Abstract
Natural selection and genetic linkage cause DNA segments to have genealogical histories resembling those of the selected sites. When a polymorphism maintained by selection is old, it will have an island of enhanced sequence variability surrounding it, which represents a detectable “signature of selection.” We investigate the structure of single-nucleotide polymorphisms (SNPs) in a 20-kb interval containing the Arabidopsis thaliana disease resistance gene RPS5, a locus containing common alleles for the presence/absence of the entire locus. The alleles are considerably diverged at surrounding sites, indicative of an old polymorphism maintained by selection. The island of “enhanced” variability extends several kilobases to either side of the RPS5 deletion junction, and these SNPs are in nearly complete linkage disequilibrium with the RPS5 insertion/deletion. At a distance of 10 kb to either side of the locus, however, we find low levels of polymorphism and the absence of linkage disequilibrium between individual SNPs and RPS5 alleles. Our results show that the interval of enhanced variability surrounding this balanced polymorphism in Arabidopsis is large enough to be readily detected, but small enough to span the focal gene and few others. For this species it should be possible to identify the complete set of genes with long-lived polymorphisms, a potentially important subset of genes segregating for functional variants.
Balanced polymorphisms are mutations maintained in populations by natural selection through heterozygote advantage, frequency-dependent selection, or spatial-temporal selection of alternative alleles. In contrast to strictly advantageous or deleterious mutations, whose persistence times as polymorphisms are generally short, balanced polymorphisms can be maintained indefinitely. They are also more likely to be segregating at intermediate frequencies, where they contribute most to population variance affecting fitness. Thus, there are good reasons to be interested in identifying balanced polymorphisms in a species.
Under favorable circumstances, it is possible to infer the existence of a balanced polymorphism by examining the distribution of single-nucleotide polymorphisms (SNPs) within and between alleles. The magnitude of interallelic divergence of selectively neutral mutations can be related to the age of alleles; statistical tests have been developed to determine whether the age of a polymorphism is unusually large relative to selectively neutral expectations and hence is a candidate for a balanced polymorphism (1–3). This approach does not require prior knowledge of a gene's function, and it is not restricted to coding regions.
Detailed studies of SNP have been conducted in both Drosophila and humans. These studies have not identified many new candidates for balanced polymorphisms, suggesting that this form of selection may contribute relatively little to the standing crop of functional variation within a species (refs. 4–7; for alternative approaches to detecting selection, see ref. 8). However, Drosophila and humans have relatively high recombination rates per adjacent base pair for coding portions of the genome (9). According to theory, the enhancement in neutral polymorphism surrounding a balanced polymorphism in these species will be confined to short regions, perhaps on the order of hundreds of base pairs or less (3, 10). Such short regions of elevated neutral polymorphism will be difficult to detect in population samples, which may explain why few genes exhibit this signature of old polymorphism (5).
Identification of a locus with an old balanced polymorphism will be facilitated when the recombination rate is low, causing the genealogical histories of adjacent SNPs to be more strongly correlated, but the ability to pinpoint the target of selection will also be reduced because larger segments of the genome will be affected. In principle it should be possible to identify a species in which the effective recombination rate is low enough to allow good statistical power to detect long-lived polymorphisms when they are present, but not so low that large segments of a chromosome share the same genealogical history.
Arabidopsis thaliana is largely self-fertilizing and has a patchy distribution of inbred populations (11–13). The effective recombination rate in this species is expected to be low because it depends on relatively rare outcrossed matings between different genotypes. But recombination between polymorphisms separated by 1 kb or less is not uncommon in SNP surveys in Arabidopsis, and the scale of linkage disequilibrium (LD) in this species does not seem to extend beyond tens or hundreds of kilobases (14). From these observations, we conjectured that the recombination structure in this species might be well suited for pinpointing old polymorphisms.
To evaluate this hypothesis, we investigated the variation surrounding RPS5, an R gene containing a polymorphism for disease resistance and susceptibility. The RPS5 locus contains a common polymorphism for the presence and absence of the entire R-gene locus. Functional RPS5+ alleles confer specific recognition of a Pseudomonas syringae strain that expresses the avirulence gene avrPph3 (15). The allele lacking the locus (16) is designated RPS5−. In a previous study, we discovered a balanced polymorphism at RPM1, another R gene in this species with a common polymorphism for the presence/absence of the locus (17). We ask whether RPS5 has a similar signature of selection and, if it does, over what physical distance this signature extends along the DNA from the site of the insertion/deletion polymorphism.
Materials and Methods
Sequence of RPS5 Gene Regions.
Accessions were chosen without prior information about their RPS5 genotypes. DNA fragment and sequence data were obtained from PCR amplification products and dye-terminator cycle-sequencing chemistry (Applied Biosystems). We were unable to amplify and sequence the distal 5′ region (5′-10kb in Fig. 1) in the accession, Bla-2. All DNA sequences have been submitted to the GenBank database (accession nos. AY062364–AY062428). Col-0 sequence (accession no. AC022522) was included in the analyses.
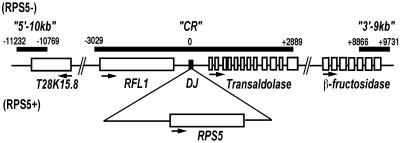
Fig 1.
The structure of the RPS5 gene regions. Open boxes depict location of known and predicted coding regions; filled boxes are regions included in this study. The numbers above solid boxes are nucleotide positions at the ends of the aligned sequences, with DJ for the RPS5 polymorphism set to 0.
Statistical Tests of Polymorphism Levels.
We used CLUSTAL X (18) for multiple alignments with minor manual corrections. Many of the statistical analyses were performed by using DNASP 3.53 (19) and PROSEQ 2.71 (http://helios.bto.ed.ac.uk/evolgen/filatov/proseq.html).
LD.
A permutation test was constructed to evaluate the statistical significance of pairwise LD among SNPs within and between regions. For bi-allelic data, a χ2 test can be used to assess the significance of the associations between any pair of sites. This test allowed us to obtain the number of significant comparisons among possible pairwise comparisons within and between regions (see ref. 20 for details). For testing the significance of this number comparing sites within a region, we randomized alleles in each of two sites and determined the significance of the associations, for all possible comparisons. The probability of obtaining the observed number of significant pairwise association tests or more under the null hypothesis of no association between alleles was obtained from 1 × 106 permutations. For testing the significance of this number comparing sites between regions, we randomized haplotypes in each of two regions. This treatment eliminates the effect of LD within a region, which can bias the number of significant comparisons between regions.
Balancing Selection.
A coalescent model with selection and recombination (3, 10) was used to model neutral variation linked to a site under balancing selection. For each sliding window, we calculated the ratio of average pairwise difference between allelic classes (observed or predicted under balancing selection) to the average pairwise difference expected under the standard infinite sites neutral model. For observed data, according to Kreitman and Hudson (3), this ratio is calculated for each window as the average pairwise difference between RPS5+ and RPS5− allelic classes in the window divided by divergence between species in the window, divided by the species-wide average ratio of polymorphism to divergence (0.08), based on eight genes in Arabidopsis (Adh, Adh upstream region, ChiA, ChiB, FAH1 and F3H, PgiC, CHI; refs. 21–27). For selection predictions, this ratio is the expected coalescence time for two lineages linked to different selected alleles, divided by 2, the expected coalescence time for two random lineages without selection (10), averaged over all sites in the window. The model we used (10) assumes that two alleles at site 0 are maintained by strong selection at a fixed frequency p, with population mutation rate β = 2Neus between selected alleles, and population recombination rate ρ = 2Ner, where Ne is the effective population size, us is the symmetric mutation rate interconverting resistance and susceptibility alleles, and r is the recombination rate per meiosis between adjacent bases. For one curve, an estimate of ρ = 2Ne(1 − s)r (28) = 6 × 10−4 is obtained from an estimate of r obtained from linear regression of six recombinant inbred markers near RPS5 (m488, mi372, mi443, SGCSNP246, ve006, and EG17G9; see www.arabidopsis.org/). Ne was estimated by dividing a genome-wide estimate of θ = 2Neu = 9 × 10−3 obtained as the average for the same eight Arabidopsis genes cited above, by u = 1.5 × 10−8 estimated for Brassicaceae (29), and selfing rate s = 0.996 (30).
Disease Resistance and Susceptibility.
The underside of plant leaves were infiltrated with 15 μl of a 10 mM MgSO4 solution containing 107 colony-forming units (DC3000:avrPph3; ref. 15) by using a blunt-end syringe. Plants were scored for the presence of a hypersensitive response (HR) after 24 h in the greenhouse. This method resulted in an unambiguous HR or disease in all but three accessions. For Pog-0, Tamm-07, and Lip-0, bacterial growth curves showed them to be resistant.
Results
Eleven of the 22 ecotypes constituting our “random” sample (Table 1) were resistant and the other 11 ecotypes were susceptible. As expected, sequencing around RPS5 gene regions revealed that all 11 resistant ecotypes contained the RPS5 locus, whereas 10 of 11 susceptible ecotypes were missing the whole RPS5 coding sequence. A single exceptional line, Tsu-0, contained the RPS5 locus with a frameshift mutation in the coding region; this RPS5 allele is likely to be nonfunctional, thus explaining this line's susceptibility to infection. An additional survey of 69 ecotypes representing worldwide samples yielded RPS5+ frequency of 0.55 (data not shown). We also surveyed 213 lines from 22 North American and European population samples. Nine of the population samples contained both RPS5+ and RPS5− alleles; the average RPS5+ frequency within populations across the 22 samples was 0.42.
Table 1.
Accessions for the RPS5 study
| Accession | Origin | RPS5 | Phenotype |
|---|---|---|---|
| Col-0 | Missouri | + | Resistant |
| Ang-0 | Belgium | + | Resistant |
| Bla-2 | Spain | + | Resistant |
| Bur-0 | Ireland | + | Resistant |
| Ct-1 | Italy | + | Resistant |
| Kz-13 | Kazakhstan | + | Resistant |
| Lip-0 | Poland | + | Resistant |
| Pog-0 | British Columbia, CA | + | Resistant |
| Tamm-07 | Finland | + | Resistant |
| Wu-0 | Germany | + | Resistant |
| Zu-0 | Switzerland | + | Resistant |
| Tsu-0 | Japan | + | Susceptible |
| Ab-27 | Indiana | − | Susceptible |
| Fm-15 | New York | − | Susceptible |
| Hs-12 | Massachusetts | − | Susceptible |
| Kas-1 | India | − | Susceptible |
| Mt-0 | Libya | − | Susceptible |
| Nfc-5 | England | − | Susceptible |
| Up-14 | Michigan | − | Susceptible |
| Rf-4 | Indiana | − | Susceptible |
| Cvi-0 | Cape Verdi Island | − | Susceptible |
| Ler-0 | Germany | − | Susceptible |
+/− indicate the presence/absence of RPS5 coding sequence in their genome, respectively.
To determine whether the polymorphism around the RPS5 deletion junction (DJ) is old, and hence a candidate for a balanced polymorphism, we examined SNP in 5,825 bp (CR) centered on the DJ of RPS5. We also examined SNP in a 966-bp segment located 10 kb upstream of the DJ (5′-10kb) and a 1,137-bp segment located 9 kb downstream of the DJ (3′-9kb). The CR sequence encompasses the complete coding sequence of RFL1 (the 5′ side of the DJ), a distantly related paralog of RPS5, and the partial sequence of the predicted gene encoding transaldolase (the 3′ side of the DJ). The tandem arrangement of RPS5 and RFL1 is present in the sister species, Arabidopsis lyrata, indicating that the RPS5 polymorphism arose as a deletion, just as observed for RPM1 (31). The 5′-10kb and 3′-9kb sequences encompass partial sequence of “T28K15.8, Hypothetical Protein” and β-fructosidase, respectively, as illustrated in Fig. 1.
SNP.
We found 222 segregating sites in 5,825 bp comprising the CR, a high level of SNP (Table 2; Fig. 2). Nucleotide diversity (π) overall is π = 0.017; this estimate increases to π = 0.025 excluding sites where mutations would cause amino acid substitutions; nucleotide diversity further increases to π = 0.033 and 0.036, for synonymous sites only in RFL1 and transaldolase, respectively, Such a high level of SNP is greater than the levels found in other Arabidopsis genes, where nucleotide diversity among ecotype accessions averages around 0.001–0.01 (12, 32). The CR, therefore, qualifies as a region of “enhanced” variability.
Table 2.
Genetic variation and tests of neutrality for the RPS5 gene regions
| Length, bp | n | S | π | Tajima's D | Wall's B | |
|---|---|---|---|---|---|---|
| CR | 5825 | 22 | 222 | 0.0171 | 2.53 | 0.61 |
| Silent | 2943.4 | 22 | 157 | 0.0246 | 2.81 | 0.69 |
| 5′-10kb | 944 | 21 | 20 | 0.0041 | −1.18 | 0.26 |
| Silent | 547.9 | 21 | 13 | 0.0050 | −0.89 | 0.25 |
| 3′-9kb | 1045 | 22 | 21 | 0.0076 | 1.41 | 0.40 |
| Silent | 524.3 | 22 | 13 | 0.0108 | 2.05 | 0.75 |
| RPS5 coding | 2670 | 8 | 7 | 0.0008 | −1.04 | 0.50 |
| Silent | 595.3 | 8 | 1 | 0.0004 | −1.05 | n.a. |
, P < 0.05;
, P < 0.01.
n, number of sequences used.
S, number of segregating sites.
π, the average number of nucleotide differences per site between two sequences with Jukes and Cantor correction (39).
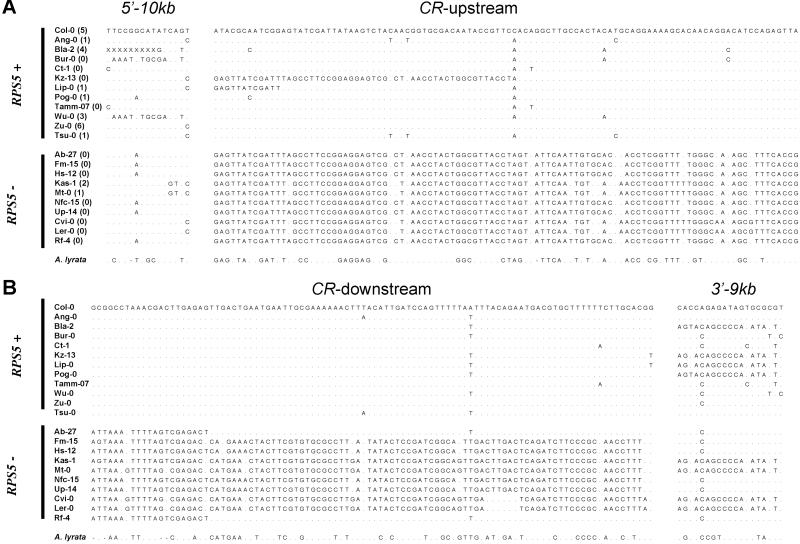
Fig 2.
SNPs within and between RPS5 allelic classes, excluding singletons, and sequence at these positions in A. lyrata. Differences from the reference sequence Col-0 (GenBank accession no. AC022522) are presented. The numbers of singleton polymorphisms are given in parentheses alongside accession names. (A) 5′-10kb and CR (upstream of DJ). (B) CR (downstream of DJ) and 3′-9kb. Some recombinational shuffling is evident within the CR, as indicated by long tracks of SNPs present in the 5′ region of the resistance alleles Kz-3 and Lip-0 that are characteristic of SNPs belonging to the susceptibility allele class, and tracks of SNPs present in the 3′ region of the susceptibility alleles Ab-27 and Rf-4 that carry SNPs characteristic of resistance alleles.
RPS5+ and RPS5− alleles form two distinct clades (Fig. 2 A and B). With the exclusion of four sequences (Kz-13, Lip-0, Ab-27, and Rf-4) that are construed to be recent recombinants between the two haplotypes (as explained in the Fig. 2 legend), 168 of the 220 remaining polymorphisms are between the two classes of alleles. As expected for a balanced polymorphism in this species, only a low level of SNP is segregating among members within each of the two allelic classes (P = 0.0013 and 0.0023 for RPS5+ and RPS5− classes, respectively, excluding the putative recombinant sequences). This configuration of polymorphism, two divergent haplotypes at a similar frequency in our sample, is incompatible with the standard equilibrium neutral model, as evidenced by tests of neutrality using frequency spectral criteria (Tajima's D = 2.46, P < 0.01 and Wall's B = 0.61, P < 0.05; refs. 33 and 34). Strongly positive values of these test statistics are consistent with a selectively maintained polymorphism.
The balancing selection hypothesis allows us to make two predictions about variation segregating further upstream and downstream from the locus. First, the density of SNP, as measured by nucleotide diversity, is expected to decrease as a function of the recombination distance between the site under selection (presumed to be the DJ) and the neutral site (10). Second, the strong association observed between SNPs in the CR and the DJ will also decrease as a function of the recombination distance. These two predictions are not independent, but rather are the joint consequence of recombination decreasing the genealogical correlation of linked DNA segments.
To test these predictions, we surveyed variation in 1-kb stretches 10 kb upstream (5′-10kb) and 9 kb downstream (3′-9kb) on either side of the DJ. We found relatively low levels of variation segregating in the 5′-10kb and 3′-9kb regions (Table 2, π = 0.005 and 0.011, respectively, for silent sites); these values are compatible with genome-wide estimates of polymorphism. Despite the relatively low levels of variability, SNP in 3′-9kb region, like that in the CR, is found on two common haplotypes, and as a result, tests of this frequency spectrum are also not compatible with the neutral equilibrium model (Fig. 2D; Table 2). No evidence, however, exists of association between RPS5 and 3′-9kb haplotypes; the two 3′-9kb haplotypes are equally present on both RPS5+ and RPS5− alleles (Fig. 2D). Whether this nonneutral pattern of polymorphism is a residual effect of the selection acting on RPS5 or whether it is indicative of other independent forces will require additional data and analyses.
LD.
We used permutation tests to investigate the significance of LD between informative SNPs within and between the CR, 5′-10kb, and 3′-9kb regions (see Materials and Methods). As might be expected in a species with a high selfing rate (and therefore a low effective recombination rate), we observed a high proportion of significant pairwise LD within each of the three investigated regions (Table 3, within region). This nonindependence of segregating SNPs within each region reflects the strong haplotype structure already described.
Table 3.
Linkage disequilibrium within and between regions
| Region | Informative sites | n (% significant) | P |
|---|---|---|---|
| Within region | |||
| CR | 201 | 17,245 (85.8) | <0.001 |
| 5′-10kb | 15 | 47 (44.8) | <0.001 |
| 3′-9kb | 18 | 92 (60.1) | <0.001 |
| Between regions | |||
| CR & 5′-10kb | 216 | 142 (4.71) | 0.128 |
| CR & 3′-9kb | 219 | 248 (6.85) | 0.124 |
| 5′-10kb and 3′-9kb | 33 | 60 (22.2) | 0.102 |
The proportion of the significant comparisons among possible pairwise comparisons.
Between the regions, however, we found no evidence for a statistical excess of significant pairwise LD (Table 3, between regions), indicating that the variation present on haplotypes within each region has been recombinationally shuffled between regions. Evidence also exists for some recombination within the CR (Fig. 2).
Our findings in the two flanking regions, a lack of elevated polymorphism and the lack of LD between these regions and the CR, suggest that the genealogical histories of the CR and the flanking regions 10 kb away are effectively decoupled. It also suggests that the target of selection maintaining the two old allelic lineages in the CR region must be at or near the DJ, the center of the region of enhanced variability.
Compatibility with Balancing Selection Model.
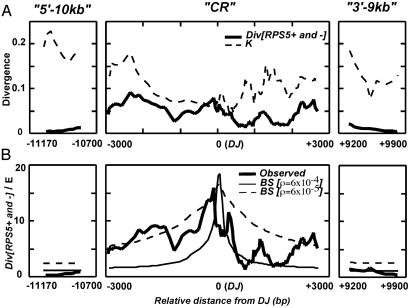
We investigated the compatibility of the data with a model of balancing selection by performing a graphical sliding window analysis of SNP and divergence between A. thaliana and A. lyrata sequences in the three regions (Fig. 3). We restricted our analysis to silent sites in an attempt to reduce the impact of selective constraint against amino acid replacement changes on the estimate of neutral variability. We also chose to compare the nucleotide diversity between RPS5+ and RPS5− alleles only, rather than overall nucleotide diversity, to sharpen specific predictions of the balancing selection model. A sliding window of 250 silent sites (Fig. 3A) shows that, relative to divergence between species, SNP is highest immediately surrounding the DJ and it remains high throughout the CR, which stands in contrast to the distal regions, where the divergence between RPS5+ and RPS5− alleles is considerably reduced relative to the divergence between species.
Fig 3.
Sliding window analysis of silent (synonymous and noncoding) sites in RPS5 flanking regions. Window size is 250 silent sites with a 25-site increment. Abscissa is base position relative to DJ in aligned sequences. (A) Observed nucleotide diversity between RPS5+ and RPS5− alleles and average divergence between A. thaliana and A. lyrata sequences, with Jukes-Cantor correction (39). (B) Results of a coalescent model with selection and recombination (3, 10). For each sliding window, plotted is the ratio of average pairwise difference between allelic classes (observed or predicted under balancing selection) to average pairwise difference expected under the standard neutral model (see Materials and Methods for details). Results for two scaled recombination rates are shown for balancing selection (BS) predictions: ρ = 6 × 10−4, our best estimate of this parameter, and ρ = 6 × 10−5, a 10-fold lower rate. Both expected curves assume an equilibrium frequency of the two allelic classes, P = 0.5; The scaled mutation rate (β) between RPS5+ and RPS5− is fitted for each curve to bring the predictions close to data at the site under selection (assumed to be the DJ).
Modeling a balanced polymorphism allows us to investigate the predicted falloff with distance from the site under selection in interallelic neutral diversity, and to ask whether such a model, when reasonably parameterized for Arabidopsis, is compatible with the SNP data. In other words, are the lower SNP levels seen in the 5′-10kb and 3′-9kb regions compared with the region surrounding the DJ, the presumed site of selection, compatible with balancing selection? Under an equilibrium model of balancing selection, four parameters control the extent of the enhanced neutral variability surrounding a site under selection, the rate of decay of this variability with physical distance, and the decay of LD between SNPs with physical distance. These parameters are the population mutation rate 2Neu, where Ne is the effective population size and u is the mutation rate (per base per generation), the population recombination rate ρ = 2Ner, where r is the recombination rate (per adjacent base pair), the equilibrium allele frequency p of the two selected alleles, and the scaled mutation rate β for the site under selection (i.e., mutations that interconvert resistance and susceptibility alleles).
For any specific combination of parameter values, it is possible to calculate the expected density of SNP at any point in the window scaled to the observed divergence in that window (see Materials and Methods). The scaled mutation rate 2Neu was chosen by averaging estimates of this parameter from previously published studies of SNP. We found that substituting different values of p (0.2 < p < 0.8) has relatively little influence on the shape of the expected distribution of SNP, and so we set this value to p = 0.5, the observed frequency of RPS5+. Rather than choosing arbitrary values of ρ, we estimated r and Ne independently from physical and genetic maps of the RPS5 region of Arabidopsis, and from genomewide measures of SNP, respectively, as described in Materials and Methods. With this estimate in hand, ρ = 6 × 10−4, we then chose a value of the scaled interallelic mutation rate to produce an expected interallelic diversity peak at the DJ similar to that seen in the actual data.
This procedure yields a reasonable fit of the observed and expected divergence between alleles across the CR, although the expected falloff of polymorphism in this region is somewhat faster than indicated by the observed data. At a distance of 9 or 10 kb away from the DJ, the expected interallelic divergence is only slightly elevated (less than 2-fold) above the expected equilibrium neutral levels for unlinked sites, and it shows a good fit to the observed interallelic divergence (Fig. 3B). We also investigated smaller values of ρ to improve the fit of the expected and observed interallelic divergence in the CR. A 10-fold decrease in ρ yielded a satisfactory fit between the observed and expected interallelic divergence in the CR, but this result produced a slightly elevated expectation for the distal regions compared with the observed data. None of the differences between the observed and expected levels of polymorphism (for both values of the population recombination rate) are statistically significant when considering average polymorphism levels across each of the three regions, suggesting a satisfactory fit of the model to the data.
Discussion
The levels and configuration of SNP in the CR region do not fit an equilibrium neutral model in two respects. First, the overall level of SNP surrounding the RPS5 DJ is higher than seen in most other similarly fashioned studies of SNP in A. thaliana. Second, nearly all of the SNP is segregating between the two RPS5 allelic classes. Because RPS5 is presumed to confer an important fitness trait, disease resistance, we believe it is reasonable to hypothesize that the polymorphism is the consequence of natural selection that maintains both resistance and susceptibility alleles.
Investigating only the expected behavior of the balancing selection model prevents us from quantitatively assessing the fit of the model and data, because individual realizations of the balancing selection model for a given set of parameters vary considerably (35). In addition, our assumptions of equal mutation rates between the two allele classes, and uniform recombination rates across the intervals studied may be unrealistic simplifications (36). Rather, this analysis is intended only to investigate whether the apparent decline in variability and LD between the CR and flanking regions is a reasonable approximation of what might be expected under balancing selection in this species. The analysis allows us to answer this question in the affirmative. Despite the fact that LD between SNPs in Arabidopsis ecotypes can extend on the order of 100 kb (14), our present state of knowledge of the genetics and population genetics of the species suggests that enhanced interallelic divergence that is characteristic of a long-lived balanced polymorphism may not have a measurable influence much further than approximately 10 kb on average from the site of selection.
The presence of a deep genealogical split between functional classes of alleles and the near-symmetrical falloff in levels of linked polymorphism on either side of RPS5 is not compatible with models of geographic subdivision or hypermutation. If in the history of A. thaliana, the species was split into isolated subpopulations that diverged into RPS5+ and RPS5− haplotypes, then the divergence between the two alleles would not be expected to decrease symmetrically around the DJ, but rather should extend genome-wide. The data are also not compatible with a model in which the original deletion event was accompanied by hypermutation in neighboring DNA. Under the hypermutation hypothesis, the divergence between A. lyrata and RPS5− alleles should be greater than between A. lyrata and RPS5+ alleles. Instead, we find roughly the same number of mutations on the phylogenetic branches leading to the two alleles relative to the sequence of A. lyrata.
Ten accessions have stop codons in the RFL1-coding region (three of 12 RPS5+ and seven of 10 RPS5−) because of frameshift and point mutations. Stop codons in RFL1 occur in both RPS5+ and RPS5− genotypes, making it unlikely that a dichotomy between potentially functional and nonfunctional RFL1 alleles could be the target of balancing selection.
Modeling selection that yields a stable balanced polymorphism, such as we observe for RPS5, will undoubtedly require inclusion of local population dynamics, with gene flow resulting from migration. The patchy distribution of A. thaliana and the fact that it is a selfer means that local populations will often be genetically differentiated. But local populations are also likely to be ephemeral, and long-range dispersal of the small seeds this species produces (including transport associated with human activity) will tend to homogenize populations across larger geographic scales. Local populations can be found that are segregating for both RPS5+ and RPS5− alleles, and epidemiological models of disease resistance can produce protected polymorphism within populations (17). But between-population dynamics can also be critical for maintaining R-gene polymorphism, and the relative importance of intra- vs. interpopulation selection for disease resistance remains to be investigated.
The evidence presented here for an old polymorphism at RPS5 is nearly identical with our previous finding at RPM1, another R-gene insertion/deletion polymorphism (17). In that study, we presented a frequency-dependent demographic model that assumed a fitness cost of the resistance genotype in the absence of a pathogen. A similar cost may be present for the functional RPS5+ allele. One other study of SNP in an R gene, RPS2 (37), also suggests a selectively maintained polymorphism. Thus, all three R genes, RPS5, RPM1, and RPS2, show relatively deep splits between resistance and susceptibility alleles. Extensive polymorphism may also be present in A. thaliana between members of R genes belonging to tandem arrays, raising the possibility that these are also subject to selection (38). Thus, we can now entertain the hypothesis that the functional variability of R genes as a class may be a general adaptive mechanism to combat pathogens, and that additional studies of SNP in other R genes will find deep genealogical splits between functional alleles.
Our analysis of RPS5 polymorphism shows that mutation and recombination in A. thaliana occur at rates that are near optimal for finding long-lived polymorphisms, simply by searching for islands of enhanced SNP. Specifically, this species exhibits a relatively low base level of SNP, making regions of elevated polymorphism easier to detect. And it has a low enough effective recombination rate to allow correlated histories to extend several but not several tens of kilobases. Because the average distance between genes in A. thaliana is on the order of the length of segments with correlated histories, signatures of balancing selection, when they occur, will cover only one or at most only a handful of genes. Thus, in principle, it should be possible to identify many of the loci in the genome segregating for long-lived polymorphisms, and to identify functionally distinct alleles on the basis of the observed genealogies.
Not only will this knowledge allow partial resolution of one of the oldest problems in population genetics, the number of genes with stable polymorphism maintained by natural selection, but it will also identify allelic variants that must be functionally important. These polymorphisms will constitute a useful set of candidates for possible involvement in complex traits, a subject of great current interest. Whole-genome SNP analysis of species such as Arabidopsis has the potential of yielding valuable insights about evolutionary mechanisms underlying variation in individual fitness, insights that almost certainly will be applicable to other species, including our own.
Acknowledgments
R. Innes kindly provided DC3000:avrPph3. The funding for this work was provided by National Institutes of Health Grant GM57994 (to J.B.).
Abbreviations
SNP, single-nucleotide polymorphism
LD, linkage disequilibrium
DJ, RPS5 deletion junction
CR, central RPS5 region
References
- 1.Hudson R. R., Kreitman, M. & Aguadé, M. (1987) Genetics 116, 153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kreitman M. & Hudson, R. R. (1991) Genetics 127, 565-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudson R. R. (1990) in Oxford Series in Ecology and Evolution, eds. Futuyma, D. & Antonovics, J. (Oxford Univ. Press, Oxford), Vol. 7, pp. 1–44. [Google Scholar]
- 4.Kreitman M. (2000) Annu. Rev. Genomics Hum. Genet. 1, 539-559. [DOI] [PubMed] [Google Scholar]
- 5.Moriyama E. N. & Powell, J. R. (1996) Mol. Biol. Evol. 12, 261-277. [DOI] [PubMed] [Google Scholar]
- 6.Stephens J. C., Schneider, J. A., Tanguay, D. A., Choi, J., Acharya, T., Stanley, S. E., Jaing, R., Messer, C. J., Chew, A., Han, J. H., et al. (2001) Science 293, 489-493. [DOI] [PubMed] [Google Scholar]
- 7.Patil N., Berno, A. J., Hinds, D. A., Barrett, W. A., Doshi, J. M., Hacker, C. R., Kautzer, C. R., Lee, D. H., Marjoribanks, C., McDonough, D. P., et al. (2001) Science 294, 1719-1723. [DOI] [PubMed] [Google Scholar]
- 8.Watt W. B. & Dean, A. M. (2000) Annu. Rev. Genet. 34, 593-622. [DOI] [PubMed] [Google Scholar]
- 9.Przeworski M., Wall, J. D. & Andolfatto, P. (2001) Mol. Biol. Evol 18, 291-298. [DOI] [PubMed] [Google Scholar]
- 10.Hudson R. R. & Kaplan, N. L. (1988) Genetics 120, 831-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abbott R. J. & Gomes, M. F. (1989) Heredity 62, 411-418. [Google Scholar]
- 12.Bergelson J., Stahl, E. A., Dudek, S. & Kreitman, M. (1998) Genetics 148, 1311-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todokoro S., Terauchi, R. & Kawano, S. (1995) Jpn. J. Genet. 70, 543-554. [Google Scholar]
- 14.Nordborg M., Borevitz, J. O., Bergelson, J., Berry, C. C., Chory, J., Hagenblad, J., Kreitman, M., Maloof, J. N., Noyes, T., Oefner, P. J., et al. (2002) Nat. Genet. 30, 190-193. [DOI] [PubMed] [Google Scholar]
- 15.Simonich M. T. & Innes, R. W. (1995) Mol. Plant–Microbe Interact. 8, 637-640. [DOI] [PubMed] [Google Scholar]
- 16.Warren R. F., Henk, P., Mowery, E., Holub, E. & Innes, R. W. (1998) Plant Cell 10, 1439-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stahl E. A., Dwyer, G., Mauricio, R., Kreitman, M. & Bergelson, J. (1999) Nature (London) 400, 667-671. [DOI] [PubMed] [Google Scholar]
- 18.Thompson J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. (1997) Nucleic Acids Res. 25, 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozas J. & Rozas, R. (1999) Bioinformatics 15, 174-175. [DOI] [PubMed] [Google Scholar]
- 20.Hudson R. R. (2001) in Handbook of Statistical Genetics, eds. Balding, D. J., Bishop, M. & Cannings, C. (Wiley, New York), pp. 309–324.
- 21.Savolainen O., Langley, C. H., Lazzaro, B. P. & Fréville, H. (2000) Mol. Biol. Evol. 17, 645-655. [DOI] [PubMed] [Google Scholar]
- 22.Miyashita N. T. (2001) Mol. Biol. Evol. 18, 164-171. [DOI] [PubMed] [Google Scholar]
- 23.Kawabe A., Innan, H., Terauchi, R. & Miyashita, N. T. (1997) Mol. Biol. Evol. 14, 1303-1315. [DOI] [PubMed] [Google Scholar]
- 24.Kawabe A. & Miyashita, N. T. (1999) Genetics 153, 1445-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aguadé M. (2001) Mol. Biol. Evol. 18, 1-9. [DOI] [PubMed] [Google Scholar]
- 26.Kawabe A., Yamane, K. & Miyashita, N. T. (2000) Genetics 156, 1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuittinen H. & Aguadé, M. (2000) Genetics 155, 863-872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordborg M. (2000) Genetics 154, 923-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch M. A., Haubold, B. & Mitchell-Olds, T. (2000) Mol. Biol. Evol. 17, 1483-1498. [DOI] [PubMed] [Google Scholar]
- 30.Bergelson J., Purrington, C. B. & Wichmann, G. (1998) Nature (London) 395, 25. [DOI] [PubMed] [Google Scholar]
- 31.Grant M. R., McDowell, J. M., Sharpes, A. G., deTorres, Z. M., Lydiate, D. J. & Dangl, J. L. (1998) Proc. Natl. Acad. Sci. USA 95, 15843-15848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashita N. T., Kawabe, A. & Innan, H. (1999) Genetics 152, 1723-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima F. (1989) Genetics 123, 585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wall J. D. (1999) Genet. Res. 74, 65-79. [Google Scholar]
- 35.Donnelly P., Nordborg, M. & Joyce, P. (2001) Genetics 159, 853-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao H., Zhou, Q., Li, J., Smith, H., Yandeau, M., Nikolau, B. J. & Schnable, P. S. (2002) Proc. Natl. Acad. Sci. USA 99, 6157-6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caicedo A. L., Schaal, B. A. & Kunkel, B. N. (1999) Proc. Natl. Acad. Sci. USA 96, 302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergelson J., Kreitman, M., Stahl, E. A. & Tian, D. (2001) Science 292, 2281-2285. [DOI] [PubMed] [Google Scholar]
- 39.Jukes T. H. & Cantor, C. R. (1969) in Mammalian Protein Metabolism, ed. Munro, H. N. (Academic, New York), pp. 21–132.