Abstract
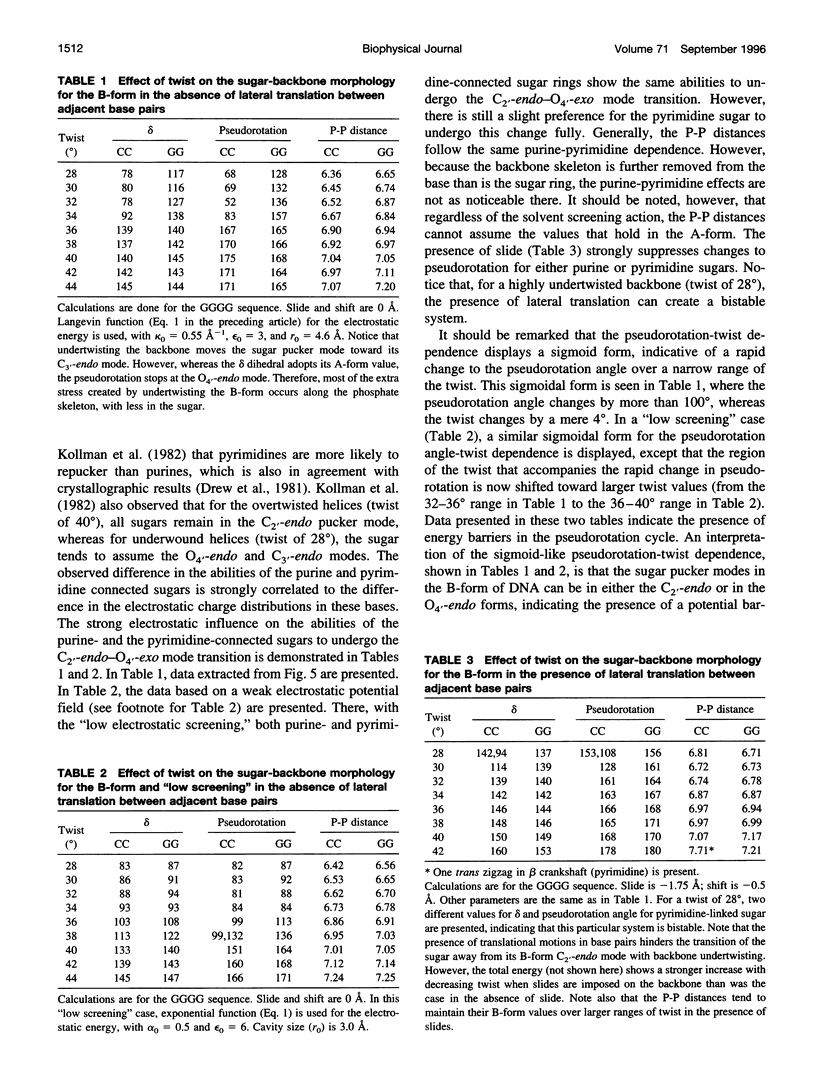
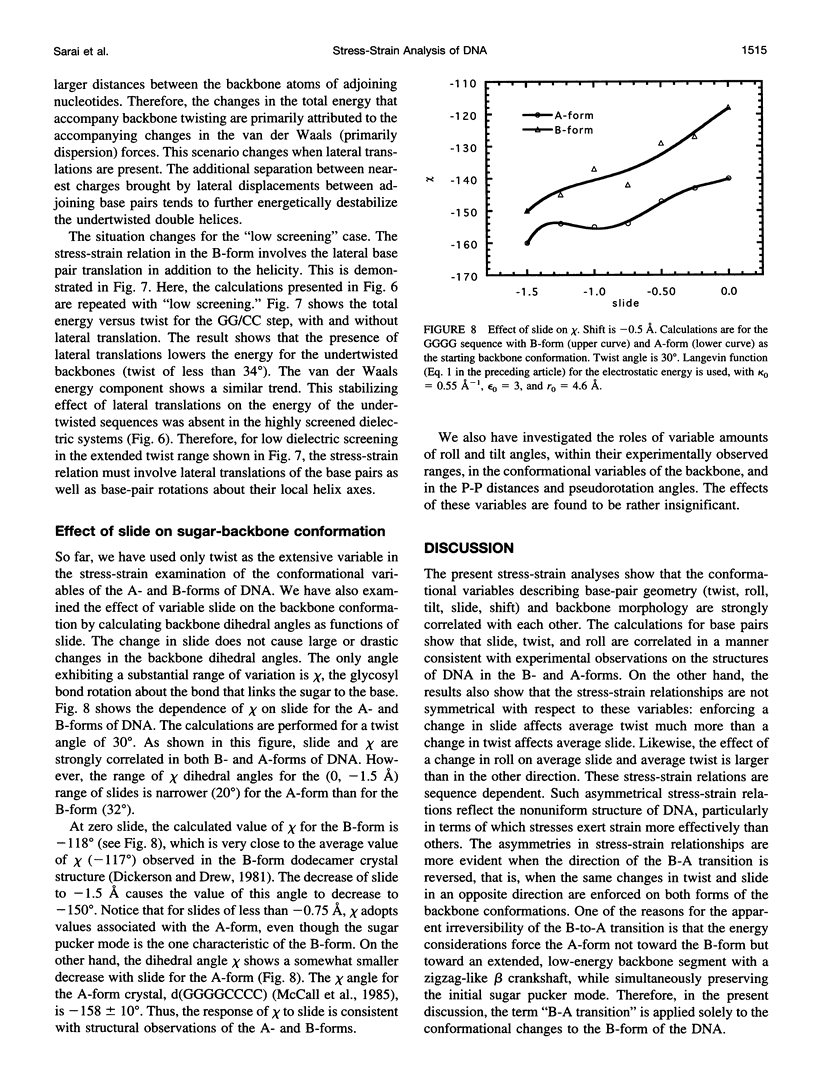
DNA exhibits conformational polymorphism, with the details depending on the sequence and its environment. To understand the mechanisms of conformational polymorphism and these transitions, we examine the interrelationships among the various conformational variables of DNA. In particular, we examine the stress-strain relation among conformational variables, describing base-pair morphology and their effects on the backbone conformation. For the calculation of base pairs, we use the method previously developed to calculate averages over conformational variables of DNA. Here we apply this method to calculate the Boltzmann averages of conformational variables for fixed values of one particular conformational variable, which reflects the strain in the structure responding to a particular driving stress. This averaging over all but one driving variable smooths the usual rough energy surface to permit observation of the effects of one conformational variable at a time. The stress-strain analyses of conformational variables of base pair slide, twist, and roll, which exhibit characteristic changes during the conformational transition of DNA, have shown that the conformational changes of base pairs are strongly correlated with one another. Furthermore, the stress-strain relations are not symmetrical with respect to these variables, i.e., the response of one coordinate to another is different from the reverse direction. We also examine the effect of conformational changes in base-pair variables on the sugar-backbone conformation by using the minimization method we developed. The conformational changes of base pairs affect the sugar pucker and other dihedral angles of the backbone of DNA, but each variable affects the sugar-backbone differently. In particular, twist is found to have the most influence in affecting the sugar pucker and backbone conformation. These calculated conformational changes in base pairs and backbone segments are consistent with experimental observations and serve to validate the calculation method.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden C. J., Kim S. H. Solvent-accessible surfaces of nucleic acids. J Mol Biol. 1979 Aug 15;132(3):411–434. doi: 10.1016/0022-2836(79)90268-7. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Optimised parameters for A-DNA and B-DNA. Biochem Biophys Res Commun. 1972 Jun 28;47(6):1504–1509. doi: 10.1016/0006-291x(72)90243-4. [DOI] [PubMed] [Google Scholar]
- Arnott S., Selsing E. Letter: The structure of polydeoxyguanylic acid with polydeoxycytidylic acid. J Mol Biol. 1974 Sep 15;88(2):551–552. doi: 10.1016/0022-2836(74)90502-6. [DOI] [PubMed] [Google Scholar]
- Briki F., Genest D. Canonical analysis of correlated atomic motions in DNA from molecular dynamics simulation. Biophys Chem. 1994 Sep;52(1):35–43. doi: 10.1016/0301-4622(94)00063-8. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R. A base-centred explanation of the B-to-A transition in DNA. J Mol Biol. 1984 Sep 25;178(3):773–782. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Dickerson R. E. Structure of a B-DNA dodecamer. III. Geometry of hydration. J Mol Biol. 1981 Sep 25;151(3):535–556. doi: 10.1016/0022-2836(81)90009-7. [DOI] [PubMed] [Google Scholar]
- Drew H. R., Wing R. M., Takano T., Broka C., Tanaka S., Itakura K., Dickerson R. E. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch V., Ravishanker G., Beveridge D. L., Westhof E. Molecular dynamics simulations of poly(dA).poly(dT): comparisons between implicit and explicit solvent representations. Biopolymers. 1993 Oct;33(10):1537–1552. doi: 10.1002/bip.360331005. [DOI] [PubMed] [Google Scholar]
- Gorin A. A., Zhurkin V. B., Olson W. K. B-DNA twisting correlates with base-pair morphology. J Mol Biol. 1995 Mar 17;247(1):34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Schyolkina A. K., Poletayev A. I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12(1):89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- Leslie A. G., Arnott S., Chandrasekaran R., Ratliff R. L. Polymorphism of DNA double helices. J Mol Biol. 1980 Oct 15;143(1):49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- Mazur J., Jernigan R. L. Distance-dependent dielectric constants and their application to double-helical DNA. Biopolymers. 1991 Nov;31(13):1615–1629. doi: 10.1002/bip.360311316. [DOI] [PubMed] [Google Scholar]
- Mazur J., Sarai A., Jernigan R. L. Sequence dependence of the B-A conformational transition of DNA. Biopolymers. 1989 Jul;28(7):1223–1233. doi: 10.1002/bip.360280704. [DOI] [PubMed] [Google Scholar]
- McCall M., Brown T., Kennard O. The crystal structure of d(G-G-G-G-C-C-C-C). A model for poly(dG).poly(dC). J Mol Biol. 1985 Jun 5;183(3):385–396. doi: 10.1016/0022-2836(85)90009-9. [DOI] [PubMed] [Google Scholar]
- Olson W. K., Flory P. J. Spatial configuration of polynucleotide chains. II. Conformational energies and the average dimensions of polyribonucleotides. Biopolymers. 1972 Jan;11(1):25–56. doi: 10.1002/bip.1972.360110103. [DOI] [PubMed] [Google Scholar]
- Pilet J., Brahms J. Dependence of B-A conformational change in DNA on base composition. Nat New Biol. 1972 Mar 29;236(65):99–100. doi: 10.1038/newbio236099a0. [DOI] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Saenger W., Hunter W. N., Kennard O. DNA conformation is determined by economics in the hydration of phosphate groups. 1986 Nov 27-Dec 3Nature. 324(6095):385–388. doi: 10.1038/324385a0. [DOI] [PubMed] [Google Scholar]
- Sarai A., Mazur J., Nussinov R., Jernigan R. L. Origin of DNA helical structure and its sequence dependence. Biochemistry. 1988 Nov 1;27(22):8498–8502. doi: 10.1021/bi00422a030. [DOI] [PubMed] [Google Scholar]
- Sarai A., Mazur J., Nussinov R., Jernigan R. L. Sequence dependence of DNA conformational flexibility. Biochemistry. 1989 Sep 19;28(19):7842–7849. doi: 10.1021/bi00445a046. [DOI] [PubMed] [Google Scholar]
- Sarma M. H., Gupta G., Sarma R. H. 500-MHz 1H NMR study of poly(dG).poly(dC) in solution using one-dimensional nuclear Overhauser effect. Biochemistry. 1986 Jun 17;25(12):3659–3665. doi: 10.1021/bi00360a028. [DOI] [PubMed] [Google Scholar]
- Srinivasan A. R., Olson W. K. Nucleic acid model building: the multiple backbone solutions associated with a given base morphology. J Biomol Struct Dyn. 1987 Jun;4(6):895–938. doi: 10.1080/07391102.1987.10507690. [DOI] [PubMed] [Google Scholar]
- Tung C. S. A reduced set of coordinates for modeling DNA structures: (I). A B-to-A transition pathway driven by pseudorotational angle. J Biomol Struct Dyn. 1992 Jun;9(6):1185–1194. doi: 10.1080/07391102.1992.10507986. [DOI] [PubMed] [Google Scholar]
- Tung C. S., Harvey S. C. Computer graphics program to reveal the dependence of the gross three-dimensional structure of the B-DNA double helix on primary structure. Nucleic Acids Res. 1986 Jan 10;14(1):381–387. doi: 10.1093/nar/14.1.381. [DOI] [PMC free article] [PubMed] [Google Scholar]


