Abstract
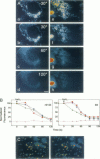
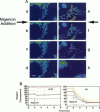
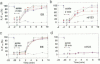
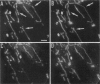
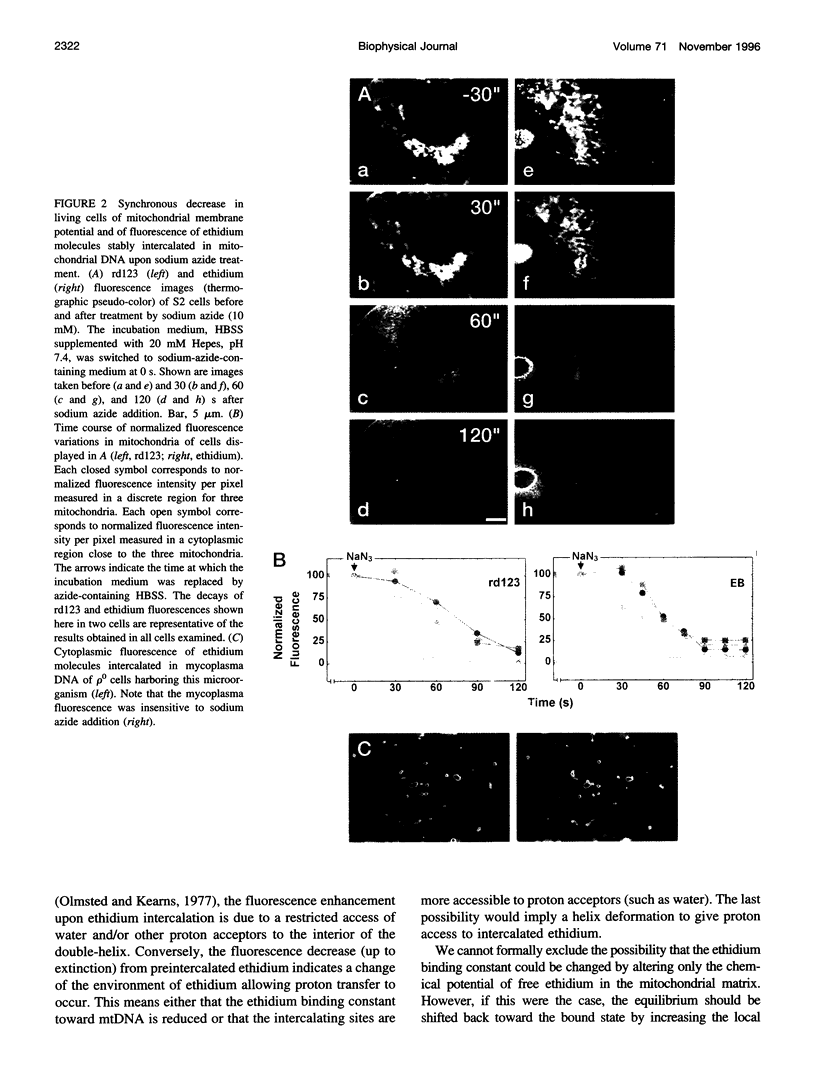
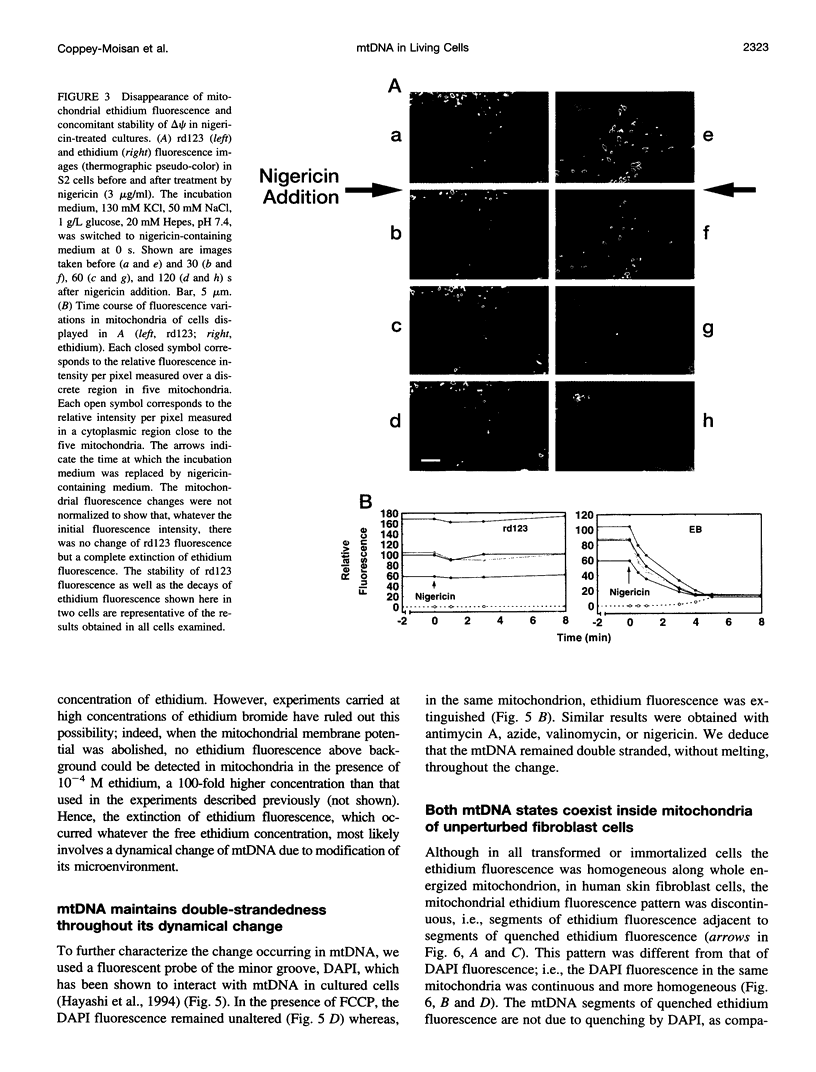
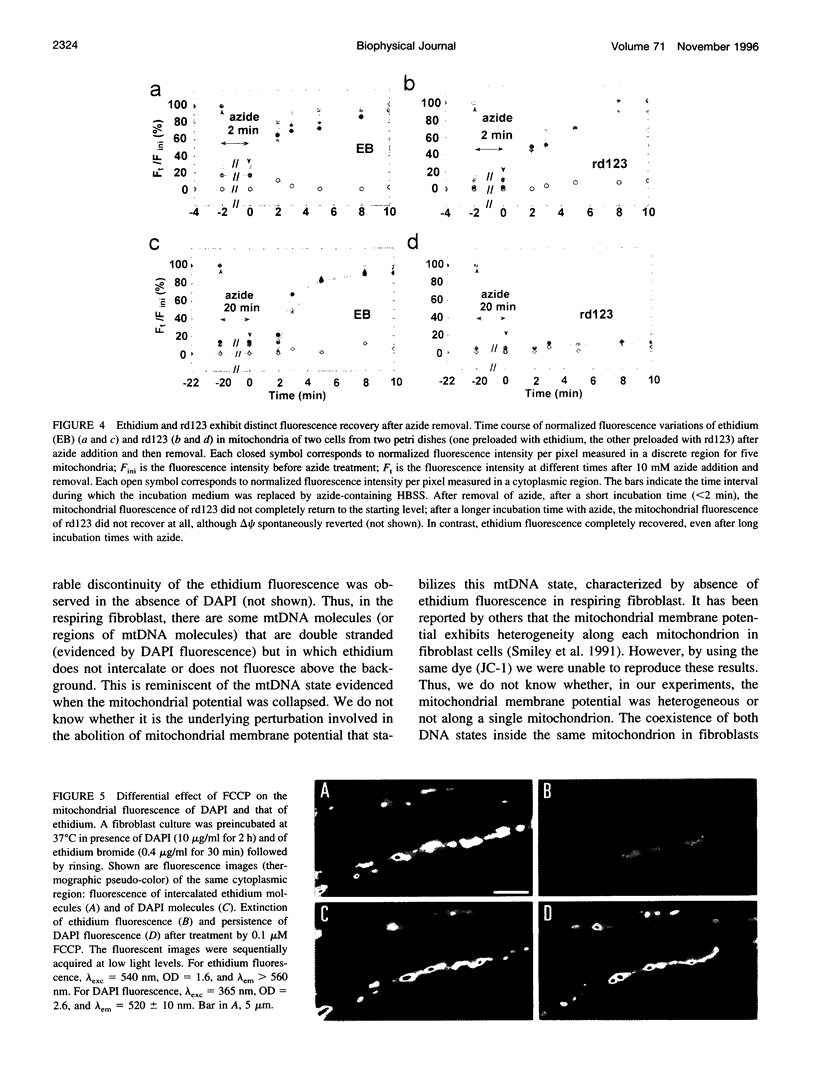
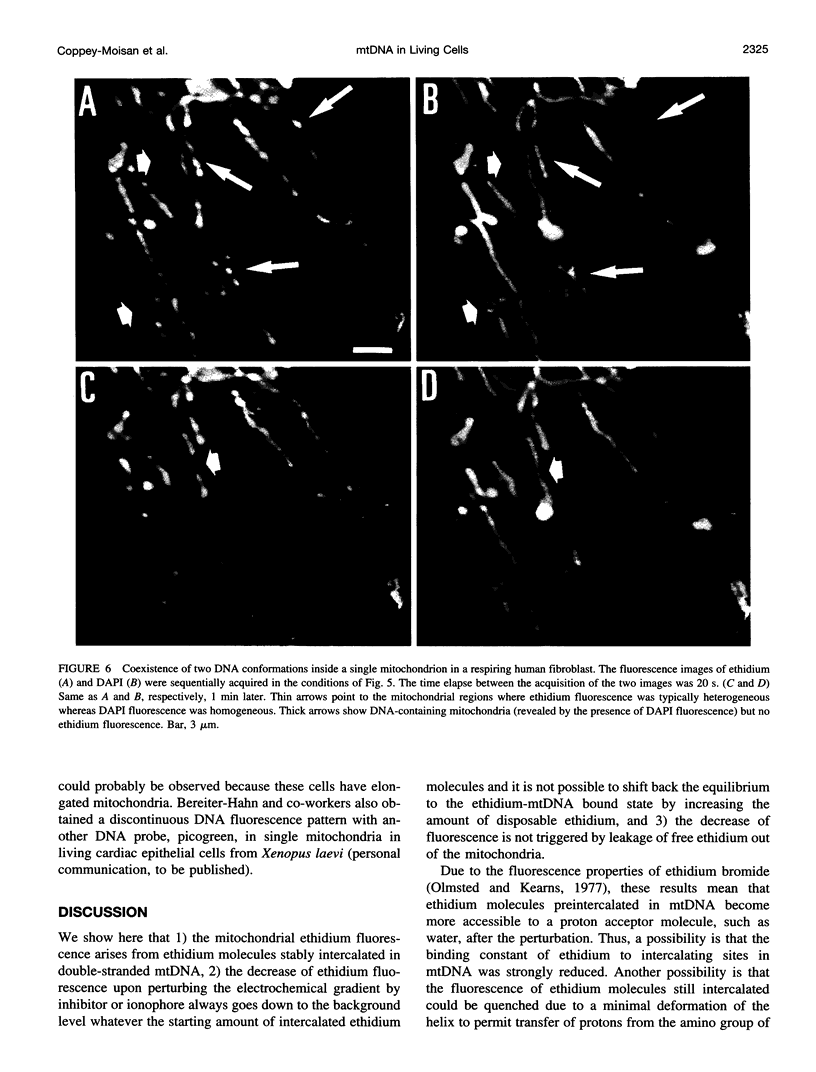
Digital-imaging microscopy was used in conditions that allowed the native state to be preserved and hence fluorescence variations of specific probes to be followed in the real time of living mammalian cells. Ethidium bromide was shown to enter into living cells and to intercalate stably into mitochondrial DNA (mtDNA), giving rise to high fluorescence. When the membrane potential or the pH gradient across the inner membrane was abolished by specific inhibitors or ionophores, the ethidium fluorescence disappeared from all mtDNA molecules within 2 min. After removal of the inhibitors or ionophores, ethidium fluorescence rapidly reappeared in mitochondria, together with the membrane potential. The fluorescence extinction did not result from an equilibrium shift caused by leakage of free ethidium out of mitochondria when the membrane potential was abolished but was most likely due to a dynamical mtDNA change that exposed intercalated ethidium to quencher, either by weakening the ethidium binding constant or by giving access of a proton acceptor (such as water) to the interior of mtDNA. Double labeling with ethidium and with a minor groove probe (4',6-diamino-2-phenylindole) indicated that mtDNA maintains a double-stranded structure. The two double-stranded DNA states, revealed by the fluorescence of mitochondrial ethidium, enhanced or quenched in the presence of ethidium, seem to coexist in mitochondria of unperturbed fibroblast cells, suggesting a spontaneous dynamical change of mtDNA molecules. Therefore, the ethidium fluorescence variation allows changes of DNA to be followed, a property that has to be taken into consideration when using this intercalator for in vivo as well as in vitro imaging studies.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albring M., Griffith J., Attardi G. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1348–1352. doi: 10.1073/pnas.74.4.1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer L. M., Moudrianakis E. N. Interaction of ethidium bromide with whole and selectively deproteinized deoxynucleoproteins from calf thymus. J Mol Biol. 1972 Feb 14;63(3):505–521. doi: 10.1016/0022-2836(72)90444-5. [DOI] [PubMed] [Google Scholar]
- Arbuckle N. D., Luisi B. A recipe for specificity. Nat Struct Biol. 1995 May;2(5):341–346. doi: 10.1038/nsb0595-341. [DOI] [PubMed] [Google Scholar]
- Arscott P. G., Ma C., Wenner J. R., Bloomfield V. A. DNA condensation by cobalt hexaammine (III) in alcohol-water mixtures: dielectric constant and other solvent effects. Biopolymers. 1995 Sep;36(3):345–364. doi: 10.1002/bip.360360309. [DOI] [PubMed] [Google Scholar]
- Baxevanis A. D., Arents G., Moudrianakis E. N., Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995 Jul 25;23(14):2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomfield V. A. Condensation of DNA by multivalent cations: considerations on mechanism. Biopolymers. 1991 Nov;31(13):1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- Bunting J. R., Phan T. V., Kamali E., Dowben R. M. Fluorescent cationic probes of mitochondria. Metrics and mechanism of interaction. Biophys J. 1989 Nov;56(5):979–993. doi: 10.1016/S0006-3495(89)82743-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel P., Lebrun A., Heller C., Lavery R., Viovy J. L., Chatenay D., Caron F. DNA: an extensible molecule. Science. 1996 Feb 9;271(5250):792–794. doi: 10.1126/science.271.5250.792. [DOI] [PubMed] [Google Scholar]
- Coppey-Moisan M., Delic J., Magdelenat H., Coppey J. Principle of digital imaging microscopy. Methods Mol Biol. 1994;33:359–393. doi: 10.1385/0-89603-280-9:359. [DOI] [PubMed] [Google Scholar]
- DeFrancesco L., Attardi G. In situ photochemical crosslinking of HeLa cell mitochondrial DNA by a psoralen derivative reveals a protected region near the origin of replication. Nucleic Acids Res. 1981 Nov 25;9(22):6017–6030. doi: 10.1093/nar/9.22.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic J., Coppey J., Magdelenat H., Coppey-Moisan M. Impossibility of acridine orange intercalation in nuclear DNA of the living cell. Exp Cell Res. 1991 May;194(1):147–153. doi: 10.1016/0014-4827(91)90144-j. [DOI] [PubMed] [Google Scholar]
- Eickbush T. H., Moudrianakis E. N. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978 Feb;13(2):295–306. doi: 10.1016/0092-8674(78)90198-8. [DOI] [PubMed] [Google Scholar]
- Emaus R. K., Grunwald R., Lemasters J. J. Rhodamine 123 as a probe of transmembrane potential in isolated rat-liver mitochondria: spectral and metabolic properties. Biochim Biophys Acta. 1986 Jul 23;850(3):436–448. doi: 10.1016/0005-2728(86)90112-x. [DOI] [PubMed] [Google Scholar]
- Farkas D. L., Wei M. D., Febbroriello P., Carson J. H., Loew L. M. Simultaneous imaging of cell and mitochondrial membrane potentials. Biophys J. 1989 Dec;56(6):1053–1069. doi: 10.1016/S0006-3495(89)82754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. P., Lisowsky T., Parisi M. A., Clayton D. A. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J Biol Chem. 1992 Feb 15;267(5):3358–3367. [PubMed] [Google Scholar]
- Gao M., Knipe D. M. Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA-binding protein. J Virol. 1989 Dec;63(12):5258–5267. doi: 10.1128/jvi.63.12.5258-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Rau D. C. Water release associated with specific binding of gal repressor. EMBO J. 1995 Mar 15;14(6):1257–1263. doi: 10.1002/j.1460-2075.1995.tb07109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand R., Attardi G. Synthesis and turnover of mitochondrial ribonucleic acid in HeLa cells: the mature ribosomal and messenger ribonucleic acid species are metabolically unstable. Mol Cell Biol. 1981 Jun;1(6):497–511. doi: 10.1128/mcb.1.6.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest D., Wahl P. Fluorescence anisotropy decay due to rotational brownian motion of ethidium intercalated in double strand DNA. Biochim Biophys Acta. 1978 Dec 21;521(2):502–509. doi: 10.1016/0005-2787(78)90292-7. [DOI] [PubMed] [Google Scholar]
- Hackenbrock C. R. Chemical and physical fixation of isolated mitochondria in low-energy and high-energy states. Proc Natl Acad Sci U S A. 1968 Oct;61(2):598–605. doi: 10.1073/pnas.61.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenbrock C. R. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J Cell Biol. 1968 May;37(2):345–369. doi: 10.1083/jcb.37.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi J., Takemitsu M., Goto Y., Nonaka I. Human mitochondria and mitochondrial genome function as a single dynamic cellular unit. J Cell Biol. 1994 Apr;125(1):43–50. doi: 10.1083/jcb.125.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Härd T., Kearns D. R. Reduced DNA flexibility in complexes with a type II DNA binding protein. Biochemistry. 1990 Jan 30;29(4):959–965. doi: 10.1021/bi00456a017. [DOI] [PubMed] [Google Scholar]
- Macgregor R. B., Jr, Clegg R. M., Jovin T. M. Viscosity dependence of ethidium-DNA intercalation kinetics. Biochemistry. 1987 Jun 30;26(13):4008–4016. doi: 10.1021/bi00387a040. [DOI] [PubMed] [Google Scholar]
- Markovits J., Ramstein J., Roques B. P., Le Pecq J. B. Effect of B-Z transition and nucleic acid structure on the conformational dynamics of bound ethidium dimer measured by hydrogen deuterium exchange kinetics. Nucleic Acids Res. 1985 May 24;13(10):3773–3788. doi: 10.1093/nar/13.10.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray C. T., van Holde K. E. Binding of ethidium to the nucleosome core particle. 1. Binding and dissociation reactions. Biochemistry. 1991 Jun 11;30(23):5631–5643. doi: 10.1021/bi00237a001. [DOI] [PubMed] [Google Scholar]
- Morais R., Zinkewich-Péotti K., Parent M., Wang H., Babai F., Zollinger M. Tumor-forming ability in athymic nude mice of human cell lines devoid of mitochondrial DNA. Cancer Res. 1994 Jul 15;54(14):3889–3896. [PubMed] [Google Scholar]
- Naimushin A. N., Clendenning J. B., Kim U. S., Song L., Fujimoto B. S., Stewart D. W., Schurr J. M. Effect of ethidium binding and superhelix density on the apparent supercoiling free energy and torsion constant of pBR322 DNA. Biophys Chem. 1994 Nov;52(3):219–226. doi: 10.1016/0301-4622(94)00037-k. [DOI] [PubMed] [Google Scholar]
- Olmsted J., 3rd, Kearns D. R. Mechanism of ethidium bromide fluorescence enhancement on binding to nucleic acids. Biochemistry. 1977 Aug 9;16(16):3647–3654. doi: 10.1021/bi00635a022. [DOI] [PubMed] [Google Scholar]
- Parsegian V. A., Rand R. P., Rau D. C. Macromolecules and water: probing with osmotic stress. Methods Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Preisler R. S., Chen H. H., Colombo M. F., Choe Y., Short B. J., Jr, Rau D. C. The B form to Z form transition of poly(dG-m5dC) is sensitive to neutral solutes through an osmotic stress. Biochemistry. 1995 Nov 7;34(44):14400–14407. doi: 10.1021/bi00044a017. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Yanagi K., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991 Jan 5;217(1):177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- Rau D. C., Parsegian V. A. Direct measurement of the intermolecular forces between counterion-condensed DNA double helices. Evidence for long range attractive hydration forces. Biophys J. 1992 Jan;61(1):246–259. doi: 10.1016/S0006-3495(92)81831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reers M., Smith T. W., Chen L. B. J-aggregate formation of a carbocyanine as a quantitative fluorescent indicator of membrane potential. Biochemistry. 1991 May 7;30(18):4480–4486. doi: 10.1021/bi00232a015. [DOI] [PubMed] [Google Scholar]
- Robert-Nicoud M., Arndt-Jovin D. J., Zarling D. A., Jovin T. M. Immunological detection of left-handed Z DNA in isolated polytene chromosomes. Effects of ionic strength, pH, temperature and topological stress. EMBO J. 1984 Apr;3(4):721–731. doi: 10.1002/j.1460-2075.1984.tb01875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley S. T., Reers M., Mottola-Hartshorn C., Lin M., Chen A., Smith T. W., Steele G. D., Jr, Chen L. B. Intracellular heterogeneity in mitochondrial membrane potentials revealed by a J-aggregate-forming lipophilic cation JC-1. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3671–3675. doi: 10.1073/pnas.88.9.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. B., Cui Y., Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996 Feb 9;271(5250):795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Jain S. C., Gilbert S. G. Visualization of drug-nucleic acid interactions at atomic resolution. III. Unifying structural concepts in understanding drug-DNA interactions and their broader implications in understanding protein-DNA interactions. J Mol Biol. 1977 Aug 15;114(3):333–365. doi: 10.1016/0022-2836(77)90254-6. [DOI] [PubMed] [Google Scholar]
- Viegas-Pequignot E., Dutrillaux B., Magdelenat H., Coppey-Moisan M. Mapping of single-copy DNA sequences on human chromosomes by in situ hybridization with biotinylated probes: enhancement of detection sensitivity by intensified-fluorescence digital-imaging microscopy. Proc Natl Acad Sci U S A. 1989 Jan;86(2):582–586. doi: 10.1073/pnas.86.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M. H., Gronenborn A. M., Clore G. M. Intercalation, DNA kinking, and the control of transcription. Science. 1996 Feb 9;271(5250):778–784. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- Widom J., Baldwin R. L. Inhibition of cation-induced DNA condensation by intercalating dyes. Biopolymers. 1983 Jun;22(6):1621–1632. doi: 10.1002/bip.360220613. [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Small E. W. Fluorescence anisotropy decay of ethidium bound to nucleosome core particles. 2. The torsional motion of the DNA is highly constrained and sensitive to pH. Biochemistry. 1991 May 28;30(21):5304–5313. doi: 10.1021/bi00235a025. [DOI] [PubMed] [Google Scholar]
- Zylber E., Vesco C., Penman S. Selective inhibition of the synthesis of mitochondria-associated RNA by ethidium bromide. J Mol Biol. 1969 Aug 28;44(1):195–204. doi: 10.1016/0022-2836(69)90414-8. [DOI] [PubMed] [Google Scholar]
- van de Sande J. H., McIntosh L. P., Jovin T. M. Mn2+ and other transition metals at low concentration induce the right-to-left helical transformation of poly[d(G-C)]. EMBO J. 1982;1(7):777–782. doi: 10.1002/j.1460-2075.1982.tb01247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]