Abstract
Inbred mouse strains exhibit significant differences in their susceptibility to viruses in the genus Flavivirus, which includes human pathogens such as yellow fever, Dengue, and West Nile virus. A single gene, designated Flv, confers this differential susceptibility and was mapped previously to a region of mouse chromosome 5. A positional cloning strategy was used to identify 22 genes from the Flv gene interval including 10 members of the 2′-5′-oligoadenylate synthetase gene family. One 2′-5′-oligoadenylate synthetase gene, Oas1b, was identified as Flv by correlation between genotype and phenotype in nine mouse strains. Susceptible mouse strains produce a protein lacking 30% of the C-terminal sequence as compared with the resistant counterpart because of the presence of a premature stop codon. The Oas1b gene differs from all the other murine Oas genes by a unique four-amino acid deletion in the P-loop located within the conserved RNA binding domain. Expression of the resistant allele of Oas1b in susceptible embryo fibroblasts resulted in partial inhibition of the replication of a flavivirus but not of an alpha togavirus.
Innate resistance to flavivirus-induced morbidity and mortality was demonstrated in mice in the 1920s (1) and showed monogenic autosomal dominant inheritance (2). The alleles that determined resistance and susceptibility were designated Flvr and Flvs, respectively (3). Resistant mice are susceptible to infections with other types of viruses but are resistant to all flaviviruses (4). Resistant mice can be infected by flaviviruses, but the virus titers in their tissues are lower by 1,000–10,000 times than those in the tissues of susceptible animals, and the spread of the infection in resistant mice is slower (5, 6). Cell cultures derived from many different tissues of resistant mice also produce lower yields of virus; peak titers from resistant cultures are 100–1,000 times lower than those from susceptible cultures (7–9). Previous studies indicate that the Flv gene product acts intracellularly on flavivirus replication.
The flavivirus-resistant allele was demonstrated in wild Mus musculus domesticus populations in both the U.S. and Australia, and flavivirus genetic resistance was reported in other Mus species (10–12). Most commonly used inbred laboratory mouse strains were derived from a small number of progenitors, and the majority of them have a homozygous flavivirus-susceptible genotype. Only the Det, BSVR, BRVR, CASA/Rk, CAST/Ei, MOLD/Rk, and PRI inbred strains have the resistant allele (13). The characteristics of a resistant-like allele (designated Flvr-like) in CASA/Rk and CAST/Ei strains were similar to those of the PRI Flvr allele. The MOLD/Rk animals carry an allele designated minor resistance, Flvmr, that can protect carriers from disease after infection with the attenuated 17D strain of yellow fever virus but not from the virulent Murray Valley encephalitis virus (10).
The resistant allele from donor PRI mice was introduced onto the susceptible C3H/He background to produce the congenic inbred C3H.PRI-Flvr (formerly C3H.RV) strain by a standard backcross protocol followed by brother-sister matings with selection at each generation for the resistance phenotype (12). These congenic strains also carry different alleles of the Ric gene, which controls susceptibility to Rickettsia tsutsugamushi and is located on mouse chromosome 5 (14). These data suggested linkage between the Flv and the Ricloci, and the congenic strains were subsequently used to map the Flv locus on mouse chromosome 5 by linkage with the Ric and rd loci (15). Subsequently, 12 microsatellite markers from mouse chromosome 5 were genotyped relative to the Flv gene in 1,325 backcross animals. Two of the microsatellite markers, D5Mit408 and D5Mit242, exhibited map distances with the Flv locus of 0.30 and 0.15 centimorgans, respectively, whereas one additional marker, D5Mit159, showed no recombination with Flv, indicating linkage of <0.15 centimorgans (16).
To isolate the Flv gene we used a positional cloning strategy. The loci located near the D5Mit159 marker were identified first, and then their sequences were compared in cells from congenic resistant and susceptible mice. We report identification of the Flv gene as mouse 2′-5′-oligoadenylate synthetase 1B (Oas1b).
Materials and Methods
Cell Cultures and Virus Stocks.
Cell lines were established previously by simian virus 40 transformation of embryo fibroblasts obtained from congenic C3H.PRI-Flvr and C3H/He (9). Baby hamster kidney (BHK-21/WI2) cells (referred to hereafter as BHK cells) were used for virus plaque assays (8).
The cDNAs corresponding to the ORFs of the C3H.PRI-Flvr alleles of the Oas1b (GenBank accession no. AF328926) and Na+/Ca2+-exchanger (GenBank accession no. AF261233) genes were cloned separately into the pEF6/V5-His-TOPO mammalian expression vector (Invitrogen), and these plasmids were transfected into susceptible C3H/He cells with Lipofectamine 2000 (Life Technologies, Grand Island, NY). Stable integrants were selected by using blasticidin S, and cells from individual foci were isolated with cloning rings and propagated. Expression of the recombinant proteins, which contained C-terminal 6×His and V5 tags, was analyzed by Western blotting of cell lysates using V5 antibody (Invitrogen). Stable cell lines expressing a low level (1×), intermediate level (8×), or high level (20×) of Oas1b protein were obtained.
A stock of West Nile virus (WNV), strain Eg101, was prepared as a 10% (wt/vol) newborn mouse brain homogenate (titer = 2 × 108 plaque-forming units/ml). A stock of Sindbis virus, strain SAAR 339, was prepared as a 10% (wt/vol) newborn mouse brain homogenate (titer = 7 × 109 plaque-forming units/ml). For virus-growth experiments, confluent monolayers in T25 flasks were infected with WNV or Sindbis virus at a multiplicity of infection of 0.5. Both viruses were titered on BHK cells by plaque assay.
Construction of a Bacterial Artificial Chromosome (BAC) Contig.
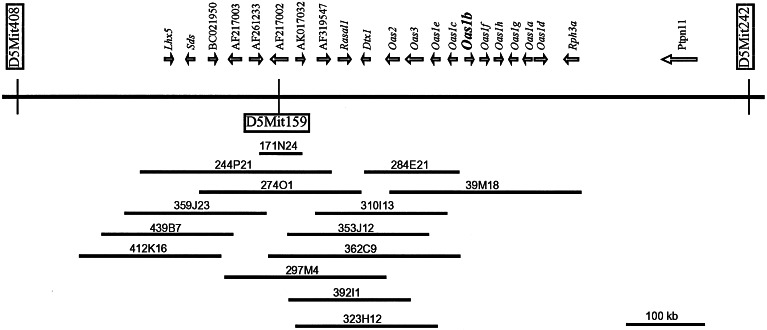
A single genomic clone, 171N24, that contained the D5Mit159 marker and was ≈40 kb in length was isolated from a mouse BAC library (Baylor College of Medicine, Houston) with a unique probe derived from D5Mit159. The terminal sequence obtained from the T7 promoter side of the 171N24 clone did not match any of the DNA sequences in the GenBank database, but the sequence located next to the SP6 promoter was part of a large interspersed repeat. Additional BAC clones were subsequently isolated from the RPCI-23 C57BL/6 mouse BAC library (Roswell Park Cancer Institute, Buffalo, NY) with a probe designed from the 171N24 clone sequence adjacent to the T7 promoter. Four positive signals were detected, and the clones 244P21, 274O1, 297M4, and 359J23 were purchased and analyzed. The size of the insertion in each clone was estimated from restriction patterns observed after pulse-field gel electrophoresis. The terminal DNA sequences for each of the BAC clones were determined and used to design eight new primer pairs for PCR amplification of fragments from each end of each BAC clone. Each BAC clone DNA was tested subsequently as a template in a PCR with each primer pair, and the data obtained were used to align the clones into a single BAC contig of 300 kb.
Two sequences, 297M4T7 and 244P21SP6, that were the most distal in the initial contig were used to rescreen the library and eight additional BAC clones were isolated. These clones were sequenced partially and aligned into the contig by screening them with other BAC-clone terminal sequences. A GenBank search against the 5′ and 3′ BAC insert sequences identified one additional BAC clone, 39M18 (GenBank accession no. AC015535).
Isolation of Transcription Units from the BAC Contig.
Direct cDNA selection, exon trapping, and searches of genes annotated in the Celera database (www.celera.com) were used to identify transcripts from the Flv gene interval. Direct cDNA selection was performed according to the protocol of Lovett (17) with adaptor-ligated double-stranded cDNA prepared from C3H.PRI-Flvr cells. Exon trapping was performed with an Exontrap kit (MoBitec, Gottingen, Germany). The cDNAs obtained after each exon trapping or cDNA-selection experiment were tested by hybridization with the different BAC clone DNAs, and those that showed specific hybridization with the initial BAC clone DNA were cloned into a pCR-XL-TOPO vector (Invitrogen) and sequenced.
The length of the mRNA corresponding to a partial cDNA isolated by cDNA selection or exon trapping was estimated by Northern blotting using the method of Sambrook et al. (18). The partial cDNA sequences were extended by rapid amplification of cDNA ends using a Marathon cDNA amplification kit (CLONTECH). The expression patterns of the candidate genes were analyzed by using mouse multiple-tissue poly(A)+ (Stratagene) and total RNA (Seegene, Seoul, Korea) Northern blots hybridized to probes excised with endonucleases from the cDNAs and labeled with the RTS RadPrime kit (Life Technologies).
Partial and full-length cDNA sequences were used to search the Celera mouse genome database to identify additional transcripts from closely linked genes in the Flv region. Primer pairs designed from sequenced cDNAs and from gene sequences obtained from the Celera database were used to amplify cDNAs from the congenic C3H.PRI-Flvr and C3H/He mouse strains. The primer sequences are listed in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
Amplification and Sequencing of the Oas1b Exons from Genomic DNA.
Genomic DNAs for eight mouse strains, 129/SvJ, BALB/c, BRVR, C57BL/6, CASA/Rk, CAST/Ei, CBA/J, and MOLD/Rk, were purchased from The Jackson Laboratory and used for PCR amplification of Oas1b exons. The primers (see Table 3, which is published as supporting information on the PNAS web site) used for amplification and direct sequencing were designed from the genomic DNA sequence of the Oas1b gene (GenBank accession no. AC015535).
Phylogenetic and Domain Architecture Analysis of Oas Proteins.
Protein sequences of mouse and human 2′-5′-oligoadenylate synthetases 2 and 3 were divided into fragments corresponding to a single functional unit (19). Multiple sequence alignments were constructed by using CLUSTAL X (20). Phylogenetic trees were built from multiple alignments by using the neighbor-joining method (21). The bootstrapping procedure (22) was applied to the PHYLIP format tree output. Known and putative domains in Oas sequences were revealed by searches against Pfam (23) and ProDom (24) databases.
Results
Physical Map of the Flv Gene Interval.
To create a genomic contig, two independent mouse BAC libraries were screened, and 14 BAC clones were isolated. Alignment of these clones provided a BAC contig with an estimated length of more than 700 kb (Fig. 1). Two flanking microsatellite markers, D5Mit408 and D5Mit242, were mapped outside the contig according to the Celera mouse database. The D5Mit159 marker was located in the central part of the contig (Fig. 1).
Figure 1.
Physical and transcript maps of the Flv gene interval. Genes are represented by their accepted abbreviations or GenBank accession numbers of their transcripts. The arrows represent the direction of gene transcription. The centromere is oriented toward the left of the figure. The Oas1b gene is indicated in bold. The flanking microsatellite markers are shown inside vertical rectangles, and the D5Mit159 marker is shown inside a horizontal rectangle. The horizontal bars beneath the genes represent the BAC clones listed by their library name.
Transcript Map of the Flv Gene Region.
Direct cDNA selection and exon-trapping techniques as well as searches of the GenBank and Celera mouse databases were used to identify candidate genes. Four transcripts, a Ca2+ channel (GenBank accession no. AF217002), an unknown mRNA (AF217003), an ATP-dependent helicase (AF319547), and a serine dehydratase (AF328927), were identified by direct cDNA selection. Three transcripts, a Na+/Ca2+ exchanger (AF261233), the Oas2 (AF418010), and the Oas3(AF453830), were detected by exon trapping. The partial sequences obtained were extended to full-length cDNAs by 5′ and 3′ rapid amplification of cDNA ends techniques. Two previously identified genes, Oas1a and Oas1b (AF328926), were cloned by the exon-trapping technique (Fig. 1).
Although small (42-kDa), medium (70-kDa), and large (105-kDa) forms of 2′-5′-oligoadenylate synthetases had been detected previously in mice by various biochemical techniques (25, 26), only the cDNAs of some of the mouse 42-kDa proteins had been cloned previously (27–29). Three DNA sequences of the Oas1a gene were reported in the Mouse Genome Informatics (MGI) database (www.informatics.jax.org) under the accession ID 97429. Two of these sequences, M33863 and X04958, are almost identical to each other and to the Celera transcript, mCT15312, whereas the third sequence, X58077, is similar to the Celera transcript mCT15074, which maps to a different genomic region. We designated this gene Oas1g (see below). The sequence, AF328926, cloned in this study was identical to the previously isolated partial (576-bp) Oas1b sequence, X55982 (28). Two other sequences, M63849 and M63850, deposited in the MGI database under the accession ID 97430 and also designated Oas1b were similar to each other but not to X55982 and AF328926 or to any mouse transcripts, expressed sequence tags, or genomic sequences in neither the NCBI nor the Celera databases. However, M63849 and M63850 showed identity with the human OAS1 sequences, and it is likely that these two sequences were cloned from a mouse cDNA library contaminated with human clones.
Twelve additional genes were identified in the Flvregion by searching the Celera mouse database with sequences from the nine loci detected by cDNA selection and exon trapping. The LIM homeobox 5 (Lhx5), a threonine dehydratase (BC021950), an unknown protein (AK017032), the RAS protein activator-like 1 (Rasal1), the deltex 1 (Dtx1), the Oas1c(AB067528), the rabphilin 3A (Rph3a), and the protein tyrosine phosphatase, nonreceptor type 11 (Ptpn11) sequences were available in GenBank. Genomic and cDNA sequences of four genes annotated by the Celera database (subsequently named Oas1d, Oas1e, Oas1f, and Oas1g, see Table 1) were identified in the Flvregion, and their sequences were used to search the NCBI mouse expressed sequence-tag database. The expressed sequence-tag sequences obtained were used to generate cDNA consensus sequences as well as to design PCR primers for the amplification of each previously uncharacterized gene from mRNA. One additional gene, Oas1h (GenBank accession no. AB067530), not annotated in Celera, was subsequently identified by a BLAST search of the NCBI database with the Oas1b sequence as a query. The Oas1h cDNA sequence was used to search the Celera database, and this gene was mapped on the Flv gene interval between Oas1f and Oas1g (Fig. 1).
Table 1.
Mouse 2′–5′-oligoadenylate synthetase genes and their orthologs
| Mouse gene | GenBank accession no(s). (cDNA clone name) | Celera transcript no(s). | Orthologous sequences (GenBank) |
|---|---|---|---|
| Oas1a | X04958, M33863 (L3), BC013715 | mCT15312 | Rat, Z18877; pig, AJ225090; marmot, AF082498; OAS1*, NM_002534, NM_016816 |
| Oas1b | X55982 (L1), AF328926, AB067529, BC012877, AF418004–AF418009 | mCT15306 | Rat, AF068268OAS1*, NM_002534, NM_016816 |
| Oas1c | AB067528, AF459815 | mCT15073 | OAS1*, NM_002334, NM_016816 |
| Oas1d | AB067532, AY055829 | mCT15317 | OAS1*, NM_002334, NM_016816 |
| Oas1e | AB067531, AY055830, AY055831 | mCT15075 | OAS1*, NM_002334, NM_016816 |
| Oas1f | AF481733 | mCT15304 | OAS1*, NM_002334, NM_016816 |
| Oas1g | X58077 (L2), BC018470 | mCT15074 | OAS1*, NM_002334, NM_016816 |
| Oas1h | AB067530 | None | OAS1*, NM_002334, NM_016816 |
| Oas2 | AB067535, AF418010 | mCT15077 | OAS2*, NM_002535, NM_016817 |
| Oas3 | AB067534, AF453830 | mCT15081 | OAS3*, NM_006187 |
| Oasl1 | AB067533, AY057107 | mCT18390 | OASL*, NM_003733 |
| Oasl2 | AF068835 | mCT118383 | Unknown |
| mCT18449 |
Human genes.
Identification of Candidate Genes.
Full-length cDNAs were amplified by reverse transcription–PCR from congenic flavivirus-resistant (C3H.PRI-Flvr) and -susceptible (C3H/He) mouse strains for each gene identified in the Flv region with the primers listed in Table 2, sequenced, and compared. The sequences of the majority of the genes in the two mouse strains were either identical or very similar (with only a few silent substitutions). In contrast, two genes, Na+/Ca2+ exchanger and Oas1b, were polymorphic and differed by several missense mutations. The Na+/Ca2+-exchanger cDNA from the C3H.PRI-Flvr and C3H/He mouse strains differed by five nonsynonymous substitutions (data not shown). cDNAs for this gene were subsequently sequenced from two additional susceptible (BALB/c and C57BL/6) and one additional resistant (BRVR) mouse strains. A random distribution of substitutions in the Na+/Ca2+-exchanger cDNAs were observed between the five mouse strains studied.
A total of 31 substitutions in Oas1b cDNA was found between the congenic C3H.PRI-Flvr and C3H/He mouse strains. Most of these substitutions were silent, but the C820T transversion in the susceptible C3H/He strain resulted in a premature stop codon. The C3H/He Oas1b gene product therefore lacked 30% of its C-terminal sequence as compared with the C3H.PRI-Flvr product (Fig. 2A). Two additional nonsynonymous mutations resulted in a threonine-to-alanine substitution at position 65 and an arginine-to-glutamine substitution at position 190 in the susceptible C3H/He gene product.
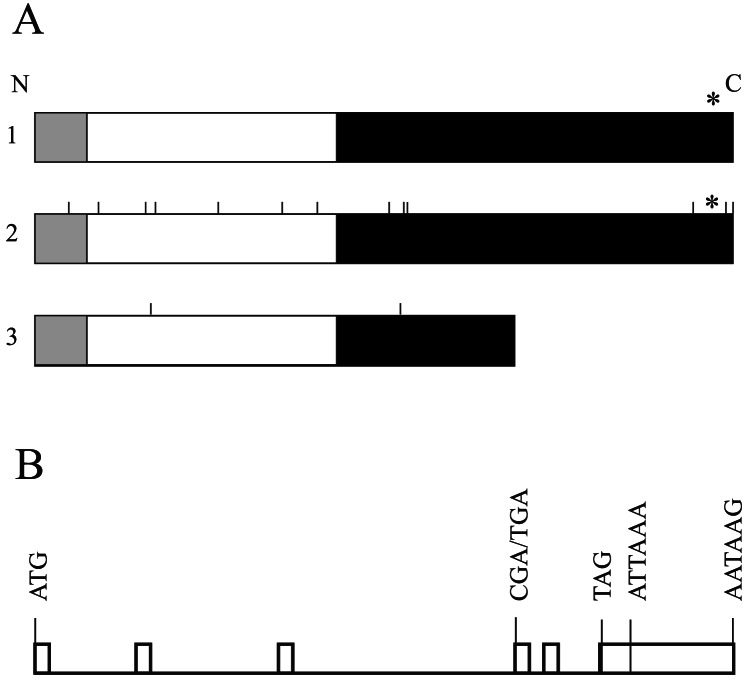
Figure 2.
Structures of the Oas1b gene and protein. (A) Domain architecture of Oas1b proteins. The N-terminal domain (≈30 amino acids, shown in gray) and the C-terminal domain (shown in black) are specific to the Oas protein family (generated with the ProDom tool). The nucleotidyltransferase domain (Pfam 01909) is shown in white. The CFK tetramerization motif is indicated by an asterisk. 1, products of the Flvr and Flvr-like alleles; 2, product of the Flvmr allele; 3, product of the Flvs allele. The positions of amino acid substitutions between the Flvv and Flvmr or the Flvs proteins are shown as vertical bars. (B) Exon-intron structure of the mouse Oas1b gene. Exons are shown as open boxes. The positions of the start (ATG) and stop (TAG) codons, the substitution (CGA/TGA) that results in a premature stop codon, and the two potential polyadenylation sites are indicated by vertical lines.
Comparison of Oas1b genomic (GenBank accession no. AC015535) and cDNA (GenBank accession no. AF328926) sequences revealed six exons. Based on the results of the 5′ rapid amplification of cDNA ends experiments, the size of the first exon was determined to be 243 bp in length and included 64 bp of the 5′ noncoding region (NCR). The lengths of the second, third, fourth, and fifth exons were 277, 185, 233, and 154 bp, respectively. The fourth exon of Oas1b in the susceptible strain contained a premature stop codon (Fig. 2B). All exon-intron boundaries contained conventional splicing sites. In the resistant strain, the sixth exon included the last 102 bp of the ORF and the 3′ NCR, which contained two potential polyadenylation signals separated by ≈2 kb.
The individual exons of the Oas1b genes from eight additional mouse strains were amplified from genomic DNA and sequenced. The Oas1b gene encodes an identical full-length protein in all resistant strains (BRVR, C3H.PRI-Flvr, CASA/Rk, and CAST/Ei), whereas the homologous gene from all susceptible strains (129/SvJ, BALB/c, C3H/He, C57BL/6, and CBA/J) encodes an identical truncated protein. The flavivirus susceptibility phenotype correlated with the Oas1b genotype in all nine mouse strains studied.
The Oas1b protein contains three domains (Fig. 2A). The N- and C-terminal domains are unique to the 2′-5′-oligoadenylate synthetase family, whereas the central domain has a distinct nucleotidyltransferase fold. Several motifs were detected previously in murine 2′-5′-oligoadenylate synthetases (see Fig. 6, which is published as supporting information on the PNAS web site). An N-terminal LXXXP motif is required for 2′-5′-oligoadenylate synthetase activity (30), whereas the P-loop motif is responsible for double-stranded RNA binding (31). It also has been shown that a DAD Mg2+ binding motif is required for normal functioning of the murine 2′-5′-oligoadenylate synthetase (32). Although the LXXXP and DAD motifs were conserved in the products of both the resistant and susceptible alleles of the Oas1b gene, the P-loop motif contained a 4-aa deletion that was not found in the other murine 2′-5′-oligoadenylate synthetases (Fig. 6). A C-terminal CFK motif seems to be critical for tetramerization of the small form of mouse 2′-5′-oligoadenylate synthetase (33). The truncated susceptible Oas1b protein does not contain the CFK motif and thus could not form the tetramer structure required for 2′-5′-oligoadenylate synthetase activity.
Although the Oas1b cDNA sequence from the MOLD/Rk strain (intermediate Flv phenotype) also encodes a full-length protein, it differs from the proteins of the other resistant strains by 14 amino acid substitutions (F26L, S45F, G63C, T65A, S83Y, C103Y, F110C, H118Q, P176L, S183L, I184T, T322A, G347A, and M350T) distributed randomly across the protein (Fig. 2A). The MOLD/Rk Oas1b protein sequence contains alanine at position 65 similar to the proteins encoded by the susceptible strains. The MOLD/Rk Oas1b protein sequence differs by two substitutions, L26F and R206H, from the recently released sequence (GenBank accession no. AAH12877) derived from the CZECH II mouse strain, which has an unknown Flv phenotype. Both MOLD/Rk and CZECH II contain the same 4-aa deletion in the P-loop motif found in all Oas1b proteins.
Constitutive Expression of the Oas1b Gene in Different Mouse Tissues.
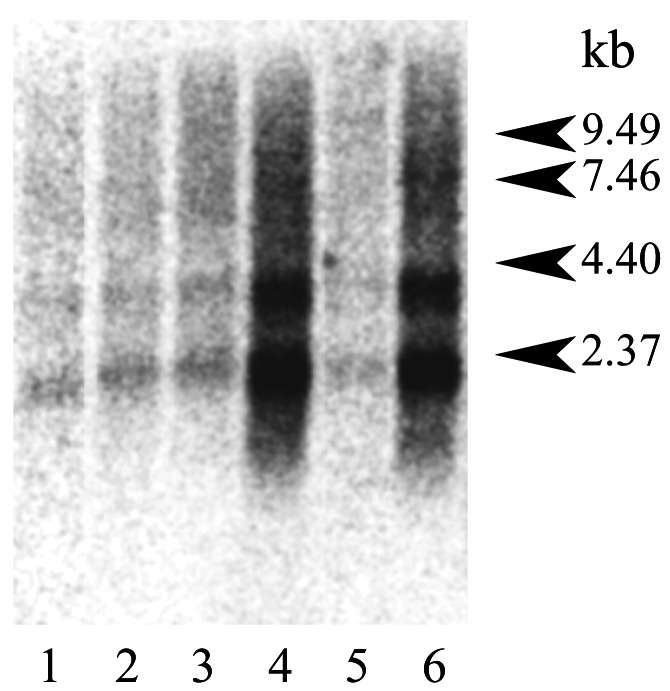
Although α/β-IFN treatment up-regulated the transcription of the murine Oas1b gene (data not shown), constitutive expression of this locus was detected by Northern blotting in all 14 adult murine tissues tested. Two transcripts of the expected sizes, ≈2 and ≈4 kb (Fig. 3), were identified with a labeled cDNA probe derived from the 3′ NCR of Oas1b (cDNA positions 1,384–1,691 bp). The highest levels of constitutive expression were detected in lung and spleen (Fig. 3), thymus, placenta, and uterus (data not shown). Lower levels of expression were observed with brain, stomach, small intestine, skin, testis (data not shown), heart, kidney, liver, and skeletal muscle (Fig. 3).
Figure 3.
Constitutive expression of mouse Oas1b in different mouse tissues. A labeled cDNA derived from the 3′ NCR of Oas1bwas used to probe a BALB/c Northern blot (Stratagene) containing poly(A)+ RNA (2 μg per lane) extracted from: 1, heart; 2, kidney; 3, liver; 4, lung; 5, skeletal muscle; or 6, spleen.
Effect of Expression of the C3H.PRI-Flvr Protein in C3H/He Cells on Flavivirus Replication.
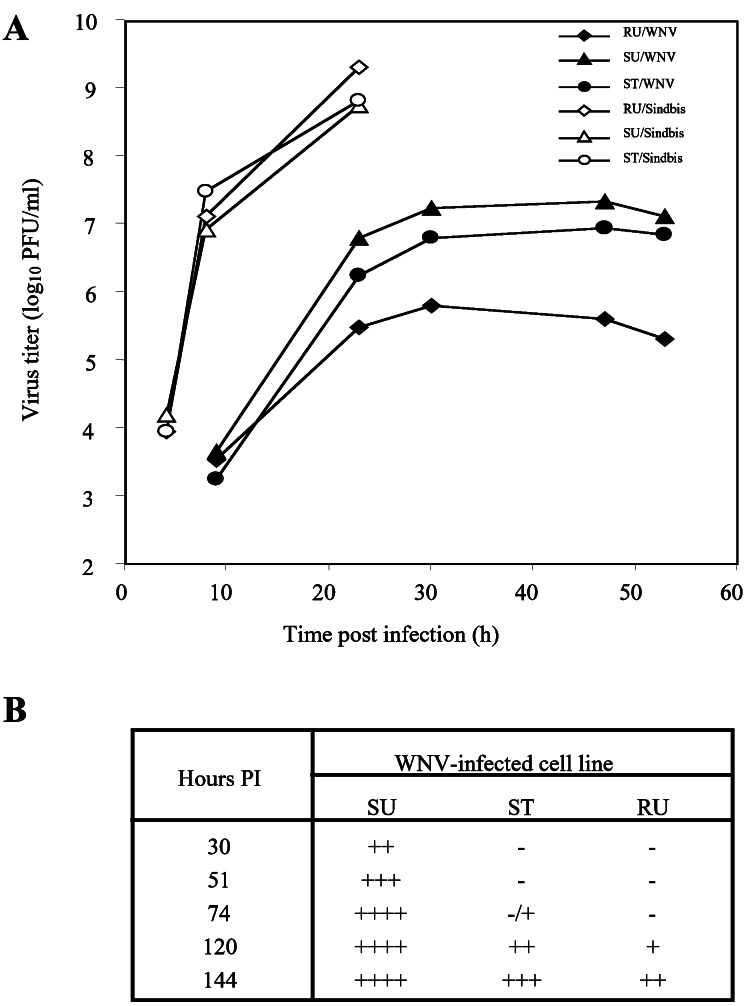
Because the Flvr allele is dominant, its expression in susceptible cells was expected to have a dominant negative effect on flavivirus replication. C3H/He cells were transfected with the mammalian expression vector pEF6/V5-His-TOPO containing either the Oas1b or the Na+/Ca2+-exchanger cDNA from C3H.PRI-Flvr. Stable cell lines were established by selection and cloning of transfected cells. The growth of the flavivirus WNV in susceptible C3H/He cell lines expressing either the Na+/Ca2+ exchanger or the Oas1b protein from resistant C3H.PRI-Flvr was compared with that in untransfected C3H/He and C3H.PRI-Flvr cells. No differences were observed either in the yields of WNV or the time of appearance of cytopathic effect (CPE) between cell lines expressing the Na+/Ca2+-exchanger protein and untransfected C3H/He cells (data not shown). However, in C3H/He cell lines expressing a low level of the resistant Oas1b protein, viral titers were lower than those observed in untransfected cells but not as low as in untransfected C3H.PRI-Flvr cells (Fig. 4A). The appearance of CPE in C3H.PRI-Flvr cells after WNV infection was delayed significantly as compared with that in C3H/He cells. The appearance of CPE was delayed also in C3H/He cells expressing the resistant Oas1b protein (Fig. 4B). In contrast, the growth kinetics and the time of appearance of CPE for an alpha togavirus, Sindbis, were similar in the three types of cells. The recombinant Oas1b protein contained both C-terminal 6×His and V5 tags that may have interfered with the activity of the 2′-5′-oligoadenylate synthetase by reducing the efficiency of tetramer formation. Surprisingly, cell lines expressing intermediate levels (8×) of resistant Oas1b protein showed lower levels of WNV suppression, whereas those expressing high levels (20×) of the protein showed no suppression (data not shown). The reasons for a negative correlation between suppression of WNV replication and the level of resistant protein expressed are not currently understood. C3H/Hc cells expressing an N-terminally tagged resistant Oas1b protein died by 48 h after transfection.
Figure 4.
Effect of the low level expression of the resistant Oas1b protein in C3H/He cells on the growth of a flavivirus, WNV, and an alpha togavirus, Sindbis. (A) Virus growth curves. Cells were infected with either WNV or Sindbis virus at a multiplicity of infection of 0.5. Samples of culture fluid were taken at the indicated times and titered by plaque assay on BHK cells. RU, untransfected resistant C3H.PRI-Flvr cells; SU, untransfected susceptible C3H/He cells; ST, susceptible C3H/He cells stably transfected with Oas1b cDNA from resistant C3H.PRI-Flvr cells. (B) Time course of the development of CPE after infection of SU, RU, and ST cells with WNV. −, no obvious CPE; +, rounding or detachment of ≈25% of the cells in the monolayer; ++, rounding or detachment of ≈50% of the cells in the monolayer; +++, rounding or detachment of ≈75% of the cells in the monolayer; ++++, complete destruction of the monolayer. PI, postinfection.
Phylogenetic Analysis of the 2′-5′-Oligoadenylate Synthetase Family.
The N-terminal 346 amino acids of the OAS1 protein represent one functional unit, whereas OAS2 and OAS3 contain two and three functional units, respectively (19, 34–36). The murine Oas gene sequences obtained previously by different laboratories were named without knowledge of the entire gene family and designated by different symbols. We propose a simplified nomenclature for the murine Oas gene family (Table 1). The single functional unit genes were designated Oas1a through Oas1h, whereas the two- and three-unit genes were designated Oas2 and Oas3, respectively (Fig. 1 and Table 1).
The 2′-5′-oligoadenylate synthetase-like genes, OASL and Oasl2, have been cloned recently from humans (36, 37) and mice (38), respectively. A Celera database search revealed an additional murine gene, Oasl1, which was located close to Oasl2 and was ≈6 Mb upstream of the Oas2 locus. All the human and mouse 2′-5′- oligoadenylate synthetase-like genes contained C-terminal ubiquitin-like domains. Although the Oasl1 gene was mapped outside of the Flvinterval, the cDNA of this gene was cloned and sequenced so that it could be included in the comparative analysis of 2′-5′-oligoadenylate synthetase motifs.
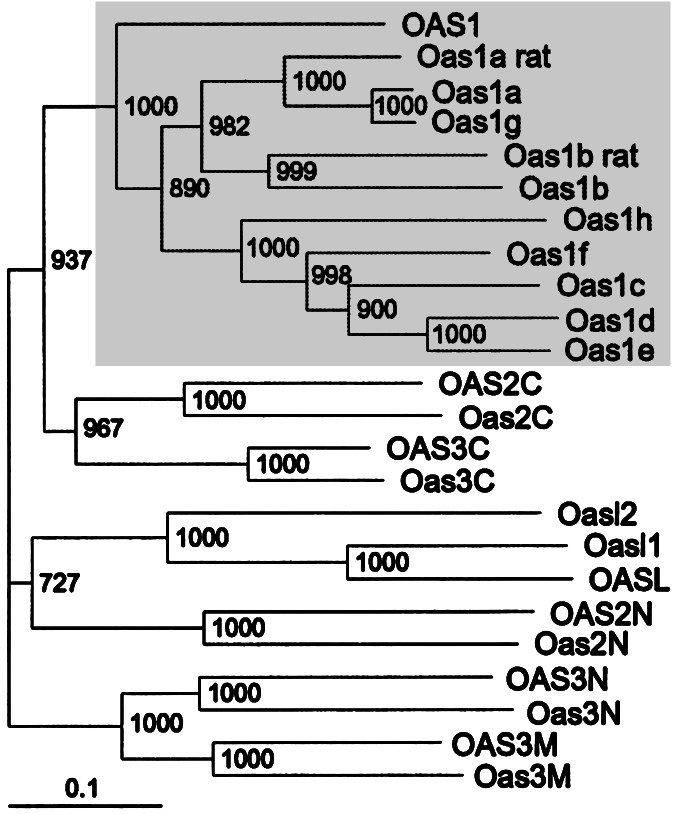
Available protein sequences for murine, rat, and human 2′-5′- oligoadenylate synthetases were aligned, and a phylogenetic tree was constructed (Fig. 5). The known rat proteins, GenBank accession nos. AAC19135 and CAA79317, are encoded by two genes orthologous to murine Oas1b and Oas1a, respectively. It is possible that there are additional Oas1 genes in rats that have not been identified to date. All rodent Oas1 sequences clustered with the single human ortholog, OAS1. This clustering was supported fully by bootstrap analysis. The existence of eight apparent Oas1 paralogs in mice likely resulted from a series of gene duplication events. The one-to-many orthologous relationship between human and murine genes is unique to Oas1 and was not observed for other members of the Oas family (Fig. 5).
Figure 5.
Unrooted neighbor-joining distance-based phylogenic tree of murine, rat, and human Oas protein sequences. Human genes are designated by capital letters, but only the first letter is capitalized for the mouse genes. The sequences of the Oas2 and OAS2 proteins were divided into N- and C-terminal domains according to ref. 19. The sequences of Oas3 and OAS3 proteins were divided into N-terminal (N), middle (M), and C-terminal (C) domains. The indicated bootstrap values were obtained with 1,000 pseudoreplicates. The Oas1 cluster is shown on a gray background. The bar indicates the number of substitutions per site.
Discussion
Twenty-two loci including thirteen previously uncharacterized genes were detected in a region of mouse chromosome 5 during positional cloning of the Flv gene. The D5Mit159 microsatellite sequence used for the initial BAC library screening was detected in the second intron of the Ca2+-channel gene (GenBank accession no. AF217002). By correlation of a polymorphism in the Oas1b gene with the susceptibility phenotypes of nine strains of flavivirus-resistant and susceptible mice, the Flv gene was identified as Oas1b, a member of the 2′-5′-oligoadenylate synthetase gene family. 2′-5′-oligoadenylate synthetases bind double-stranded RNA or particular secondary structures within single-stranded RNA and catalyze the synthesis of 2′-5′-oligoadenylates from ATP (39). A known function of 2′-5′-oligoadenylate is to bind and activate a latent endoribonuclease, RNase L, responsible for the degradation of viral and cellular single-stranded RNAs (40). 2′-5′-oligoadenylate synthetases also are involved in other cellular processes such as apoptosis, cell growth and differentiation, regulation of gene expression, DNA replication, and RNA splicing (19).
Data obtained with the three types of human 2′-5′-oligoadenylate synthetases, OAS1, OAS2, and OAS3, indicate that OAS3 functions as a monomer, whereas OAS2 and OAS1 are enzymatically active only as a homodimer and a homotetramer, respectively (19). The Oas1b genes from resistant mice encode full-length proteins, whereas those from susceptible mice encode C-terminally truncated proteins. Because the C-terminal region of the single-unit proteins is required for tetramerization, which is crucial for 2′-5′- oligoadenylate synthetase activity, it is likely that the Oas1b proteins produced by susceptible mice are not functional synthetases. The OAS1, OAS2, and OAS3 genes are differentially induced by IFN-α, -β, and -γ in various tissues (19). Although the expression of the mouse Oas1b gene was up-regulated after incubation with α/β IFN, it was found to be constitutively expressed at low levels in both resistant and susceptible cells (data not shown). These results are consistent with the previous observation that flavivirus resistance was not diminished in resistant mice after injection of anti-α/β IFN antibody (41).
The effect of the Flv gene product is virus-specific, because it suppresses the replication of the members of the genus Flavivirus but has no effect on the replication of other types of viruses. The functions of 2′-5′-oligoadenylate and the latent endoribonuclease, RNase L, both are nonspecific. The Oas1b proteins from both resistant and susceptible mice differ from other 2′-5′-oligoadenylate synthetases by one unique change, a 4-aa deletion within the P-loop motif. The P-loop region is involved in RNA recognition and binding and may allow the Oas1b protein to specifically recognize and bind a specific conserved RNA structure unique to flavivirus RNAs. In support of this hypothesis, Urosevic et al. (42) reported that the OR156 strain of Murray Valley encephalitis virus, which had a 62-nt deletion in its 3′ NCR (43), replicated more efficiently in resistant mice than did strains of this flavivirus with full-length RNAs. Results from previous sucrose gradient analyses (13) and recent RNase protection experiments (data not shown) indicate that the levels of genomic flavivirus RNA but not antigenomic RNA are reduced preferentially in flavivirus-infected resistant cells as compared with susceptible cells. Also, more flavivirus double-stranded RNA and less viral single-stranded RNA were detected in the brains of resistant mice as compared with those of susceptible animals (44). Because genomic RNA is found free in the cytoplasm, it would be more susceptible to digestion by RNase L than would antigenomic RNA, which is present only in double-stranded replication intermediate RNA structures. Because the Oas1b proteins from both resistant and susceptible mice have the same 4-aa deletion in the P-loop motif, both proteins would be expected to bind specifically to flavivirus RNAs, but RNA binding would only activate the full-length resistant Oas1b protein.
It is not known currently whether the 2′-5′-oligoadenylate synthetase activity alone is sufficient to confer the flavivirus-resistant phenotype or whether as-yet-uncharacterized activities of the Oas1b protein also contribute. Even though the Flvmr proteins also had the same deletion in the P-loop motif, the additional amino acid substitutions in these proteins apparently were responsible for the reduced level of resistance observed in this strain, which suggests that additional regions of the Oas1b protein also may be functionally important for susceptibility.
Inherited flavivirus resistance seems to be restricted to Mus species. Although rats have an Oas1bortholog, the protein produced by this gene (GenBank accession no. AF068268) does not contain a deletion in the P-loop motif, suggesting that it would not function in a flavivirus-specific manner. Humans have a single OAS1 gene that also has not been shown to have a deletion in the P-loop motif. The advantage provided to mice and possibly other rodents by the large number of Oas1 genes is not understood currently.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grants: National Institute of General Medical Sciences Grant GM54896 and National Institute of Allergy and Infectious Diseases, National Institutes of Health Grant AI45135.
Abbreviations
- Flv
flavivirus resistance gene
- Oas
2′-5′-oligoadenylate synthetase
- BHK
baby hamster kidney
- WNV
West Nile virus
- BAC
bacterial artificial chromosome
- NCR
noncoding region
- CPE
cytopathic effect
Footnotes
References
- 1.Webster L T. J Exp Med. 1923;37:231–244. doi: 10.1084/jem.37.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sabin A B. Ann NY Acad Sci. 1952;54:936–944. doi: 10.1111/j.1749-6632.1952.tb39968.x. [DOI] [PubMed] [Google Scholar]
- 3.Green M C. In: Genetic Variants and Strains of the Laboratory Mice. Green MC, editor. Stuttgart: Gustav Fischer; 1981. p. 84. [Google Scholar]
- 4.Shellam G R, Sangster M Y, Urosevic N. Rev Sci Tech. 1998;17:231–248. doi: 10.20506/rst.17.1.1083. [DOI] [PubMed] [Google Scholar]
- 5.Goodman G T, Koprowski H. Proc Natl Acad Sci USA. 1962;48:160–165. doi: 10.1073/pnas.48.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Darnell M B, Koprowski H, Lagerspetz K. J Infect Dis. 1974;129:240–247. doi: 10.1093/infdis/129.3.240. [DOI] [PubMed] [Google Scholar]
- 7.Webster L T, Johnson M S. J Exp Med. 1941;74:489–494. doi: 10.1084/jem.74.5.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vainio T. Ann Med Exp Biol Fenn. 1963;41:1–24. [PubMed] [Google Scholar]
- 9.Darnell M B, Koprowski H. J Infect Dis. 1974;129:248–256. doi: 10.1093/infdis/129.3.248. [DOI] [PubMed] [Google Scholar]
- 10.Sangster M Y, Heliams D B, MacKenzie J S, Shellam G R. J Virol. 1993;67:340–347. doi: 10.1128/jvi.67.1.340-347.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sangster M Y, Mackenzie J S, Shellam G R. Arch Virol. 1998;143:697–715. doi: 10.1007/s007050050324. [DOI] [PubMed] [Google Scholar]
- 12.Groschel D, Koprowski H. Arch Gesamte Virusforsch. 1965;17:379–391. doi: 10.1007/BF01241192. [DOI] [PubMed] [Google Scholar]
- 13.Brinton M A. In: Viral Pathogenesis. Nathanson N, editor. Philadelphia: Lippincott–Raven; 1997. pp. 303–328. [Google Scholar]
- 14.Jerrells T R, Osterman J V. Infect Immun. 1981;31:1014–1022. doi: 10.1128/iai.31.3.1014-1022.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shellam G R, Urosevic N, Sangster M Y, Mansfield J P, Mackenzie J S. Mouse Genome. 1993;91:572–574. [Google Scholar]
- 16.Urosevic N, Mansfield J P, Mackenzie J S, Shellam G R. Arbovirus Res Aust. 1997;7:296–299. [Google Scholar]
- 17.Lovett M. In: Current Protocols in Human Genetics. Seidman J, Seidman C, editors. New York: Wiley Interscience; 1994. pp. 6.3.1–6.3.15. [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 7.37–7.52. [Google Scholar]
- 19.Hovnanian A, Rebouillat D, Mattei M G, Levy E R, Marie I, Monaco A P, Hovanessian A G. Genomics. 1998;52:267–277. doi: 10.1006/geno.1998.5443. [DOI] [PubMed] [Google Scholar]
- 20.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 22.Felsenstein J. Evolution (Lawrence, Kans) 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy S R, Griffiths-Jones S, Howe K L, Marshall M, Sonnhammer E L. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corpet F, Servant F, Gouzy J, Kahn D. Nucleic Acids Res. 2000;28:267–269. doi: 10.1093/nar/28.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dougherty J P, Samanta H, Farrell P J, Lengyel P. J Biol Chem. 1980;255:3813–3816. [PubMed] [Google Scholar]
- 26.St.-Laurent G, Yoshie O, Floyd-Smith G, Samanta H, Sehgal P B, Lengyel P. Cell. 1983;33:95–102. doi: 10.1016/0092-8674(83)90338-0. [DOI] [PubMed] [Google Scholar]
- 27.Ichii Y, Fukunaga R, Shiojiri S, Sokawa Y. Nucleic Acids Res. 1986;14:10117. doi: 10.1093/nar/14.24.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford M N, Kumar A, Nissim A, Chebath J, Williams B R. Nucleic Acids Res. 1991;19:1917–1924. doi: 10.1093/nar/19.8.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh S K, Kusari J, Bandyopadhyay S K, Samanta H, Kumar R, Sen G C. J Biol Chem. 1991;266:15293–15299. [PubMed] [Google Scholar]
- 30.Ghosh A, Desai S Y, Sarkar S N, Ramaraj P, Ghosh S K, Bandyopadhyay S, Sen G C. J Biol Chem. 1997;272:15452–15458. doi: 10.1074/jbc.272.24.15452. [DOI] [PubMed] [Google Scholar]
- 31.Saraste M, Sibbald P R, Wittinghofer A. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 32.Yamamoto Y, Sono D, Sokawa Y. J Interferon Cytokine Res. 2000;20:337–344. doi: 10.1089/107999000312496. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh A, Sarkar S N, Guo W, Bandyopadhyay S, Sen G C. J Biol Chem. 1997;272:33220–33226. doi: 10.1074/jbc.272.52.33220. [DOI] [PubMed] [Google Scholar]
- 34.Benech P, Merlin G, Revel M, Chebath J. Nucleic Acids Res. 1985;13:1267–1281. doi: 10.1093/nar/13.4.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marie I, Hovanessian A G. J Biol Chem. 1992;267:9933–9939. [PubMed] [Google Scholar]
- 36.Rebouillat D, Hovnanian A, Marie I, Hovanessian A G. J Biol Chem. 1999;274:1557–1565. doi: 10.1074/jbc.274.3.1557. [DOI] [PubMed] [Google Scholar]
- 36a.Hartmann R, Olsen H S, Widder S, Jorgensen R, Justesen J. Nucleic Acids Res. 1998;26:4121–4128. doi: 10.1093/nar/26.18.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rebouillat D, Marie I, Hovanessian A G. Eur J Biochem. 1998;257:319–330. doi: 10.1046/j.1432-1327.1998.2570319.x. [DOI] [PubMed] [Google Scholar]
- 38.Tiefenthaler M, Marksteiner R, Neyer S, Koch F, Hofer S, Schuler G, Nussenzweig M, Schneider R, Heufler C. J Immunol. 1999;163:760–765. [PubMed] [Google Scholar]
- 39.Hovanessian A G, Brown R E, Kerr I M. Nature (London) 1977;268:537–540. doi: 10.1038/268537a0. [DOI] [PubMed] [Google Scholar]
- 40.Clemens M J, Williams B R. Cell. 1978;13:565–572. doi: 10.1016/0092-8674(78)90329-x. [DOI] [PubMed] [Google Scholar]
- 41.Brinton M A, Arnheiter H, Haller O. Infect Immun. 1982;36:284–288. doi: 10.1128/iai.36.1.284-288.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Urosevic N, Silvia O J, Sangster M Y, Mansfield J P, Hodgetts S I, Shellam G R. J Gen Virol. 1999;80:897–906. doi: 10.1099/0022-1317-80-4-897. [DOI] [PubMed] [Google Scholar]
- 43.Poidinger M, Hall R A, Mackenzie J S. Virology. 1996;218:417–421. doi: 10.1006/viro.1996.0213. [DOI] [PubMed] [Google Scholar]
- 44.Urosevic N, van Maanen M, Mansfield J P, Mackenzie J S, Shellam G R. J Gen Virol. 1997;78:23–29. doi: 10.1099/0022-1317-78-1-23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.