux P. C R Acad Sci Paris. 1993;316:367–373. [PubMed] [Google Scholar]
- 24.Johnston M, Andrews S, Brinkman R, Cooper J, Ding H, Dover J, Du Z, Favello A, Fulton L, Gattung S, et al. Science. 1994;265:2077–2082. doi: 10.1126/science.8091229. [DOI] [PubMed] [Google Scholar]
- 25.Fischer G, Neuveglise C, Durrens P, Gaillardin C, Dujon B. Genome Res. 2001;11:2009–2019. doi: 10.1101/gr.212701. [DOI] [PubMed] [Google Scholar]
- 26.Seoighe C, Federspiel N, Jones T, Hansen N, Bivolarovic V, Surzycki R, Tamse R, Komp C, Huizar L, Davis R W, et al. Proc Natl Acad Sci USA. 2000;97:14433–14437. doi: 10.1073/pnas.240462997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wendland J, Pohlmann R, Dietrich F, Steiner S, Mohr C, Philippsen P. Curr Genet. 1999;35:618–625. doi: 10.1007/s002940050460. [DOI] [PubMed] [Google Scholar]
- 28.Petersen R F, Nilsson-Tillgren T, Piskur J. Int J Syst Bacteriol. 1999;49:1925–1931. doi: 10.1099/00207713-49-4-1925. [DOI] [PubMed] [Google Scholar]
- 29.Seoighe C, Wolfe K H. Curr Opin Microbiol. 1999;2:548–554. doi: 10.1016/s1369-5274(99)00015-6. [DOI] [PubMed] [Google Scholar]
- 30.Cai J, Roberts I N, Collins M D. Int J Syst Bacteriol. 1996;46:542–549. doi: 10.1099/00207713-46-2-542. [DOI] [PubMed] [Google Scholar]
- 31.Kurtzman C P, Robnett C J. Antonie Van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 32.Belloch C, Querol A, Garcia M D, Barrio E. Int J Syst Evol Microbiol. 2000;50:405–416. doi: 10.1099/00207713-50-1-405. [DOI] [PubMed] [Google Scholar]
- 33.Wolfe K H. Nat Rev Genet. 2001;2:333–341. doi: 10.1038/35072009. [DOI] [PubMed] [Google Scholar]