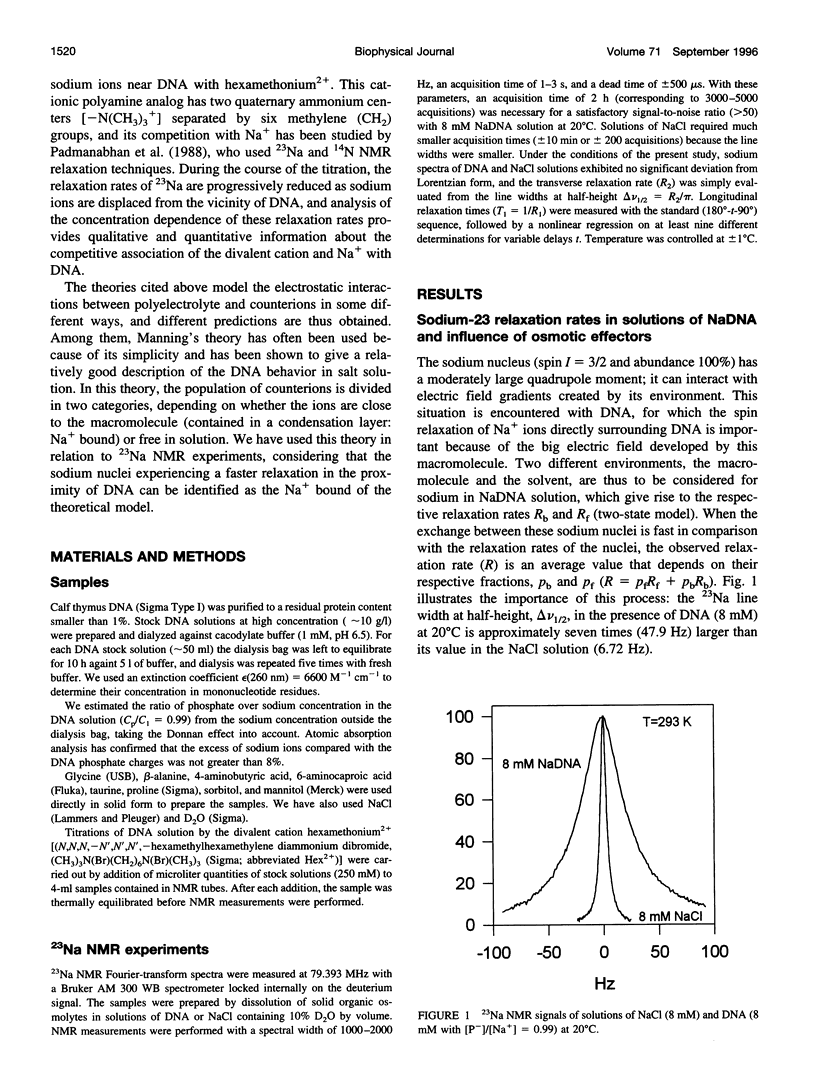
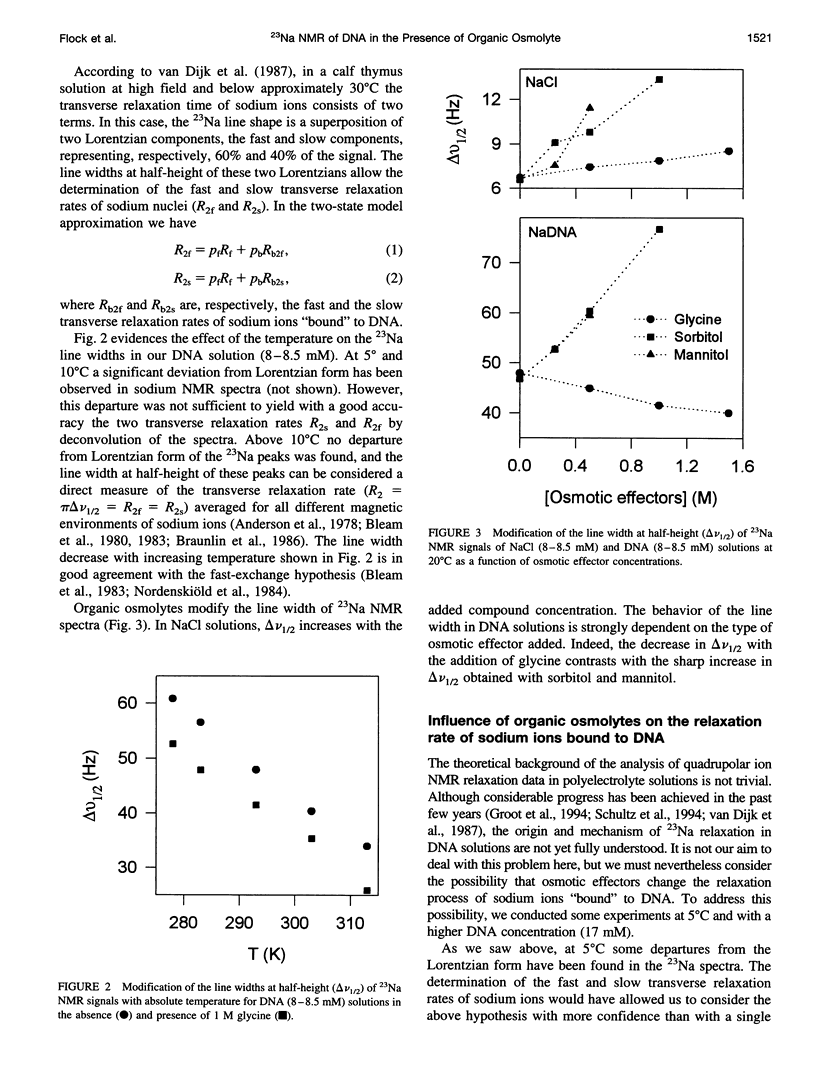
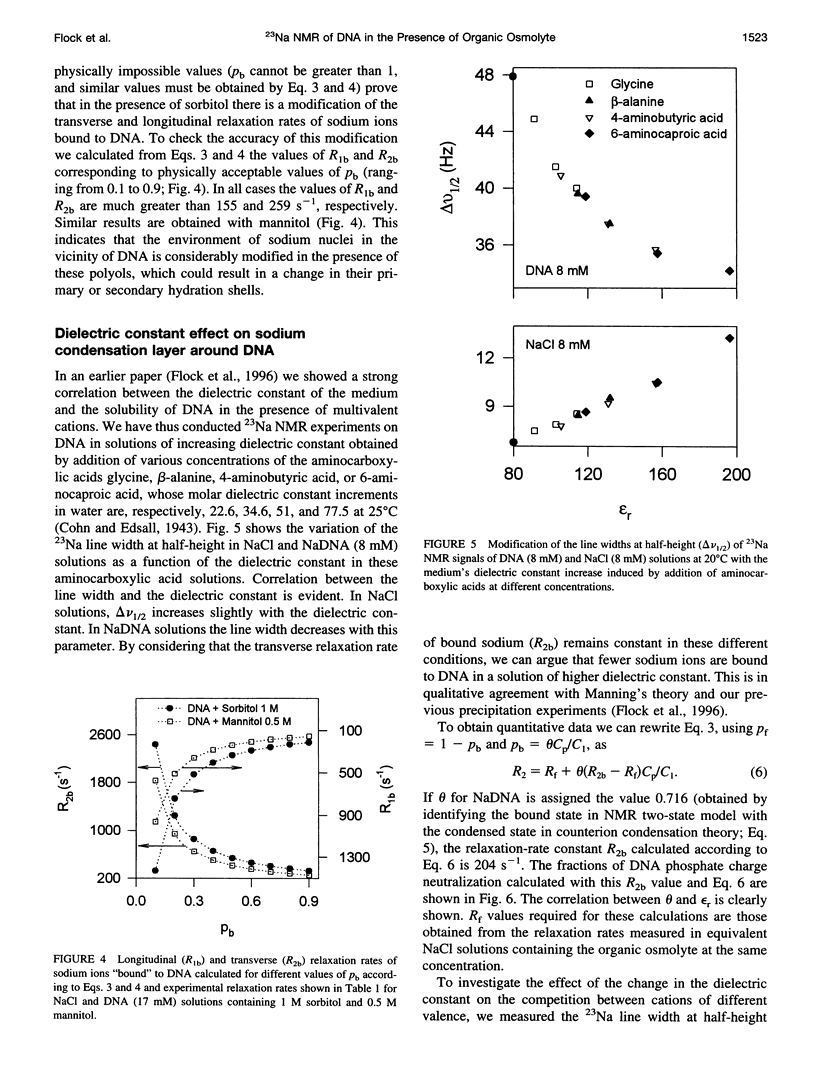
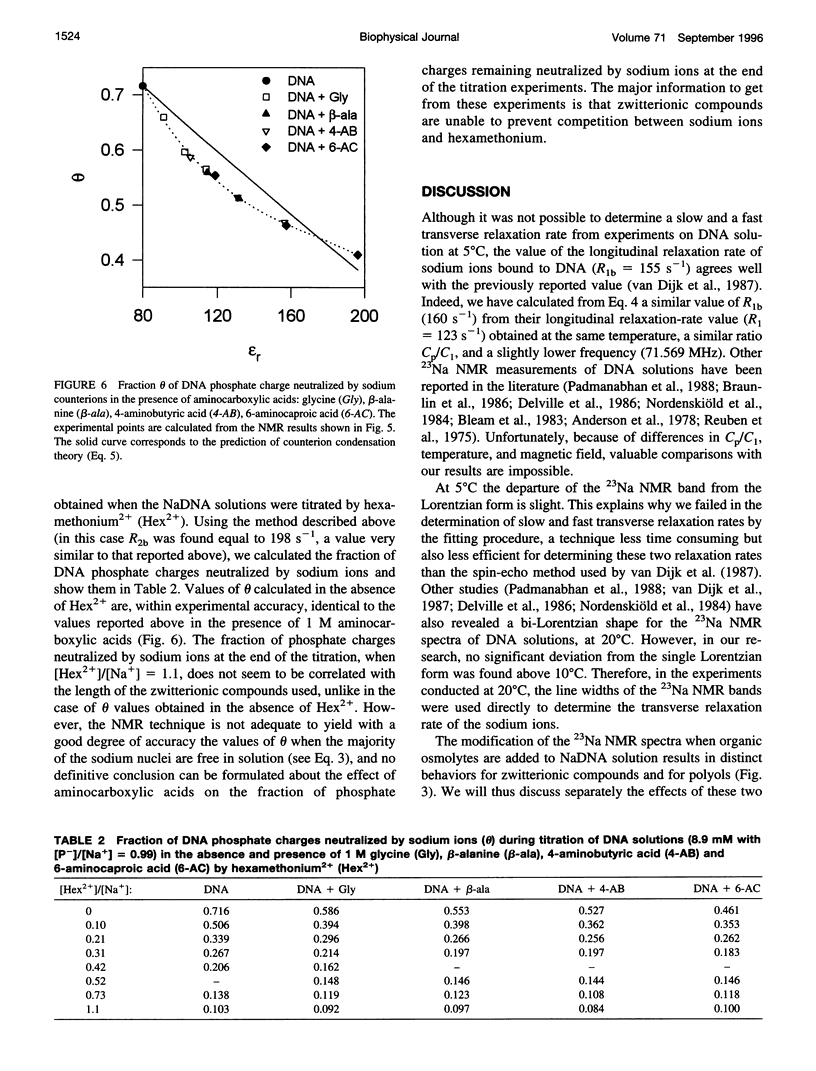
Abstract
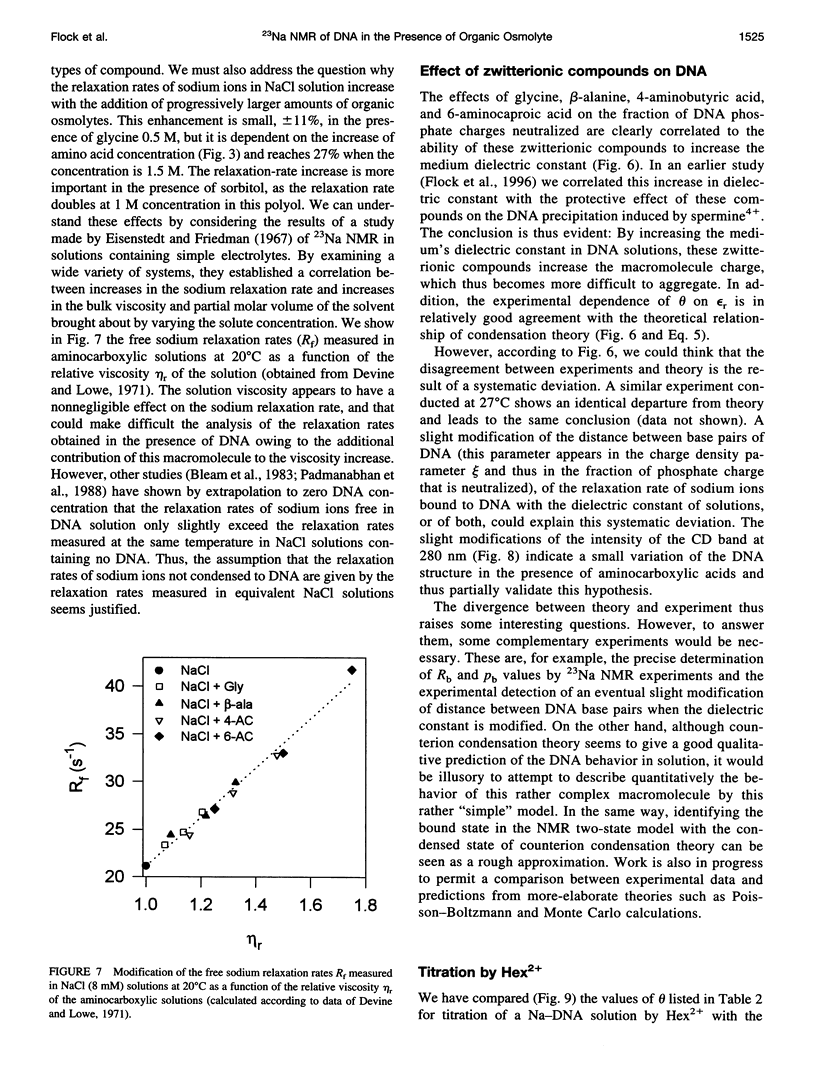
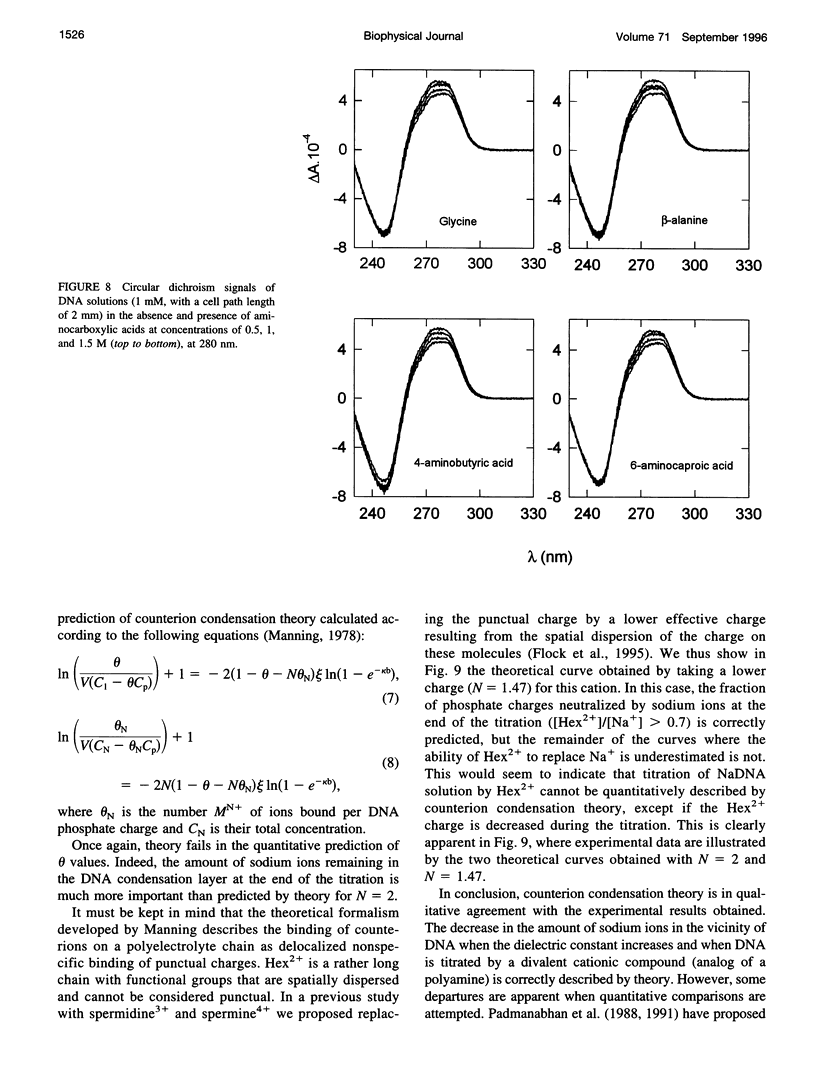
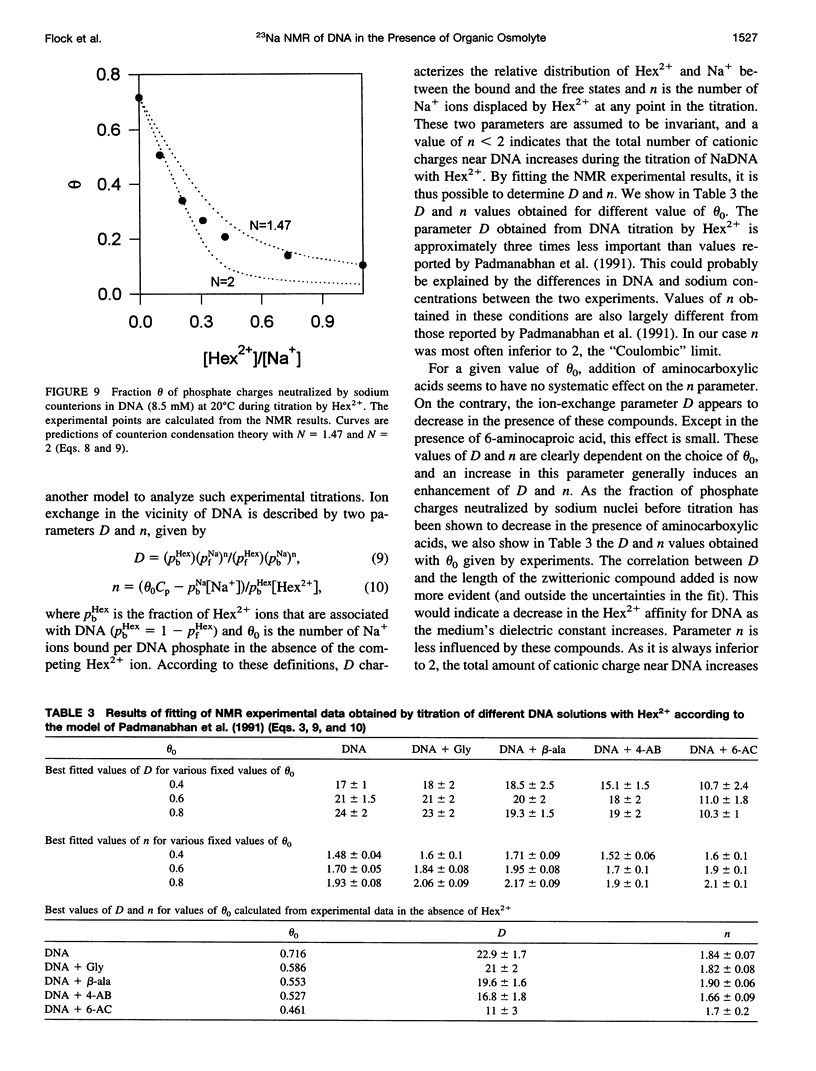
The effect of different organic osmolytes on the DNA counterion condensation layer has been investigated by 23Na NMR relaxation measurements. The zwitterionic compounds glycine, beta-alanine, 4-aminobutyric acid, and 6-aminocaproic acid have shown an increasing capacity to decrease the amount of sodium ions in the vicinity of the macromolecule. The experimental data have been correlated with the dielectric constant increase in their corresponding solutions and have been compared with the prediction of counterion condensation theory. Polyols (sorbitol and mannitol) did not display the same effect. These compounds largely increase the relaxation rate of sodium ions in the proximity of DNA, unlike the zwitterionic compounds. This probably results from a perturbation of the water dynamic around the macromolecule, of the primary or secondary hydration shell of the sodium nuclei involved, or both.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson C. F., Record M. T., Jr, Hart P. A. Sodium-23 NMR studies of cation-DNA interactions. Biophys Chem. 1978 Jan;7(4):301–316. doi: 10.1016/0301-4622(78)85007-8. [DOI] [PubMed] [Google Scholar]
- Bleam M. L., Anderson C. F., Record M. T. Relative binding affinities of monovalent cations for double-stranded DNA. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3085–3089. doi: 10.1073/pnas.77.6.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield V. A. Condensation of DNA by multivalent cations: considerations on mechanism. Biopolymers. 1991 Nov;31(13):1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- Bloomfield V. A., Wilson R. W., Rau D. C. Polyelectrolyte effects in DNA condensation by polyamines. Biophys Chem. 1980 Jun;11(3-4):339–343. doi: 10.1016/0301-4622(80)87006-2. [DOI] [PubMed] [Google Scholar]
- Braunlin W. H., Anderson C. F., Record M. T. 23Na-NMR investigations of counterion exchange reactions of helical DNA. Biopolymers. 1986 Jan;25(1):205–214. doi: 10.1002/bip.360250114. [DOI] [PubMed] [Google Scholar]
- Braunlin W. H., Anderson C. F., Record M. T., Jr Competitive interactions of Co(NH3)6(3+) and Na+ with helical B-DNA probed by 59Co and 23Na NMR. Biochemistry. 1987 Dec 1;26(24):7724–7731. doi: 10.1021/bi00398a028. [DOI] [PubMed] [Google Scholar]
- Buche A., Colson P., Houssier C. Effect of organic effectors on chromatin solubility, DNA-histone H1 interactions, DNA and histone H1 structures. J Biomol Struct Dyn. 1993 Aug;11(1):95–119. doi: 10.1080/07391102.1993.10508712. [DOI] [PubMed] [Google Scholar]
- Buche A., Colson P., Houssier C. Organic osmotic effectors and chromatin structure. J Biomol Struct Dyn. 1990 Dec;8(3):601–618. doi: 10.1080/07391102.1990.10507831. [DOI] [PubMed] [Google Scholar]
- Buche A., Ouassaidi A., Hacha R., Delpire E., Gilles R., Houssier C. Glycine and other amino compounds prevent chromatin precipitation at physiological ionic strength. FEBS Lett. 1989 Apr 24;247(2):367–370. doi: 10.1016/0014-5793(89)81372-9. [DOI] [PubMed] [Google Scholar]
- Delpire E., Duchêne C., Goessens G., Gilles R. Effects of osmotic shocks on the ultrastructure of different tissues and cell types. Exp Cell Res. 1985 Sep;160(1):106–116. doi: 10.1016/0014-4827(85)90240-x. [DOI] [PubMed] [Google Scholar]
- Delville A., Laszlo P., Schyns R. Displacement of sodium ions by surfactant ions from DNA. A 23Na-NMR investigation. Biophys Chem. 1986 Jul;24(2):121–133. doi: 10.1016/0301-4622(86)80005-9. [DOI] [PubMed] [Google Scholar]
- Flock S., Labarbe R., Houssier C. Dielectric constant and ionic strength effects on DNA precipitation. Biophys J. 1996 Mar;70(3):1456–1465. doi: 10.1016/S0006-3495(96)79705-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flock S., Labarbe R., Houssier C. Osmotic effectors and DNA structure: effect of glycine on precipitation of DNA by multivalent cations. J Biomol Struct Dyn. 1995 Aug;13(1):87–102. doi: 10.1080/07391102.1995.10508823. [DOI] [PubMed] [Google Scholar]
- Gilles R. Comparative aspects of cell osmoregulation and volume control. Ren Physiol Biochem. 1988 May-Oct;11(3-5):277–288. doi: 10.1159/000173167. [DOI] [PubMed] [Google Scholar]
- Le Bret M., Zimm B. H. Monte Carlo determination of the distribution of ions about a cylindrical polyelectrolyte. Biopolymers. 1984 Feb;23(2):271–285. doi: 10.1002/bip.360230208. [DOI] [PubMed] [Google Scholar]
- Manning G. S. Self-attraction and natural curvature in null DNA. J Biomol Struct Dyn. 1989 Aug;7(1):41–61. doi: 10.1080/07391102.1989.10507751. [DOI] [PubMed] [Google Scholar]
- Manning G. S. The molecular theory of polyelectrolyte solutions with applications to the electrostatic properties of polynucleotides. Q Rev Biophys. 1978 May;11(2):179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- Marquet R., Houssier C. Thermodynamics of cation-induced DNA condensation. J Biomol Struct Dyn. 1991 Aug;9(1):159–167. doi: 10.1080/07391102.1991.10507900. [DOI] [PubMed] [Google Scholar]
- Nordenskiöld L., Chang D. K., Anderson C. F., Record M. T., Jr 23Na NMR relaxation study of the effects of conformation and base composition on the interactions of counterions with double-helical DNA. Biochemistry. 1984 Sep 11;23(19):4309–4317. doi: 10.1021/bi00314a009. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S., Brushaber V. M., Anderson C. F., Record M. T., Jr Relative affinities of divalent polyamines and of their N-methylated analogues for helical DNA determined by 23Na NMR. Biochemistry. 1991 Jul 30;30(30):7550–7559. doi: 10.1021/bi00244a026. [DOI] [PubMed] [Google Scholar]
- Padmanabhan S., Richey B., Anderson C. F., Record M. T., Jr Interaction of an N-methylated polyamine analogue, hexamethonium(2+), with NaDNA: quantitative 14N and 23Na NMR relaxation rate studies of the cation-exchange process. Biochemistry. 1988 Jun 14;27(12):4367–4376. doi: 10.1021/bi00412a025. [DOI] [PubMed] [Google Scholar]
- Reuben J., Shporer M., Gabbay E. J. The Alkali Ion-DNA Interaction as Reflected in the Nuclear Relaxation Rates of Na and Rb. Proc Natl Acad Sci U S A. 1975 Jan;72(1):245–247. doi: 10.1073/pnas.72.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Nordenskiöld L., Rupprecht A. A study of the quadrupolar NMR splittings of 7Li+, 23Na+, and 133Cs+ counterions in macroscopically oriented DNA fibers. Biopolymers. 1992 Dec;32(12):1631–1642. doi: 10.1002/bip.360321206. [DOI] [PubMed] [Google Scholar]
- Vorontsov-Velyaminov P. N., Lyubartsev A. P. Monte-Carlo-self consistent field method in the polyelectrolyte theory. J Biomol Struct Dyn. 1989 Dec;7(3):739–747. doi: 10.1080/07391102.1989.10508517. [DOI] [PubMed] [Google Scholar]
- Wilson R. W., Bloomfield V. A. Counterion-induced condesation of deoxyribonucleic acid. a light-scattering study. Biochemistry. 1979 May 29;18(11):2192–2196. doi: 10.1021/bi00578a009. [DOI] [PubMed] [Google Scholar]
- Wright L. A., Lerner L. E. Magnesium-DNA interactions from interpretation of 25Mg-nmr relaxation rates: field and coion dependence. Biopolymers. 1994 Jun;34(6):691–700. doi: 10.1002/bip.360340602. [DOI] [PubMed] [Google Scholar]
- van Dijk L., Gruwel M. L., Jesse W., de Bleijser J., Leyte J. C. Sodium ion and solvent nuclear relaxation results in aqueous solutions of DNA. Biopolymers. 1987 Feb;26(2):261–284. doi: 10.1002/bip.360260208. [DOI] [PubMed] [Google Scholar]


