Abstract
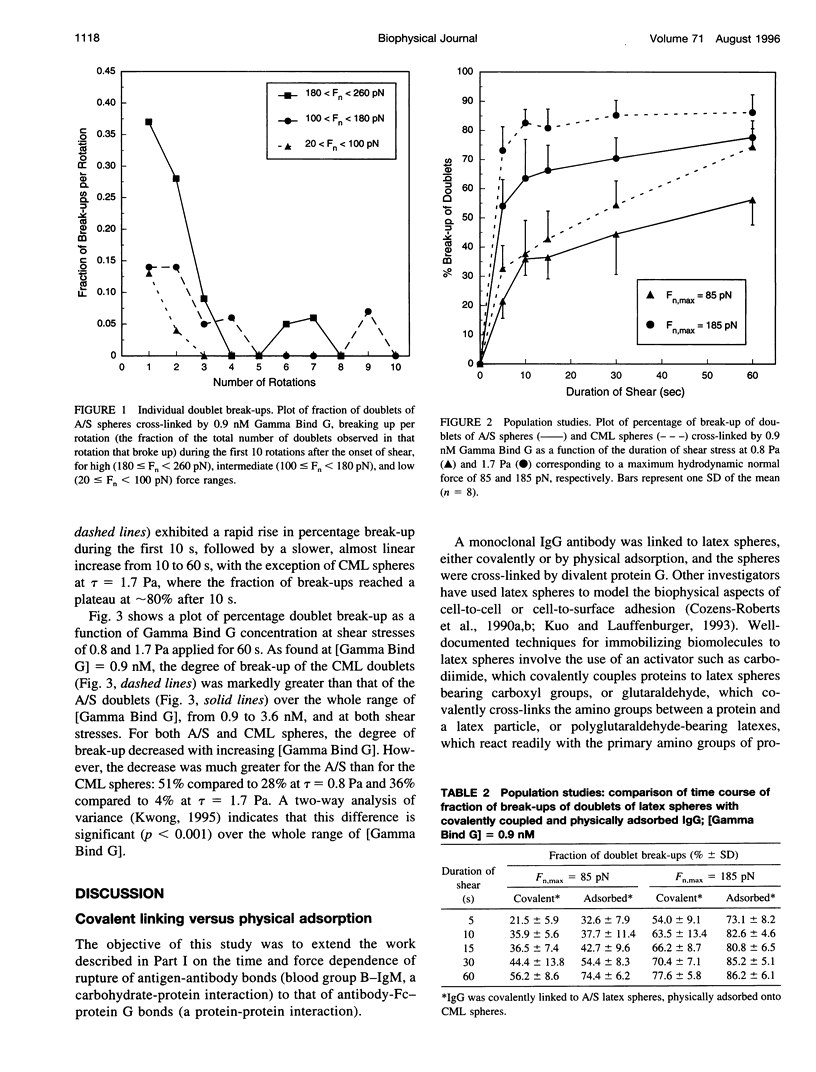
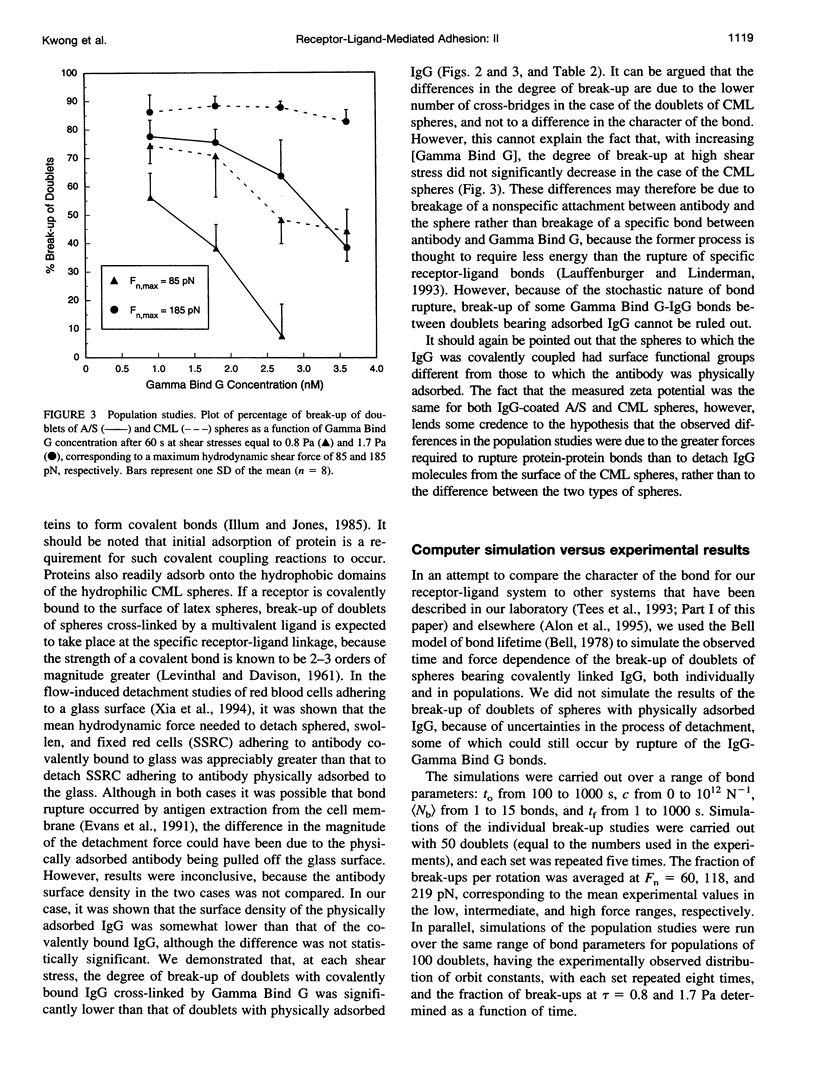
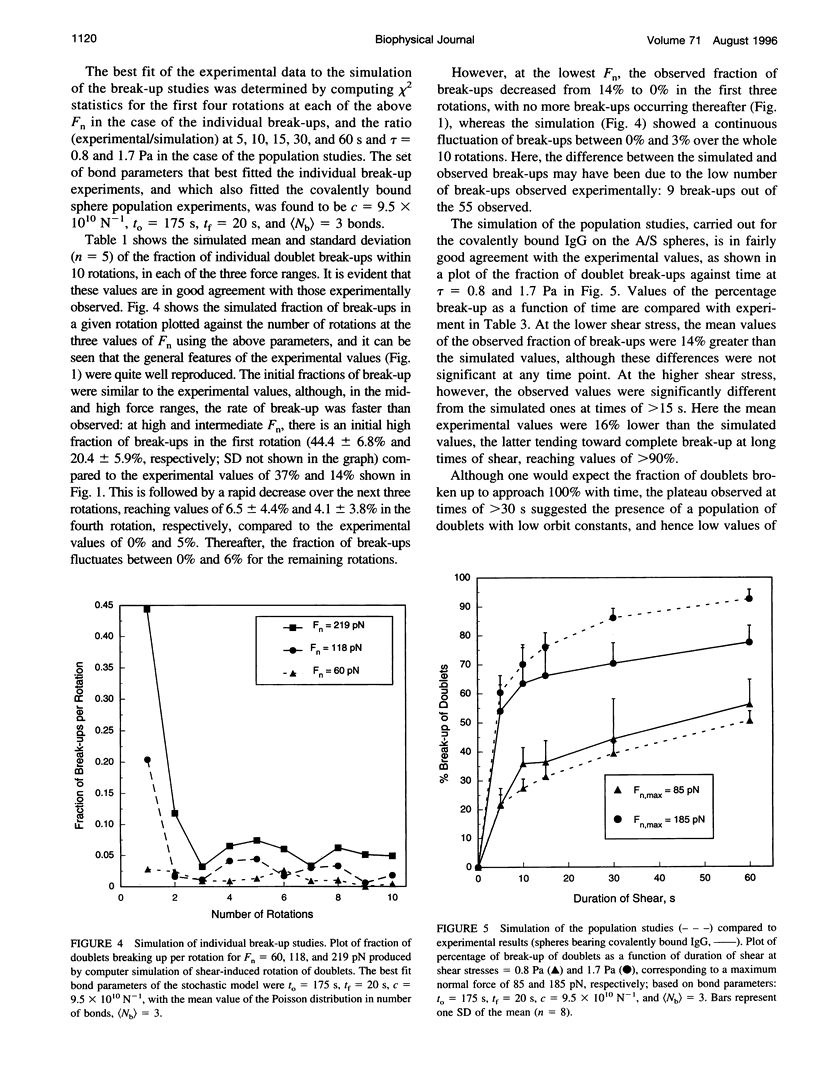
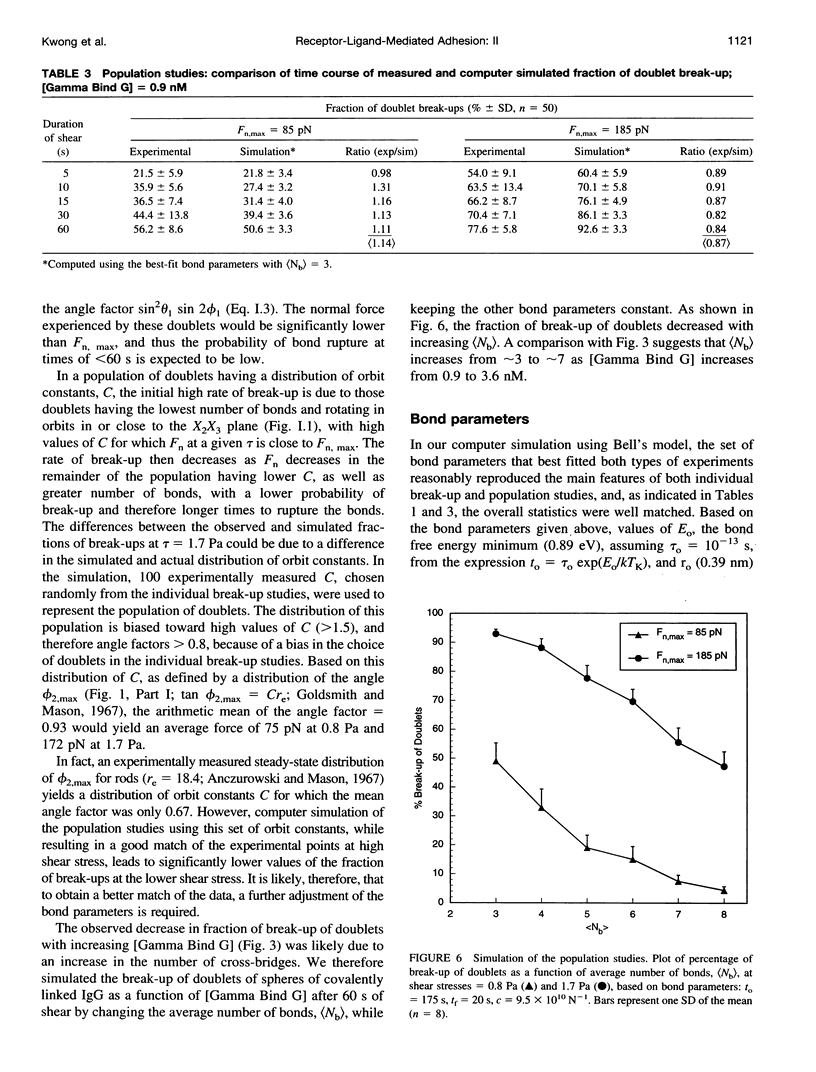
In an extension of the previous paper, we describe the force dependence of break-up of doublets of latex spheres cross-linked by protein G-IgG bonds via the Fc region of the antibody. The receptor, the monoclonal Bear-1 antibody, was either covalently linked to 4.75-microns aldehyde/sulfate (A/S) latex spheres in a one-step reaction, or physically adsorbed to the 4.63-microns carboxyl-modified latex spheres used in Part I of this paper. The spheres were suspended in 19% buffered Dextran 40 containing the ligand, the bivalent recombinant protein G (Gamma-Bind G), and observed in the counter-rotating cone and plate Rheoscope. Break-up of doublets, tracked individually under the microscope, as well as in populations of 50-150 particles, was studied over a range of normal force from 20 to 260 pN. In individual particle studies, the fraction of doublets of spheres with covalently linked IgG breaking up in the first 10 rotations, increased from 16% in the low-force to 63% in the high-force range. In population studies, the fraction broken up increased with duration and magnitude of the applied force, and decreased with increasing ligand concentration. Moreover, doublets of physically adsorbed IgG spheres required significantly lower force than doublets of covalently linked IgG spheres for the same degree of break-up, possibly because of surface detachment of IgG molecules rather than rupture of receptor-ligand bonds. Computer simulation, using the Bell stochastic model of break-up and a Poisson distribution for the number of bonds, described in Part I, showed that the parameters of the protein-protein bond differed significantly from those of the carbohydrate-protein bond studied in Part I of this paper, the former being much more responsive to force than the latter.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alon R., Hammer D. A., Springer T. A. Lifetime of the P-selectin-carbohydrate bond and its response to tensile force in hydrodynamic flow. Nature. 1995 Apr 6;374(6522):539–542. doi: 10.1038/374539a0. [DOI] [PubMed] [Google Scholar]
- Bell G. I. Models for the specific adhesion of cells to cells. Science. 1978 May 12;200(4342):618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Cozens-Roberts C., Lauffenburger D. A., Quinn J. A. Receptor-mediated cell attachment and detachment kinetics. I. Probabilistic model and analysis. Biophys J. 1990 Oct;58(4):841–856. doi: 10.1016/S0006-3495(90)82430-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozens-Roberts C., Quinn J. A., Lauffenberger D. A. Receptor-mediated adhesion phenomena. Model studies with the Radical-Flow Detachment Assay. Biophys J. 1990 Jul;58(1):107–125. doi: 10.1016/S0006-3495(90)82357-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson H. P. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc Natl Acad Sci U S A. 1994 Oct 11;91(21):10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans E., Berk D., Leung A. Detachment of agglutinin-bonded red blood cells. I. Forces to rupture molecular-point attachments. Biophys J. 1991 Apr;59(4):838–848. doi: 10.1016/S0006-3495(91)82296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illum L., Jones P. D. Attachment of monoclonal antibodies to microspheres. Methods Enzymol. 1985;112:67–84. doi: 10.1016/s0076-6879(85)12008-2. [DOI] [PubMed] [Google Scholar]
- Kuo S. C., Lauffenburger D. A. Relationship between receptor/ligand binding affinity and adhesion strength. Biophys J. 1993 Nov;65(5):2191–2200. doi: 10.1016/S0006-3495(93)81277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEVINTHAL C., DAVISON P. F. Degradation of deoxyribonucleic acid under hydrodynamic shearing forces. J Mol Biol. 1961 Oct;3:674–683. doi: 10.1016/s0022-2836(61)80030-2. [DOI] [PubMed] [Google Scholar]
- Rembaum A., Margel S., Levy J. Polyglutaraldehyde: a new reagent for coupling proteins to microspheres and for labeling cell-surface receptors. J Immunol Methods. 1978;24(3-4):239–250. doi: 10.1016/0022-1759(78)90128-x. [DOI] [PubMed] [Google Scholar]
- Tees D. F., Coenen O., Goldsmith H. L. Interaction forces between red cells agglutinated by antibody. IV. Time and force dependence of break-up. Biophys J. 1993 Sep;65(3):1318–1334. doi: 10.1016/S0006-3495(93)81180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tees D. F., Goldsmith H. L. Kinetics and locus of failure of receptor-ligand-mediated adhesion between latex spheres. I. Protein-carbohydrate bond. Biophys J. 1996 Aug;71(2):1102–1114. doi: 10.1016/S0006-3495(96)79312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Z., Goldsmith H. L., van de Ven T. G. Flow-induced detachment of red blood cells adhering to surfaces by specific antigen-antibody bonds. Biophys J. 1994 Apr;66(4):1222–1230. doi: 10.1016/S0006-3495(94)80906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


