Abstract
The trp RNA-binding attenuation protein (TRAP) regulates expression of the Bacillus subtilis trpEDCFBA operon by transcription attenuation and translation control mechanisms. Both mechanisms require the binding of tryptophan-activated TRAP to the 11 (G/U)AG-repeat segment in the trp leader transcript. To promote termination, TRAP must bind to the nascent RNA before the antiterminator structure forms. Because only 20 nucleotides separate the TRAP-binding site from the 3′ end of the antiterminator, TRAP has a short time frame to control this regulatory decision. Synchronization of factor binding and/or RNA folding with the RNA polymerase position is a major challenge in all attenuation mechanisms. Because RNA polymerase pausing allows this synchronization in many attenuation mechanisms, we performed experiments in vitro to determine whether pausing participates in the B. subtilis trp attenuation mechanism. We identified two NusA-stimulated pause sites in the trp leader region. Formation of pause hairpins participates in pausing at both positions. The first pause occurred at the nucleotide just preceding the critical overlap between the alternative antiterminator and terminator structures. TRAP binding to transcripts containing preexisting pause complexes releases RNA polymerase, suggesting that pausing provides additional time for TRAP to bind and promote termination. The second pause is downstream from the trp leader termination point, raising the possibility that this pause event participates in the trpE translation control mechanism. NusA also increases the efficiency of termination in the trp leader region and shifts termination one nucleotide upstream. Finally, NusA-stimulated termination is cooperative, suggesting that binding of multiple NusA molecules influences termination.
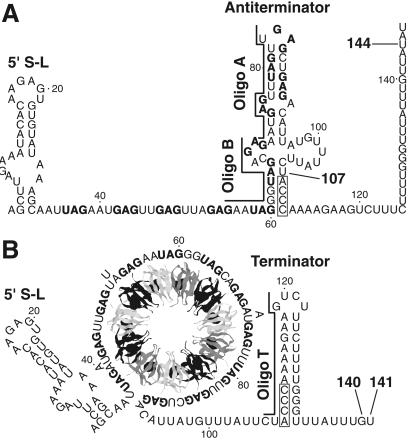
Expression of the Bacillus subtilis tryptophan biosynthetic (trpEDCFBA) operon is negatively regulated in response to tryptophan by the trp RNA-binding attenuation protein (TRAP) (reviewed in refs. 1 and 2). TRAP-mediated regulation of the trp operon includes transcription attenuation and translation control mechanisms. The untranslated trp operon leader transcript contains inverted repeats that allow folding of the transcript to form three RNA secondary structures that participate in the attenuation mechanism. Two of these structures, the antiterminator and terminator, overlap by four nucleotides and therefore are mutually exclusive (Fig. 1). The TRAP binding target in the trp leader transcript contains 11 (G/U)AG repeats, 6 of which are present within the RNA segment that folds into the antiterminator structure (3). When activated by tryptophan, TRAP interacts with the 11-triplet-repeat RNA segment, thereby preventing formation of the antiterminator. As a consequence, the formation of the overlapping terminator promotes transcription termination before RNA polymerase reaches the trp operon structural genes. In the absence of TRAP binding, formation of the antiterminator allows transcription of the entire operon (Fig. 1) (4–6). Because TRAP must bind before the antiterminator forms, the timing of TRAP binding is crucial for this regulatory decision. Lengthening the time frame in which TRAP can bind would increase the ability of active TRAP to promote termination. An additional RNA hairpin that forms at the extreme 5′ end of the B. subtilis trp leader transcript participates in the attenuation mechanism. TRAP interaction with this structure increases the affinity of TRAP for trp leader RNA and reduces the number of (G/U)AG repeats that are necessary for stable association (Fig. 1) (7, 8). Another protein, called anti-TRAP, plays a role in regulating tryptophan biosynthesis. Anti-TRAP antagonizes TRAP activity by competing with TRAP's RNA-binding surface by means of direct protein–protein interaction (9, 10). Because expression of the gene encoding anti-TRAP responds to the accumulation of uncharged tRNATrp, B. subtilis regulates tryptophan biosynthesis by sensing the levels of both tryptophan and uncharged tRNATrp in the cell (9).
Fig 1.
Model of the trpEDCFBA operon attenuation mechanism. During transcription, RNA polymerase pauses at U107. Pausing at this position is greatly stimulated by NusA. (A) In limiting tryptophan conditions, TRAP is not activated and does not bind to the nascent transcript. Eventually RNA polymerase overcomes the pause and resumes transcription. In this case, formation of the antiterminator prevents formation of the terminator, resulting in transcription readthrough into the trp structural genes. (B) In excess tryptophan conditions, TRAP is activated and binds to the (G/U)AG repeats soon after they are synthesized. NusA-stimulated pausing at U107 probably allows additional time for TRAP to bind. TRAP binding releases the paused RNA polymerase complex and transcription resumes. Bound TRAP prevents formation of the antiterminator, thereby allowing formation of the terminator, and hence the termination of transcription at G140 or U141. NusA-stimulated pausing also occurs at U144 in readthrough transcripts. This pausing event is probably involved in the trpE translation control mechanism. The 11 (G/U)AG repeats are indicated in boldface type. Numbering is from the start of transcription. The relative positions of oligonucleotide (oligo) A (nucleotides 70–84), oligo B (nucleotides 55–69), and oligo T (nucleotides 106–120) used in the pause hairpin competition studies are shown.
In addition to regulating trp operon expression by transcription attenuation, TRAP regulates translation of trpE. TRAP binding to trp operon readthrough transcripts promotes formation of an RNA hairpin that sequesters the trpE Shine–Dalgarno sequence, thereby reducing TrpE synthesis by inhibiting ribosome binding (4, 11, 12). Formation of the trpE Shine–Dalgarno blocking hairpin also reduces expression of the second gene in the operon (trpD) through translational coupling and transcriptional polarity (13).
The mechanistically distinct attenuation mechanism of the Escherichia coli trp operon requires coupled transcription and translation. NusA-stimulated RNA polymerase pausing at a specific position in the E. coli trp leader region is crucial in ensuring that coupled transcription and translation takes place (14). We carried out experiments to determine whether RNA polymerase pausing plays a role in the B. subtilis trp operon attenuation mechanism. We found that NusA stimulates the pausing of B. subtilis RNA polymerase at two sites in the trp leader region. The first pause site is at U107, the nucleotide that just precedes the overlap between the alternative antiterminator and terminator structures. The second pause site is at U144. Because U144 is just downstream from the termination sites, it is possible that pausing at this position participates in the translation control mechanism. NusA-stimulated pausing at both U107 and U144 is hairpin-dependent. NusA also stimulates TRAP-mediated termination in the trp leader and shifts the major termination point from U141 to G140.
Materials and Methods
Materials.
Plasmid pPB22 (6) was used as a template for PCR amplification of the region containing the B. subtilis trp promoter and untranslated leader region (−78 to +212 relative to the start of transcription). The PCR fragment was purified and used as a template for in vitro transcription as described (15). The RNA primer ApGpC (corresponding to the first three residues of the natural trp transcript) was obtained from Dharmacon Research, Boulder, CO. The E. coli strain M15[pREP4] (Qiagen) contains plasmid pQE30-NusA encoding N-terminal His-tagged B. subtilis NusA. The NusA protein was overproduced and purified on Ni-NTA spin columns according to the manufacturer's protocol (Qiagen). The B. subtilis σA RNA polymerase was used in these studies. TRAP was purified as described (16).
Single-Round in Vitro Transcription Assay.
Buffer containing 40 mM Tris⋅HCl (pH 8.0), 4 mM MgCl2, 0.1 mM EDTA, 4% trehalose, and 5 mM DTT was used for a two-step in vitro transcription reaction by modifying published procedures (15, 17). In the first step, a stable transcription complex was formed by initiating transcription from the trp promoter for 10 min at 37°C in a reaction containing 20 nM DNA template, 15 μM ApGpC, 8 μM ATP and UTP, 2 μM [α-32P]GTP [1 mCi/ml (1 Ci = 37 GBq)] and 62.5 μg/ml (0.16 μM) B. subtilis σA RNA polymerase. The absence of CTP made initiation primer-dependent and halted elongation after nucleotide 12 of the trp transcript. In the second step, elongation of the halted complexes was resumed at 25°C by the addition of all four NTPs (various concentrations), together with heparin (an inhibitor of reinitiation) and tryptophan in transcription buffer to final concentrations of 100 μg/ml and 1 mM, respectively. For pausing experiments, the final concentration of each NTP was either 150 or 10 μM (see text or appropriate figure legends). NusA, TRAP, and/or DNA oligonucleotides were premixed with the NTPs, heparin, and tryptophan and added together when used. Aliquots of the elongation reaction were removed at various times. The last aliquot was chased for an additional 5 min at 37°C with 0.5 mM each NTP in the transcription buffer. Elongation was stopped by mixing aliquots with an equal volume of gel loading solution (95% formamide/20 mM EDTA, pH 8.0/0.2% SDS/0.3 mg/ml each bromophenol blue and xylene cyanol). Modifications to the single-round in vitro transcription assay are described in the text or the appropriate figure legends. Single-round termination and RNA sequencing reactions were assembled essentially the same way as pausing reactions and incubated for 20 min at 25°C. For the termination assay the final concentration of each NTP was 150 μM. RNA sequencing reactions contained final concentrations of 150 μM ATP, CTP, and UTP, 10 μM GTP, and one of four 3′-dNTPs (TriLink BioTechnologies, San Diego) at the concentration equal to that of the corresponding NTP.
Sample Analysis and Quantification.
Samples were denatured for 4 min at 80°C and fractionated through 5% denaturing gels [19:1 acrylamide to bisacrylamide, 8 M urea in 1× TBE buffer) (TBE buffer = 89 mM Tris/89 mM boric acid/2 mM EDTA, pH 8.3)]. The relative amount of each RNA band was determined by using a PhosphorImager and imagequant software (Molecular Dynamics). Pause half-lives were determined by plotting relative pause band intensities versus time by using kaleidagraph and Microsoft Excel software as described (15). Pausing efficiency was calculated as the fraction of RNA polymerase molecules that pause at a particular site (15). The efficiency of termination was calculated as the fraction of a particular terminated transcript vs. the sum of all terminated and readthrough transcripts. Concentrations of NusA required to give a 50% effect on pausing or termination were calculated by using kaleidagraph software as described (16).
Results
B. subtilis RNA Polymerase Pauses in the trp Leader Region.
A multiround in vitro transcription assay containing B. subtilis σA RNA polymerase was previously used to examine TRAP-mediated transcription attenuation of the trp operon (6, 7, 11). We developed a single-round in vitro transcription assay to determine whether RNA polymerase pausing played a role in this attenuation mechanism. Transcription from the B. subtilis trp promoter was initiated with an ApGpC trinucleotide to produce a stable stalled elongation complex containing RNA that was identical to the RNA sequence of natural trp transcripts. The absence of CTP in the initiation step halted transcription after the incorporation of nucleotide 12 of the trp transcript (Fig. 1). The subsequent addition of CTP and heparin allowed synchronized single-round transcription reactions. The pausing and termination sites were identified with RNA sequencing ladders by performing single-round transcription reactions in the presence of one of four 3′-dNTPs (15).
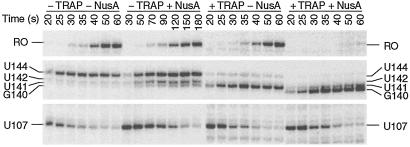
RNA species corresponding to pause complexes initially increase in abundance and subsequently chase to become longer transcripts. Because of this characteristic kinetic pattern, paused species are readily distinguished from terminated transcripts that cannot be extended. When we carried out single-round transcription experiments in the presence of 150 μM of each NTP, we observed termination at U141 and pausing at several positions in the trp leader region, including U107 and U144 (Fig. 2, 150 μM A, C, G, U, −NusA). We also carried out four parallel transcription reactions in which one of the four nucleotides was limited to decrease the rate of transcription and thereby increase the longevity of pause complexes. Pausing was more pronounced when ATP, GTP, or UTP was limiting (10 μM). When ATP was limiting we observed more pronounced pausing at U107 and U144 but not at any other position (Fig. 2, 150 μM C, G, U, 10 μM A, −NusA). When UTP was limited, pausing was observed in several regions in the trp leader that contained consecutive U residues, whereas pausing was observed at U98, U164, and U184 with limiting GTP (data not shown).
Fig 2.
NusA stimulates pausing at U107 and U144 in the trp leader region. In vitro transcription reactions were performed with the NTP concentrations listed at the top. Reactions were carried out in the absence (−) or presence (+) of 1 μM NusA. Transcription reactions were stopped at the times indicated above each lane. Reactions corresponding to “chase” were extended for an additional 5 min in the presence of 500 μM of each NTP. The positions of paused (U107 and U144), terminated (U141 and U142), and run-off (RO) transcripts are shown.
NusA Stimulates Pausing at U107 and U144.
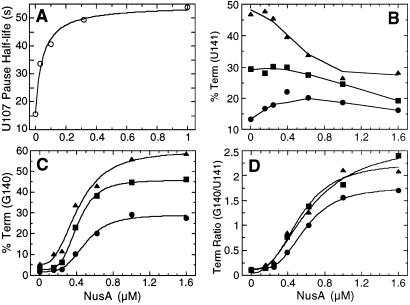
Because the E. coli transcription elongation factor NusA is known to stabilize paused transcription complexes (14, 18, 19), we examined the effect of purified B. subtilis NusA on pausing in the trp leader region of this organism. The addition of NusA to transcription reactions that contained 150 μM of each NTP stimulated pausing at U107 and U144, but not at any other position, suggesting that these pause sites are physiologically important (Fig. 2, 150 μM A, C, G, U, +NusA). These were the same pause complexes that were stabilized when transcription reactions were carried out with 10 μM ATP (Fig. 2, 150 μM C, G, U, 10 μM A, −NusA). Thus, all subsequent pausing experiments were carried out with limiting ATP to facilitate further analysis. The addition of NusA under this condition further stabilized pausing at U107 and U144 (Fig. 2, 150 μM C, G, U, 10 μM A, +NusA). When we carried out transcription reactions with saturating NusA (1 μM), we found that the U107 and U144 pause half-lives increased 4-fold and 5-fold, respectively (Table 1 and Fig. 3, compare −TRAP and −NusA with −TRAP and +NusA). The U107 pause half-life as a function of NusA concentration is shown in Table 1 and Fig. 4A. The concentration of NusA required to give a 50% effect on the U107 pause half-life was calculated to be 44 nM. The hyperbolic curve in Fig. 4A indicates that there is no cooperativity in binding or action of NusA during pausing. This result differs from the cooperative effect of NusA on termination (see below). Whereas TRAP by itself did not affect pausing at U107, TRAP did interfere with NusA-stimulated pausing when both proteins were added to the reaction simultaneously (Table 1 and Fig. 3).
Table 1.
Influence of NusA and TRAP on pausing
| Pause site | NusA, μM | TRAP, μM | Half-life, s |
|---|---|---|---|
| U107 | 0 | 1 | 15 ± 2 |
| U107 | 0 | 0 | 15 ± 1 |
| U107 | 0.032 | 0 | 34 ± 1 |
| U107 | 0.1 | 0 | 41 ± 3 |
| U107 | 0.32 | 0 | 49 ± 1 |
| U107 | 1 | 0 | 54 ± 4 |
| U107 | 1 | 0.1 | 27 ± 3 |
| U107 | 1 | 1 | 14 ± 2 |
| U144 | 0 | 0 | 36 ± 1 |
| U144 | 1 | 0 | 182 ± 23 |
The half-lives of the pause complexes. Values are the averages of at least two independent experiments ± SD.
Fig 3.
TRAP interferes with the NusA-stimulated U107 pause. Transcription reactions were carried out in the absence (−) or presence (+) of 1 μM NusA and/or TRAP. Reactions were stopped at the times indicated above each lane. Note that the time points for the −TRAP +NusA reaction differ from the other reactions. Positions of paused (U107 and U144), terminated (G140, U141 and U142), and run-off (RO) transcripts are shown. The U107 and U144 paused transcripts were completely elongated to terminated or run-off transcripts when chased (not shown on this gel). Only fragments of the gel containing bands of interest are shown.
Fig 4.
The influence of NusA on pause half-lives and termination efficiency. (A) The half-life of U107 pause complexes as a function of NusA concentration. These data are derived from Table 1. (B–D) The effect of NusA and TRAP on termination efficiency at U141 (B), G140 (C), or the G140/U141 termination ratio (D). Reactions were carried out with 0.1 μM (•), 0.2 μM (▪), or 0.5 μM (▴) TRAP and various NusA concentrations.
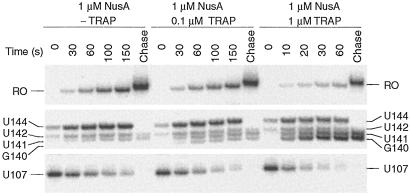
The finding that TRAP interfered with NusA-stimulated U107 pausing indicated that TRAP prevented pausing from occurring and/or TRAP binding disrupted preexisting pause complexes. Pausing efficiency measures the fraction of RNA polymerase molecules that pause at a given site and is distinct from pause half-life that measures the longevity of a pause complex. Quantitative analysis of several gels indicated that there was no significant difference in pausing efficiency at U107 in the absence or presence of TRAP and/or NusA (data not shown). Thus, our results suggested that TRAP binding disrupted preexisting pause complexes. We modified the single-round transcription assay to examine the effect of TRAP binding on preexisting pause complexes. Because our previous results established that the NusA-stimulated U107 pause was most pronounced after approximately 30 s (Fig. 3, −TRAP and +NusA), we examined the effect of TRAP binding on preexisting U107 pause complexes by adding NusA 30 s before the addition of TRAP. We found that TRAP significantly reduced the half-life of preexisting U107 pause complexes (Fig. 5). In the absence of added TRAP the U107 pause half-life was 51 ± 5 s. However, in the presence of 0.1 μM or 1.0 μM TRAP, the pause half-lives were reduced to 39 ± 4 s or 10 ± 1 s, respectively (Fig. 5). This reduction in pause half-life indicates that TRAP binding releases RNA polymerase from preexisting U107 pause complexes in such a way that transcription elongation resumes. In contrast, we found that TRAP did not influence the half-life of preexisting U144 pause complexes (data not shown).
Fig 5.
TRAP binding releases RNA polymerase from NusA-stimulated U107 pause complexes. Pause complexes were allowed to form in the presence of 1 μM NusA for 30 s before the addition of TRAP at the indicated concentration. Reactions were stopped at the times shown at the top of each lane. Note that the time points for the reaction containing 1 μM TRAP differ from the other two reactions. Positions of paused (U107 and U144), terminated (G140, U141, and U142), and run-off (RO) transcripts are shown. Reactions corresponding to “chase” were extended for an additional 5 min in the presence of 500 μM of each NTP. Only fragments of the gel containing bands of interest are shown.
NusA Stimulates Termination in the trp Leader.
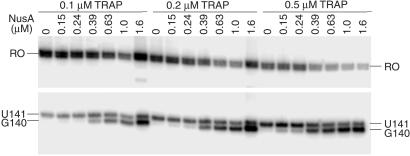
It is well established that TRAP promotes transcription termination in the trp leader region (1, 2); however, the precise point of termination was never firmly established. We found that the addition of TRAP to the single-round transcription assay resulted in increased termination at U141 (Fig. 3, compare −TRAP and −NusA with +TRAP and −NusA). In addition to its effect on pausing, NusA stimulated termination at U141 and U142 (Fig. 3, compare −TRAP and −NusA with −TRAP and +NusA). Whereas termination at U141 is an authentic termination site, it appears that termination at U142 is an artifact of limiting ATP because this species was not observed when reactions were carried out with 150 μM ATP (Fig. 2). Because A143 follows U142, it is likely that the dwell time after U142 incorporation is increased when ATP is limiting. This increased dwell time is probably responsible for this termination product. Moreover, a substantial fraction of the U142 species chased to longer transcripts (Fig. 2). When TRAP and NusA were added simultaneously we observed a further increase in termination efficiency compared with reactions containing only TRAP or NusA (Table 2 and Fig. 3). In addition to the quantitative stimulation of termination, the presence of NusA and TRAP resulted in an additional termination site at G140 (Figs. 3 and 5). As the concentration of TRAP was increased in the presence of 1 μM NusA, the predominate termination point shifted from U141 to G140 (Table 2). The termination point also gradually shifted from U141 to G140 when the concentration of NusA was increased in the presence of 0.1, 0.2, or 0.5 μM TRAP (Figs. 4 and 6). From these data it is apparent that NusA and TRAP acted together in promoting termination at G140. The concentration of NusA required for increased termination was an order of magnitude higher than the concentration that stimulated pausing at U107 (Figs. 4 and 6). The concentration of NusA required to give a 50% effect on increased termination efficiency at G140, as well as the G140/U141 termination ratio, was approximately 500 nM. The sigmoidal shape of the curves shown in Fig. 4 C and D indicates that these NusA-mediated effects were cooperative, suggesting that multiple NusA molecules participate in termination.
Table 2.
Effects of NusA and TRAP on termination
| NusA, μM | TRAP, μM | % Termination | G140/U141 |
|---|---|---|---|
| 0 | 0 | 4.9 ± 0.9 | 0.28 ± 0.02 |
| 0 | 0.1 | 12 ± 2 | 0.26 ± 0.03 |
| 0 | 1 | 64 ± 7 | 0.23 ± 0.07 |
| 1 | 0 | 12 ± 2 | 0.24 ± 0.04 |
| 1 | 0.1 | 43 ± 5 | 1.5 ± 0.2 |
| 1 | 1 | 80 ± 10 | 1.7 ± 0.3 |
Termination efficiency at both G140 and U141. Values are the averages of at least five independent experiments ± SD.
Ratio of termination at G140 to U141. Values are the averages of at least five independent experiments ± SD.
Fig 6.
NusA stimulates termination in the trp leader. The concentrations of NusA and TRAP used in each reaction are shown. The positions of terminated (G140 and U141) and run-off (RO) transcripts are indicated. Only fragments of the gel containing bands of interest are shown.
Pausing at U107 and U144 Is Hairpin-Dependent.
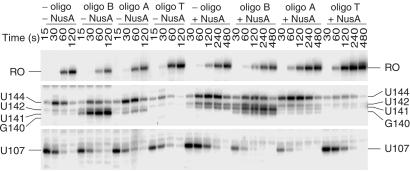
RNA hairpin-dependent and hairpin-independent pause signals have been identified in enteric bacteria. These two classes are distinguished by the 3′ end conformation of the nascent transcript, and by their responses to antisense oligonucleotides and the elongation factors NusA and NusG. NusA increased the pause half-life of hairpin-dependent pauses, whereas NusG decreased the half-life of hairpin-independent pauses (18). The existence of defined alternative secondary structures in B. subtilis trp leader RNA (Fig. 1) and NusA-stimulated pausing (Table 1 and Figs. 2, 3, 4A, and 5) suggested that the observed pause sites would involve a pause hairpin. To test this hypothesis we examined the sensitivity of pausing to antisense oligonucleotides. Two DNA oligonucleotides complementary to the base (nucleotides 55–69, oligo B) or top (nucleotides 70–84, oligo A) of the antiterminator structure (Fig. 1) were used to determine whether a pause hairpin participated in pausing at U107. Whereas neither oligonucleotide affected NusA-independent pausing, we found that both oligonucleotides antagonized the stimulatory effect of NusA by decreasing the U107 pause half-life (Fig. 7). This reduction in the NusA-stimulated pause half-life was similar to that observed for TRAP (Table 1 and Fig. 3). The inhibition of pausing at U107 by these oligonucleotides demonstrates that a hairpin is a component of this pause signal. We also found that both of these oligonucleotides were capable of disrupting preexisting NusA-stimulated U107 pause complexes (data not shown). Because both of these oligonucleotides hybridized to portions of the TRAP-binding target, our results indicate that oligonucleotide hybridization replaced the requirement of TRAP binding for releasing paused RNA polymerase. Neither of these oligonucleotides affected pausing at U144, indicating that pausing at U144 is independent of pausing at U107. Whereas both oligonucleotides A and B reduced the U107 pause half-life, their effect on termination differed. Only disruption of the base of the antiterminator by oligonucleotide B substituted for TRAP in promoting appreciable termination and in shifting termination to G140 in the presence of NusA (Fig. 7). These results indicate that the pause and antiterminator structures are distinct from one another, although it is likely that these two structures share nucleotides in common.
Fig 7.
Effect of antisense oligonucleotides on pausing and termination. Assays contained 0.1 mM oligonucleotide (oligo) B, A, or T complementary to bases 55–69, 70–84, or 106–120 of the trp leader RNA, respectively. Transcription reactions were stopped at the times indicated above each lane. The positions of paused (U107 and U144), terminated (G140, U141, and U142), and run-off (RO) transcripts are shown. Only fragments of the gel containing bands of interest are shown.
Oligonucleotide T, which is complementary to nucleotides 106–120 of the terminator hairpin (Fig. 1), inhibited termination (Fig. 7). As was previously observed (11), oligonucleotide T abolished TRAP-dependent termination (data not shown). We found that oligonucleotide T inhibited pausing at U144; however, instead of decreasing the pause half-life, oligonucleotide T caused a 3-fold reduction in pausing efficiency at this position in the absence or presence of NusA (Fig. 7). These results indicate that a portion of the terminator hairpin functions as a segment of the U144 pause hairpin. Moreover, prevention of NusA-independent pausing by oligonucleotide T indicates that RNA polymerase pausing at this position is hairpin-dependent. Oligonucleotide T did not affect pausing at U107, again indicating that the pause signals for U107 and U144 are independent from one another (Fig. 7).
Discussion
Our results establish that NusA plays a role in the B. subtilis trp operon attenuation mechanism in vitro. NusA stimulates RNA polymerase pausing after the synthesis of U107, the nucleotide just preceding the critical 4-nt overlap between the mutually exclusive antiterminator and terminator structures (Fig. 1). A reasonable explanation for why NusA-stimulated U107 pausing occurs in the B. subtilis trp leader is to ensure that tryptophan-activated TRAP binds to the nascent trp leader transcript before the antiterminator forms. Because 6 of the 11 (G/U)AG repeats that constitute the TRAP binding site are contained within the antiterminator, bound TRAP will prevent antiterminator formation. Once bound, TRAP disrupts the pause complex, resulting in the release of RNA polymerase so that transcription can resume. As a consequence, the terminator hairpin forms and transcription halts in the trp leader region (Fig. 1). Increasing the time frame in which TRAP can bind to the nascent transcript would reduce the concentration of active TRAP necessary to promote termination. The delicate balance between transcription readthrough and termination in the trp leader, and therefore the level of trp operon expression, is further modulated by the amount of anti-TRAP in the cell because this protein antagonizes TRAP's ability to bind RNA (10).
E. coli hairpin-dependent pause signals have been well characterized (18–20). NusA stimulates the basal interaction of pause hairpins with the β subunit of RNA polymerase (20). Our oligonucleotide competition studies indicate that formation of a pause hairpin plays a role in NusA-stimulated U107 pausing in the B. subtilis trp leader region (Fig. 7). We also found that TRAP (Fig. 5) and oligonucleotides A and B (data not shown) are capable of releasing RNA polymerase from preexisting NusA-stimulated U107 pause complexes. Interestingly, we found that neither TRAP nor these two oligonucleotides reduced the U107 pause half-life in the absence of NusA (Figs. 3 and 7). Our results suggest that the basal interaction between the U107 pause hairpin and RNA polymerase is insufficient to promote stable pausing, thus making NusA essential for the function of this pause hairpin. The distance between the base of the E. coli his leader pause hairpin and the 3′ end of the transcript is 11 nt. This distance was found to be important for pausing (19). Whereas we have not systematically examined the nucleotides involved in U107 pause hairpin formation, it appears that this hairpin consists of an RNA segment contained within the top of the antiterminator structure (Fig. 1). Our data also suggest that the U107 pausing mechanism contains hairpin-dependent and hairpin-independent signals. It is possible that pausing at U107 by RNA polymerase is hairpin-independent and that the role of the hairpin is the recruitment of NusA. This type of mechanism was proposed for the pausing event that takes place during transcription of the RNA component of B. subtilis RNase P (21). We found that TRAP was capable of reducing the U107 pause half-life only when NusA was a component of the pause complex. Because it is likely that some of the (G/U)AG repeats are present within the presumed U107 pause hairpin (Fig. 1), it appears that the TRAP and NusA binding sites overlap. Thus, TRAP-mediated melting of the hairpin and/or interaction of TRAP with NusA probably contributes to the release of RNA polymerase. The ability of oligonucleotide A to reduce the U107 pause half-life is consistent with this explanation. However, oligonucleotide B, which is also capable of releasing NusA-stimulated U107 pause complexes, does not hybridize to the hairpin. Apparently, oligonucleotide B is capable of displacing NusA without melting the pause hairpin.
NusA-stimulated termination at intrinsic terminators (22) and termination at residues other than U (17, 23) have been reported for E. coli. The finding that NusA changes the conformation of RNA polymerase at the binding site for the 3′ end of nascent RNA in E. coli (24) might explain why we observed a NusA-dependent shift in termination from U141 to G140. Termination at G140 requires the presence of NusA and disruption of the base of the antiterminator by TRAP (Figs. 3, 5, and 6) or artificially by an oligonucleotide (Fig. 7). Thus, the shift in the main termination point to G140 appears to be caused by TRAP-mediated conformational changes in RNA structure and possibly a NusA-dependent alteration of the conformation of the 3′ end of the transcript. We found that the concentration of NusA required to increase the termination efficiency was an order of magnitude higher than what was required for pause stimulation and that the effect of NusA on termination in the B. subtilis trp leader region is cooperative (Fig. 4). This finding suggests that more than one NusA molecule is present in the termination complex. The previous finding that two molecules of NusA are present in the λ N antitermination complex (25) is consistent with this possibility. Because our data indicate that NusA-stimulated U107 pausing is not cooperative, it is apparent that the roles of NusA in pausing and termination are mechanistically distinct. Because RNA polymerase pausing is a prerequisite for transcription termination (19), cooperative binding of an additional molecule(s) of NusA to an RNA polymerase complex paused at the trp leader terminator hairpin may contribute to the mechanism of transcript release. The mechanism of NusA-mediated pausing and termination is distinct in E. coli as well. In E. coli a substantially higher concentration of NusA is required for termination than for pausing (26). However, in this case, it was concluded that the effects of NusA on pausing and termination resulted from the binding of NusA to different sites on the transcription complex (26).
In addition to the NusA-dependent regulatory mechanisms already described, NusA changes the in vitro folding pathway of the circularly permutated ribozyme from B. subtilis RNase P (27). Thus, as previously noted (14), site-specific pausing of transcribing RNA polymerases is an important event for mechanisms that involve cotranscriptional RNA folding. We identified a NusA-stimulated pause site (U144) just downstream from where transcription terminates in the B. subtilis trp leader region (G140 and U141). TRAP binding to trp operon readthrough transcripts promotes formation of an RNA hairpin that sequesters the trpE Shine–Dalgarno sequence (4, 11, 12). Thus, pausing at U144 is probably involved in this translation control mechanism. Perhaps in the absence of bound TRAP, pausing at U144 is necessary to ensure that the RNA folds into the structure that prevents formation of the trpE Shine–Dalgarno blocking hairpin (4, 11, 12). Our oligonucleotide T competition experiments (Fig. 7) indicate that a portion of the terminator hairpin functions as the U144 pause hairpin. Despite the similarities described above, pausing at U144 appears to be mechanistically distinct from pausing at U107. Oligonucleotide T reduced the pausing efficiency, but not the half-life, in the absence or presence of NusA. Thus, the U144 pause hairpin appears to participate in pausing by RNA polymerase and in NusA recruitment. The general transcription elongation factor NusA is an essential protein in E. coli (28) and B. subtilis (29). This fact underscores the importance of pause-modulating proteins. Whereas the participation of NusA in B. subtilis trp operon regulation has not been demonstrated in vivo, our in vitro results strongly support its involvement.
Acknowledgments
We thank Tina Henkin for providing us with the nusA-overexpressing strain, Michael Chamberlin for RNA polymerase, and both Irina Artsimovitch and Robert Landick for discussions in setting up the single-round transcription assay. We also thank Janell Schaak for assistance with Fig. 1, as well as Robert Switzer, Paul Gollnick, and Janell Schaak for critical reading of the manuscript. This work was supported by Grant GM52840 from the National Institutes of Health.
Abbreviations
TRAP, trp RNA-binding attenuation protein
References
- 1.Babitzke P. & Gollnick, P. (2001) J. Bacteriol. 183, 5795-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gollnick P., Babitzke, P., Merino, E. & Yanofsky, C. (2001) in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, eds. Sonenshein, A. L., Hoch, J. A. & Losick, R. (Am. Soc. Microbiol., Washington, DC), pp. 233–244.
- 3.Babitzke P., Stults, J. T., Shire, S. J. & Yanofsky, C. (1994) J. Biol. Chem. 269, 16597-16604. [PubMed] [Google Scholar]
- 4.Kuroda M. I., Henner, D. & Yanofsky, C. (1988) J. Bacteriol. 170, 3080-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otridge J. & Gollnick, P. (1993) Proc. Natl. Acad. Sci. USA 90, 128-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke P. & Yanofsky, C. (1993) Proc. Natl. Acad. Sci. USA 90, 133-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudershana S., Du, H., Mahalanabis, M. & Babitzke, P. (1999) J. Bacteriol. 181, 5742-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du H., Yakhnin, A. V., Dharmaraj, S. & Babitzke, P. (2000) J. Bacteriol. 182, 1819-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valbuzzi A. & Yanofsky, C. (2001) Science 293, 2057-2059. [DOI] [PubMed] [Google Scholar]
- 10.Valbuzzi A., Gollnick, P., Babitzke, P. & Yanofsky, C. (2002) J. Biol. Chem. 277, 10608-10613. [DOI] [PubMed] [Google Scholar]
- 11.Merino E., Babitzke, P. & Yanofsky, C. (1995) J. Bacteriol. 177, 6362-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du H. & Babitzke, P. (1998) J. Biol. Chem. 273, 20494-20503. [DOI] [PubMed] [Google Scholar]
- 13.Yakhnin H., Babiarz, J. E., Yakhnin, A. V. & Babitzke, P. (2001) J. Bacteriol. 183, 5918-5926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landick R., Turnbough, C. L. & Yanofsky, C. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), pp. 1263–1286.
- 15.Landick R., Wang, D. & Chan, C. L. (1996) Methods Enzymol. 274, 335-353. [DOI] [PubMed] [Google Scholar]
- 16.Yakhnin A. V., Trimble, J. J., Chiaro, C. R. & Babitzke, P. (2000) J. Biol. Chem. 275, 4519-4524. [DOI] [PubMed] [Google Scholar]
- 17.Reynolds R., Bermudes-Cruz, R. M. & Chamberlin, M. J. (1992) J. Mol. Biol. 224, 31-51. [DOI] [PubMed] [Google Scholar]
- 18.Artsimovitch I. & Landick, R. (2000) Proc. Natl. Acad. Sci. USA 97, 7090-7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooney R. A., Artsimovitch, I. & Landick, R. (1998) J. Bacteriol. 180, 3265-3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toulokhonov I., Artsimovitch, I. & Landick, R. (2001) Science 292, 730-733. [DOI] [PubMed] [Google Scholar]
- 21.Artsimovitch I., Svetlov, V., Anthony, L., Burgess, R. R. & Landick, R. (2000) J. Bacteriol. 182, 6027-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmidt M. C. & Chamberlin, M. J. (1987) J. Mol. Biol. 195, 809-818. [DOI] [PubMed] [Google Scholar]
- 23.McDowell J. C., Roberts, J. W., Din, D. J. & Gross, C. (1994) Science 266, 822-825. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y. & Hanna, M. (1994) J. Bacteriol. 176, 1787-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz R. J., Li, J. & Greenblatt, J. (1987) Cell 51, 631-641. [DOI] [PubMed] [Google Scholar]
- 26.Sigmund C. D. & Morgan, E. A. (1988) Biochemistry 27, 5622-5627. [DOI] [PubMed] [Google Scholar]
- 27.Pan T., Artsimovitch, I., Fang, X. W., Landick, R. & Sosnick, T. (1999) Proc. Natl. Acad. Sci. USA 96, 9545-9550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng C. & Friedman, D. (1994) Proc. Natl. Acad. Sci. USA 91, 7543-7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingham C. J., Dennis, J. & Furneaux, P. A. (1999) Mol. Microbiol. 31, 651-663. [DOI] [PubMed] [Google Scholar]