Abstract
The grand canonical ensemble Monte Carlo molecular simulation method is used to investigate hydration patterns in the crystal hydrate structure of the dCpG/proflavine intercalated complex. The objective of this study is to show by example that the recently advocated grand canonical ensemble simulation is a computationally efficient method for determining the positions of the hydrating water molecules in protein and nucleic acid structures. A detailed molecular simulation convergence analysis and an analogous comparison of the theoretical results with experiments clearly show that the grand ensemble simulations can be far more advantageous than the comparable canonical ensemble simulations.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhat T. N., Bentley G. A., Boulot G., Greene M. I., Tello D., Dall'Acqua W., Souchon H., Schwarz F. P., Mariuzza R. A., Poljak R. J. Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc Natl Acad Sci U S A. 1994 Feb 1;91(3):1089–1093. doi: 10.1073/pnas.91.3.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar M. J., Storch D. M. Alternative view of enzyme reactions. Proc Natl Acad Sci U S A. 1985 Apr;82(8):2225–2229. doi: 10.1073/pnas.82.8.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Herzyk P., Goodfellow J. M., Neidle S. Molecular dynamics simulations of dinucleoside and dinucleoside-drug crystal hydrates. J Biomol Struct Dyn. 1991 Oct;9(2):363–386. doi: 10.1080/07391102.1991.10507918. [DOI] [PubMed] [Google Scholar]
- Hummer G., García A. E., Soumpasis D. M. Hydration of nucleic acid fragments: comparison of theory and experiment for high-resolution crystal structures of RNA, DNA, and DNA-drug complexes. Biophys J. 1995 May;68(5):1639–1652. doi: 10.1016/S0006-3495(95)80381-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J., Chothia C. The structure of protein-protein recognition sites. J Biol Chem. 1990 Sep 25;265(27):16027–16030. [PubMed] [Google Scholar]
- Kim K. S., Corongiu G., Clementi E. Networks of water molecules in a proflavine deoxydinucleoside phosphate complex. J Biomol Struct Dyn. 1983 Oct;1(1):263–285. doi: 10.1080/07391102.1983.10507439. [DOI] [PubMed] [Google Scholar]
- Levitt M., Park B. H. Water: now you see it, now you don't. Structure. 1993 Dec 15;1(4):223–226. doi: 10.1016/0969-2126(93)90011-5. [DOI] [PubMed] [Google Scholar]
- Lounnas V., Pettitt B. M. A connected-cluster of hydration around myoglobin: correlation between molecular dynamics simulations and experiment. Proteins. 1994 Feb;18(2):133–147. doi: 10.1002/prot.340180206. [DOI] [PubMed] [Google Scholar]
- Lounnas V., Pettitt B. M. Distribution function implied dynamics versus residence times and correlations: solvation shells of myoglobin. Proteins. 1994 Feb;18(2):148–160. doi: 10.1002/prot.340180207. [DOI] [PubMed] [Google Scholar]
- Lounnas V., Pettitt B. M., Phillips G. N., Jr A global model of the protein-solvent interface. Biophys J. 1994 Mar;66(3 Pt 1):601–614. doi: 10.1016/s0006-3495(94)80835-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezei M., Beveridge D. L., Berman H. M., Goodfellow J. M., Finney J. L., Neidle S. Monte Carlo studies on water in the dCpG/proflavin crystal hydrate. J Biomol Struct Dyn. 1983 Oct;1(1):287–297. doi: 10.1080/07391102.1983.10507440. [DOI] [PubMed] [Google Scholar]
- Nar H., Messerschmidt A., Huber R., van de Kamp M., Canters G. W. X-ray crystal structure of the two site-specific mutants His35Gln and His35Leu of azurin from Pseudomonas aeruginosa. J Mol Biol. 1991 Mar 20;218(2):427–447. doi: 10.1016/0022-2836(91)90723-j. [DOI] [PubMed] [Google Scholar]
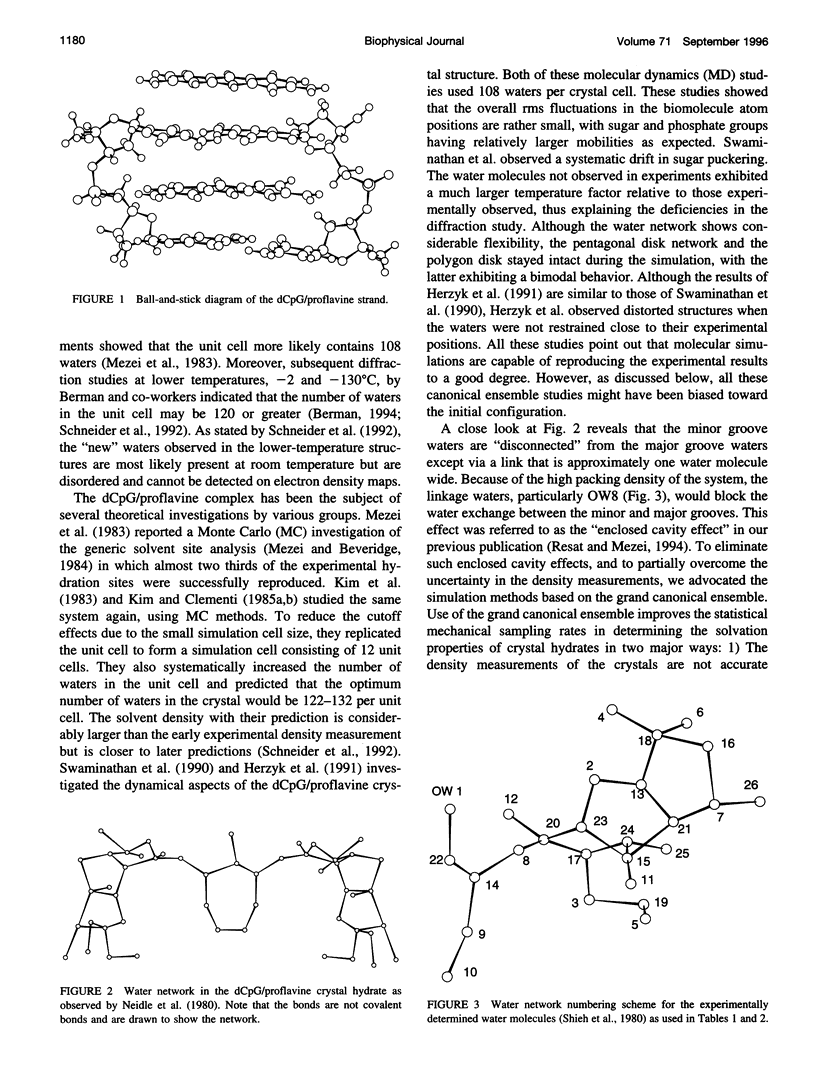
- Neidle S., Berman H. M., Shieh H. S. Highly structured water network in crystals of a deoxydinucleoside---drug complex. Nature. 1980 Nov 13;288(5787):129–133. doi: 10.1038/288129a0. [DOI] [PubMed] [Google Scholar]
- Otting G., Liepinsh E., Wüthrich K. Protein hydration in aqueous solution. Science. 1991 Nov 15;254(5034):974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
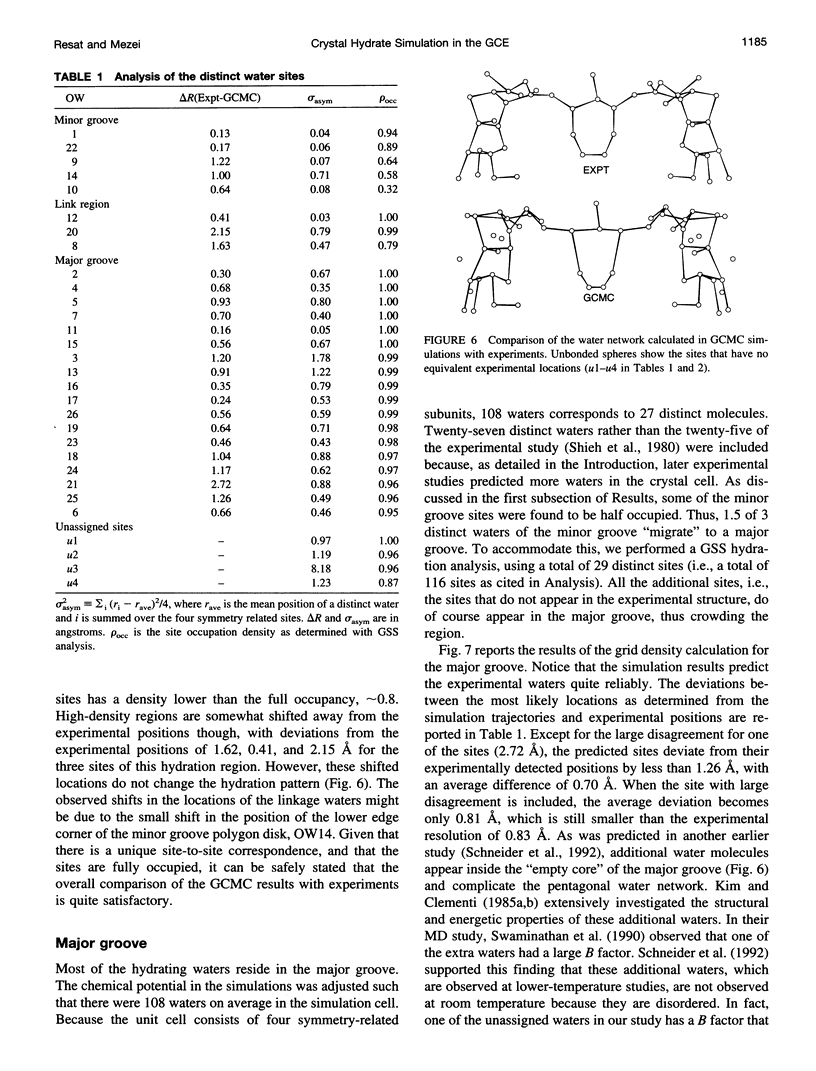
- Schneider B., Ginell S. L., Berman H. M. Low temperature structures of dCpG-proflavine. Conformational and hydration effects. Biophys J. 1992 Dec;63(6):1572–1578. doi: 10.1016/S0006-3495(92)81755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
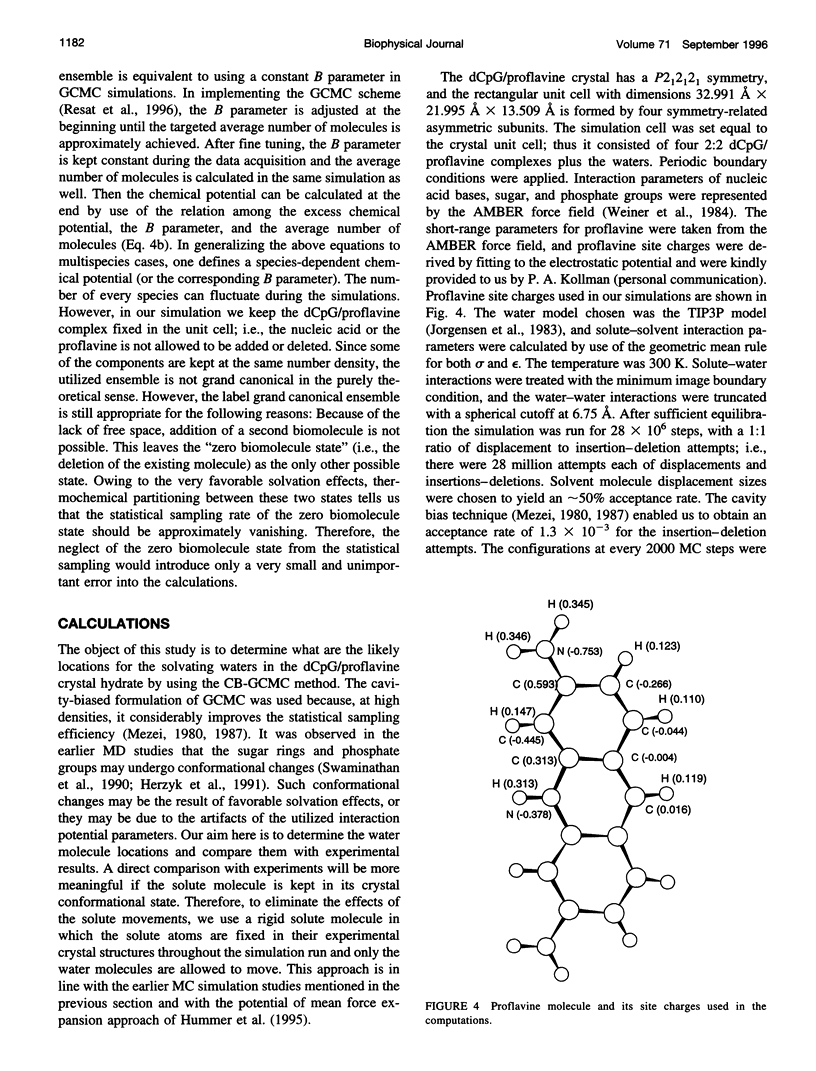
- Shieh H. S., Berman H. M., Dabrow M., Neidle S. The structure of drug-deoxydinucleoside phosphate complex; generalized conformational behavior of intercalation complexes with RNA and DNA fragments. Nucleic Acids Res. 1980 Jan 11;8(1):85–97. doi: 10.1093/nar/8.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A. Structural studies of protein-nucleic acid interaction: the sources of sequence-specific binding. Q Rev Biophys. 1990 Aug;23(3):205–280. doi: 10.1017/s0033583500005552. [DOI] [PubMed] [Google Scholar]
- Warshel A., Aqvist J., Creighton S. Enzymes work by solvation substitution rather than by desolvation. Proc Natl Acad Sci U S A. 1989 Aug;86(15):5820–5824. doi: 10.1073/pnas.86.15.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A., Naray-Szabo G., Sussman F., Hwang J. K. How do serine proteases really work? Biochemistry. 1989 May 2;28(9):3629–3637. doi: 10.1021/bi00435a001. [DOI] [PubMed] [Google Scholar]