Abstract
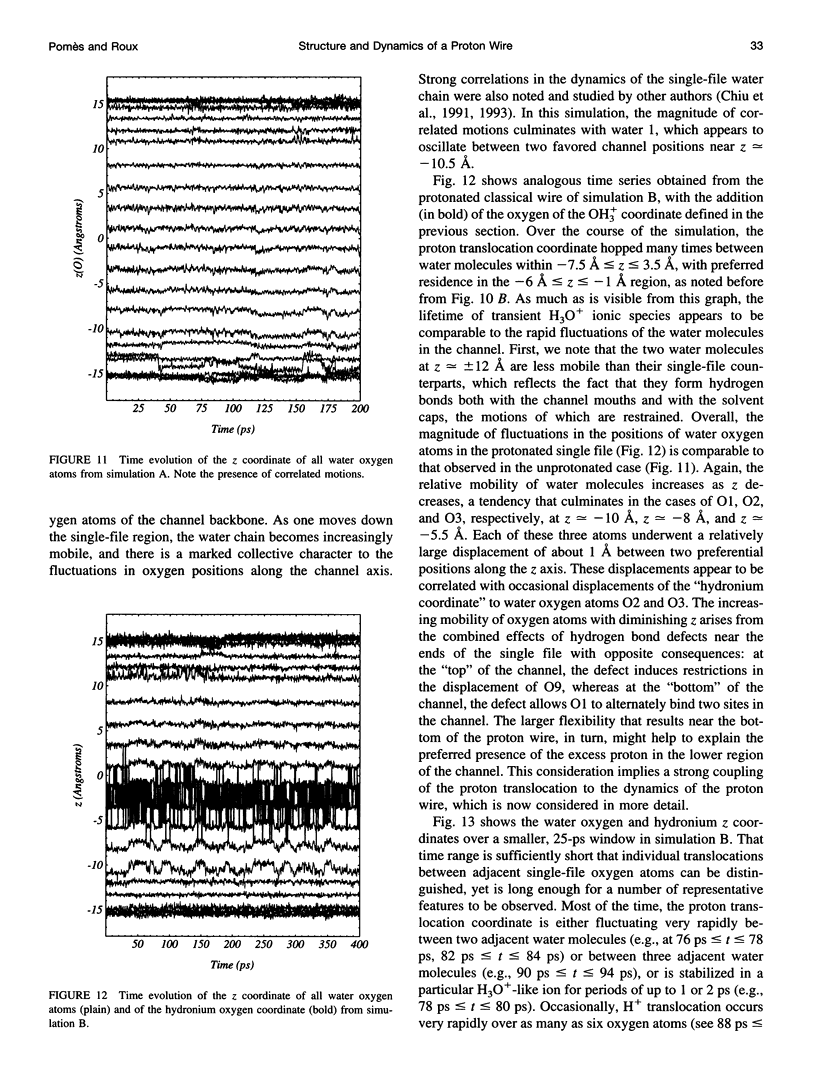
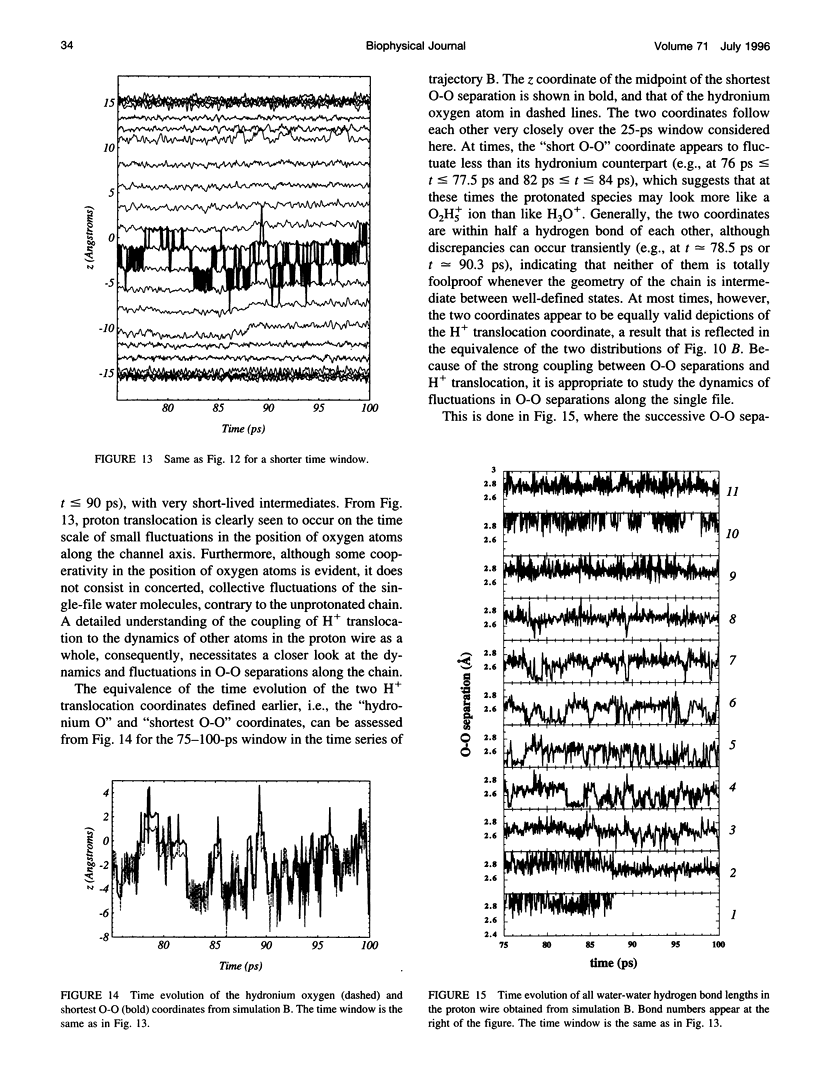
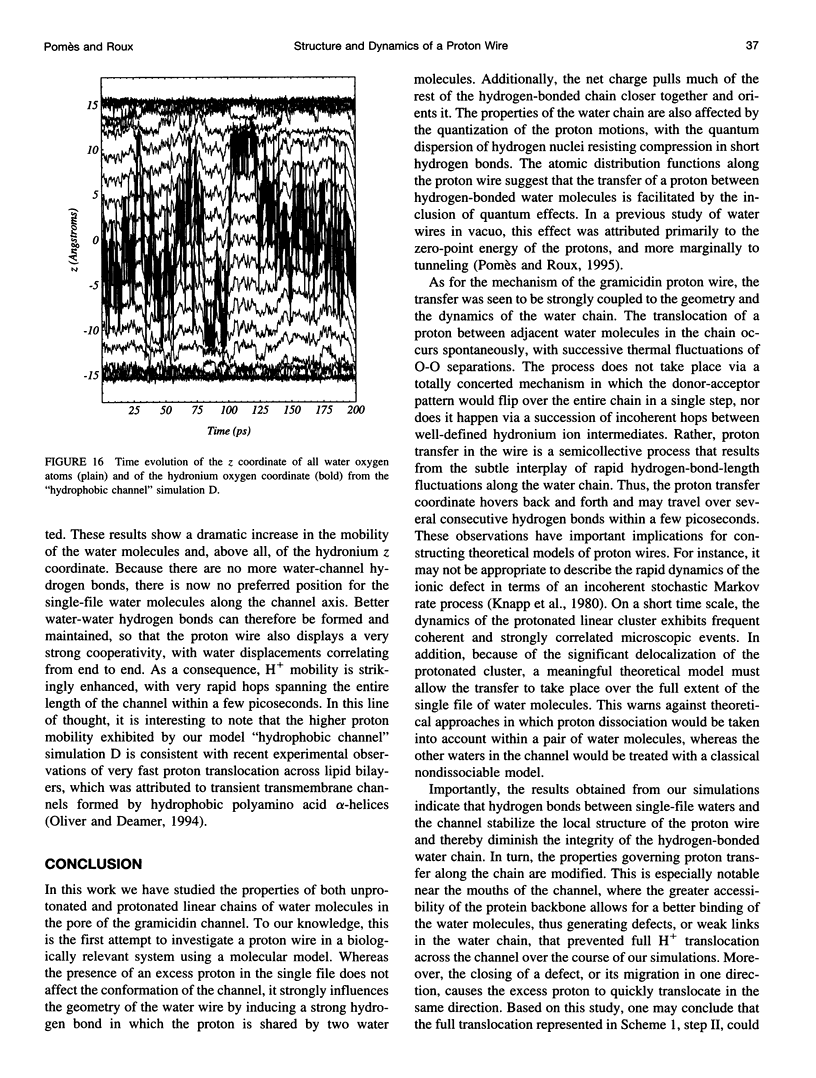
The rapid translocation of H+ along a chain of hydrogen-bonded water molecules, or proton wire, is thought to be an important mechanism for proton permeation through transmembrane channels. Computer simulations are used to study the properties of the proton wire formed by the single-file waters in the gramicidin A channel. The model includes the polypeptidic dimer, with 22 water molecules and one excess proton. The dissociation of the water molecules is taken into account by the "polarization model" of Stillinger and co-workers. The importance of quantum effects due to the light mass of the hydrogen nuclei is examined with the use of discretized Feynman path integral molecular dynamics simulations. Results show that the presence of an excess proton in the pore orients the single-file water molecules and affects the geometry of water-water hydrogen bonding interactions. Rather than a well-defined hydronium ion OH3+ in the single-file region, the protonated species is characterized by a strong hydrogen bond resembling that of O2H5+. The quantum dispersion of protons has a small but significant effect on the equilibrium structure of the hydrogen-bonded water chain. During classical trajectories, proton transfer between consecutive water molecules is a very fast spontaneous process that takes place in the subpicosecond time scale. The translocation along extended regions of the chain takes place neither via a totally concerted mechanism in which the donor-acceptor pattern would flip over the entire chain in a single step, nor via a succession of incoherent hops between well-defined intermediates. Rather, proton transfer in the wire is a semicollective process that results from the subtle interplay of rapid hydrogen-bond length fluctuations along the water chain. These rapid structural fluctuations of the protonated single file of waters around an average position and the slow movements of the average position of the excess proton along the channel axis occur on two very different time scales. Ultimately, it is the slow reorganization of hydrogen bonds between single-file water molecules and channel backbone carbonyl groups that, by affecting the connectivity and the dynamics of the single-file water chain, also limits the translocation of the proton across the pore.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akeson M., Deamer D. W. Proton conductance by the gramicidin water wire. Model for proton conductance in the F1F0 ATPases? Biophys J. 1991 Jul;60(1):101–109. doi: 10.1016/S0006-3495(91)82034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arseniev A. S., Barsukov I. L., Bystrov V. F., Lomize A. L., Ovchinnikov YuA 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single-stranded helices. FEBS Lett. 1985 Jul 8;186(2):168–174. doi: 10.1016/0014-5793(85)80702-x. [DOI] [PubMed] [Google Scholar]
- Cao Y., Váró G., Chang M., Ni B. F., Needleman R., Lanyi J. K. Water is required for proton transfer from aspartate-96 to the bacteriorhodopsin Schiff base. Biochemistry. 1991 Nov 12;30(45):10972–10979. doi: 10.1021/bi00109a023. [DOI] [PubMed] [Google Scholar]
- Chiu S. W., Jakobsson E., Subramaniam S., McCammon J. A. Time-correlation analysis of simulated water motion in flexible and rigid gramicidin channels. Biophys J. 1991 Jul;60(1):273–285. doi: 10.1016/S0006-3495(91)82049-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. W., Novotny J. A., Jakobsson E. The nature of ion and water barrier crossings in a simulated ion channel. Biophys J. 1993 Jan;64(1):98–109. doi: 10.1016/S0006-3495(93)81344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu S. W., Subramaniam S., Jakobsson E., McCammon J. A. Water and polypeptide conformations in the gramicidin channel. A molecular dynamics study. Biophys J. 1989 Aug;56(2):253–261. doi: 10.1016/S0006-3495(89)82671-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D. W. Proton permeation of lipid bilayers. J Bioenerg Biomembr. 1987 Oct;19(5):457–479. doi: 10.1007/BF00770030. [DOI] [PubMed] [Google Scholar]
- Etchebest C., Pullman A. The gramicidin A channel: energetics and structural characteristics of the progression of a sodium ion in the presence of water. J Biomol Struct Dyn. 1986 Feb;3(4):805–825. doi: 10.1080/07391102.1986.10508463. [DOI] [PubMed] [Google Scholar]
- Finkelstein A., Andersen O. S. The gramicidin A channel: a review of its permeability characteristics with special reference to the single-file aspect of transport. J Membr Biol. 1981 Apr 30;59(3):155–171. doi: 10.1007/BF01875422. [DOI] [PubMed] [Google Scholar]
- Fornili S. L., Vercauteren D. P., Clementi E. Water structure in the Gramicidin A transmembrane channel. Biochim Biophys Acta. 1984 Apr 11;771(2):151–164. doi: 10.1016/0005-2736(84)90527-3. [DOI] [PubMed] [Google Scholar]
- Gao J., Xia X. A priori evaluation of aqueous polarization effects through Monte Carlo QM-MM simulations. Science. 1992 Oct 23;258(5082):631–635. doi: 10.1126/science.1411573. [DOI] [PubMed] [Google Scholar]
- Hladky S. B., Haydon D. A. Ion transfer across lipid membranes in the presence of gramicidin A. I. Studies of the unit conductance channel. Biochim Biophys Acta. 1972 Aug 9;274(2):294–312. doi: 10.1016/0005-2736(72)90178-2. [DOI] [PubMed] [Google Scholar]
- Jeng M. F., Englander S. W. Stable submolecular folding units in a non-compact form of cytochrome c. J Mol Biol. 1991 Oct 5;221(3):1045–1061. doi: 10.1016/0022-2836(91)80191-v. [DOI] [PubMed] [Google Scholar]
- Jordan P. C. Ion-water and ion-polypeptide correlations in a gramicidin-like channel. A molecular dynamics study. Biophys J. 1990 Nov;58(5):1133–1156. doi: 10.1016/S0006-3495(90)82456-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchem R. R., Hu W., Cross T. A. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 1993 Sep 10;261(5127):1457–1460. doi: 10.1126/science.7690158. [DOI] [PubMed] [Google Scholar]
- Mackay D. H., Berens P. H., Wilson K. R., Hagler A. T. Structure and dynamics of ion transport through gramicidin A. Biophys J. 1984 Aug;46(2):229–248. doi: 10.1016/S0006-3495(84)84016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay D. H., Wilson K. R. Possible allosteric significance of water structures in proteins. J Biomol Struct Dyn. 1986 Dec;4(3):491–500. doi: 10.1080/07391102.1986.10506364. [DOI] [PubMed] [Google Scholar]
- Nagle J. F., Morowitz H. J. Molecular mechanisms for proton transport in membranes. Proc Natl Acad Sci U S A. 1978 Jan;75(1):298–302. doi: 10.1073/pnas.75.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagle J. F. Theory of passive proton conductance in lipid bilayers. J Bioenerg Biomembr. 1987 Oct;19(5):413–426. doi: 10.1007/BF00770027. [DOI] [PubMed] [Google Scholar]
- Nagle J. F., Tristram-Nagle S. Hydrogen bonded chain mechanisms for proton conduction and proton pumping. J Membr Biol. 1983;74(1):1–14. doi: 10.1007/BF01870590. [DOI] [PubMed] [Google Scholar]
- Oliver A. E., Deamer D. W. Alpha-helical hydrophobic polypeptides form proton-selective channels in lipid bilayers. Biophys J. 1994 May;66(5):1364–1379. doi: 10.1016/S0006-3495(94)80927-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullman A. Energy profiles in the gramicidin A channel. Q Rev Biophys. 1987 Nov;20(3-4):173–200. doi: 10.1017/s0033583500004170. [DOI] [PubMed] [Google Scholar]
- Roux B., Karplus M. Molecular dynamics simulations of the gramicidin channel. Annu Rev Biophys Biomol Struct. 1994;23:731–761. doi: 10.1146/annurev.bb.23.060194.003503. [DOI] [PubMed] [Google Scholar]
- Scheiner S. Ab initio studies of hydrogen bonds: the water dimer paradigm. Annu Rev Phys Chem. 1994;45:23–56. doi: 10.1146/annurev.pc.45.100194.000323. [DOI] [PubMed] [Google Scholar]
- Skerra A., Brickmann J. Structure and dynamics of one-dimensional ionic solutions in biological transmembrane channels. Biophys J. 1987 Jun;51(6):969–976. doi: 10.1016/S0006-3495(87)83424-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urry D. W. The gramicidin A transmembrane channel: a proposed pi(L,D) helix. Proc Natl Acad Sci U S A. 1971 Mar;68(3):672–676. doi: 10.1073/pnas.68.3.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf T. B., Roux B. Molecular dynamics simulation of the gramicidin channel in a phospholipid bilayer. Proc Natl Acad Sci U S A. 1994 Nov 22;91(24):11631–11635. doi: 10.1073/pnas.91.24.11631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf T. B., Roux B. Structure, energetics, and dynamics of lipid-protein interactions: A molecular dynamics study of the gramicidin A channel in a DMPC bilayer. Proteins. 1996 Jan;24(1):92–114. doi: 10.1002/(SICI)1097-0134(199601)24:1<92::AID-PROT7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]




