Abstract
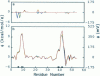
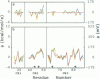
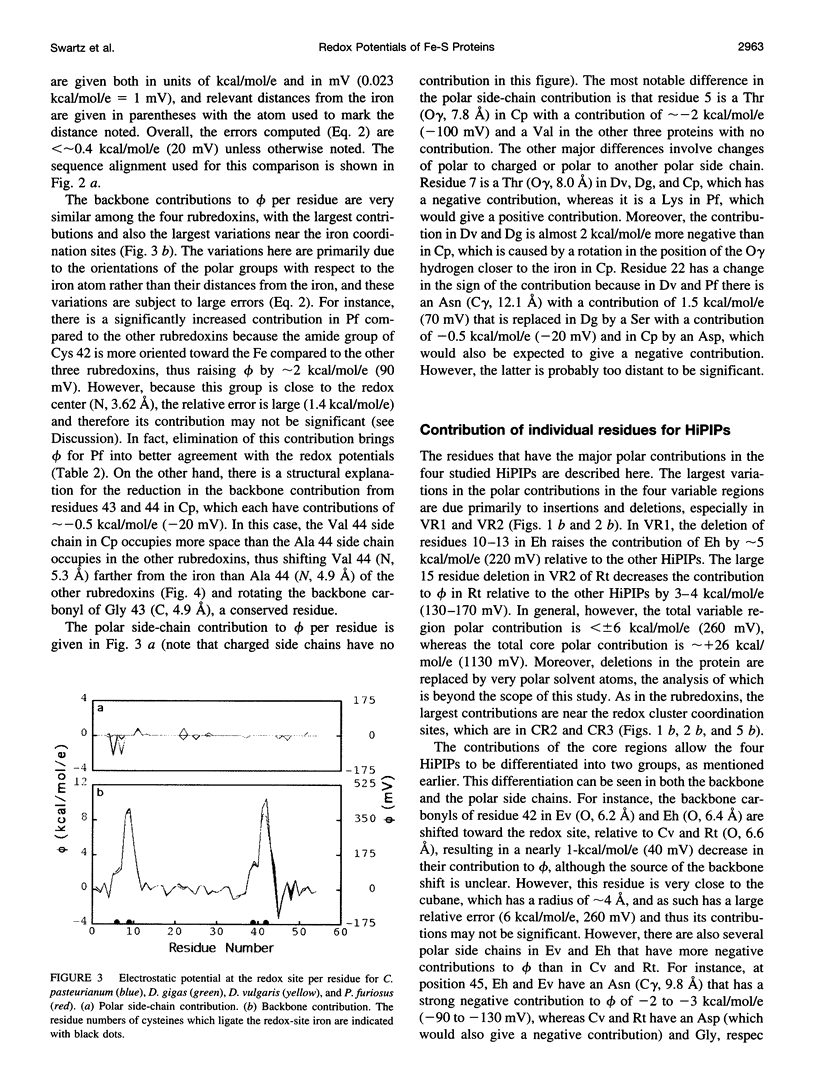
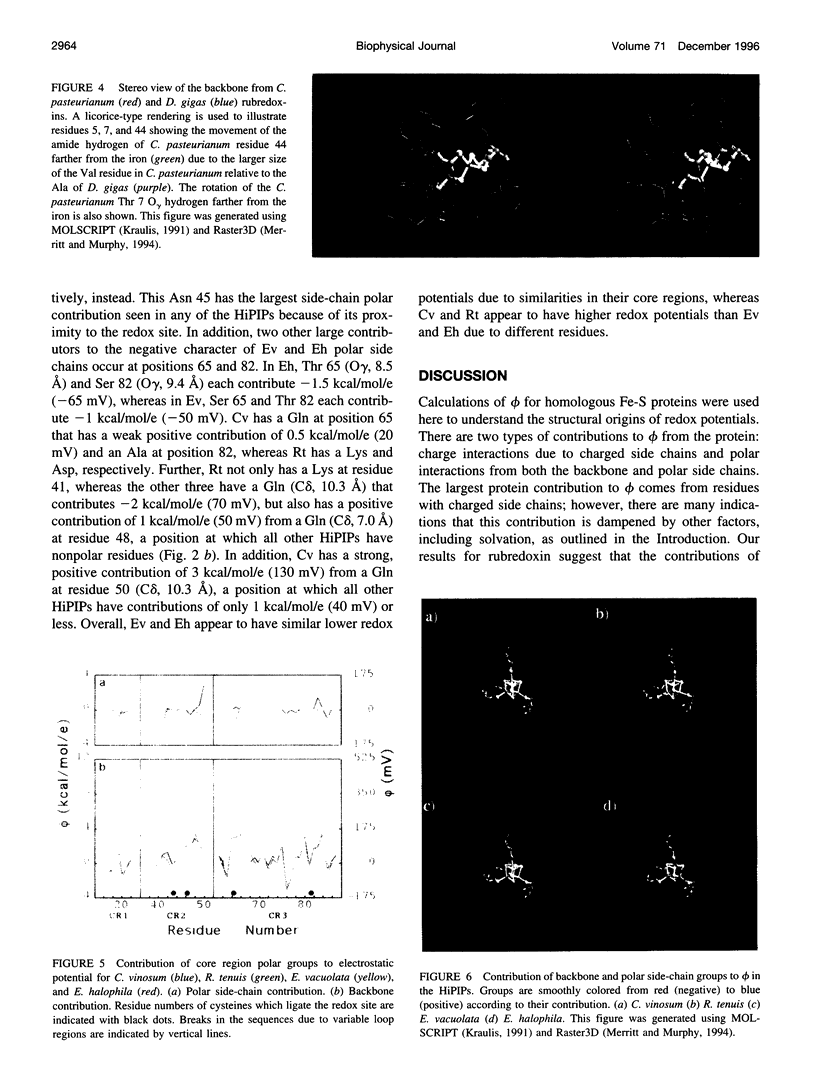
Redox potentials often differ dramatically for homologous proteins that have identical redox centers. For two types of iron-sulfur proteins, the rubredoxins and the high-potential iron-sulfur proteins (HiPIPs), no structural explanations for these differences have been found. We calculated the classical electrostatic potential at the redox site using static crystal structures of four rubredoxins and four HiPIPs to identify important structural determinants of their redox potentials. The contributions from just the backbone and polar side chains are shown to explain major features of the experimental redox potentials. For instance, in the rubredoxins, the presence of Val 44 versus Ala 44 causes a backbone shift that explains a approximately 50 mV lower redox potential in one of the four rubredoxins. This result is consistent with experimental redox potentials of five additional rubredoxins with known sequence. Also, we attribute the unusually lower redox potentials of two of the HiPIPs studied to a less positive electrostatic environment around their redox sites. Finally, molecular dynamics simulations of solvent around static rubredoxin crystal structures indicate that water alone is a major factor in dampening the contribution of charged side chains, in accord with experiments showing that mutations of surface charges produce relatively little effect on redox potentials.
Full text
PDF
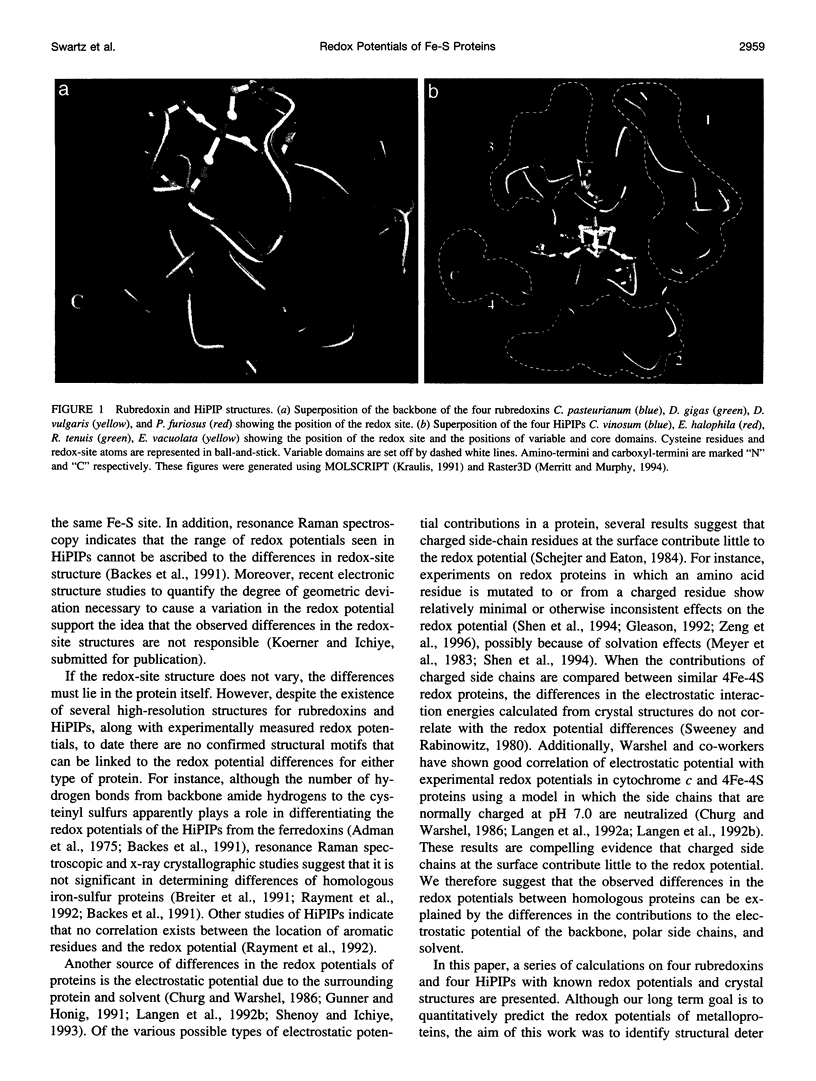
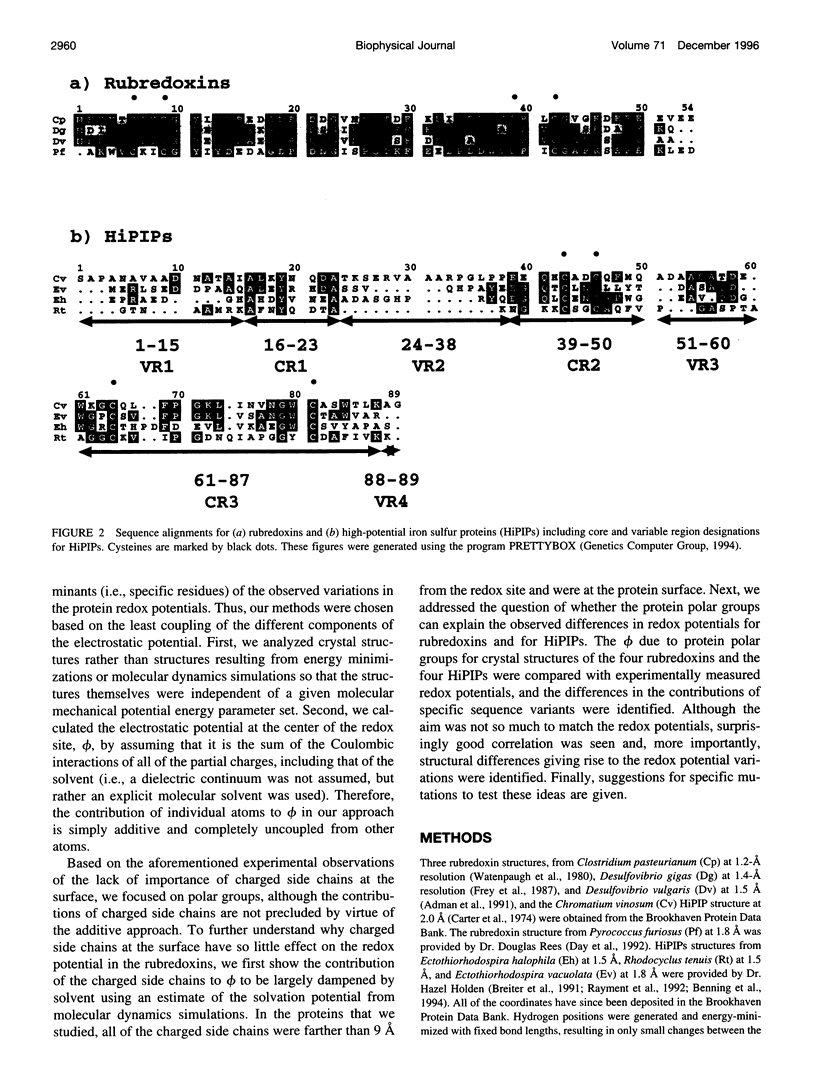

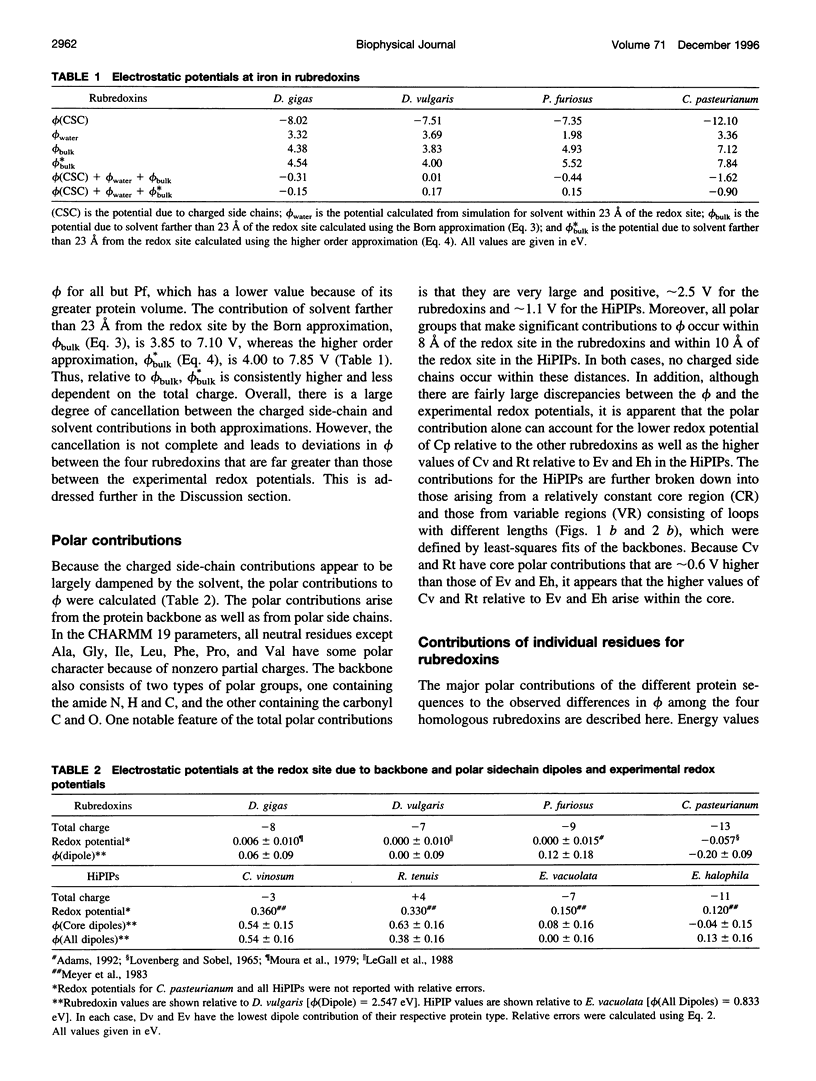





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T., Sieker L. C., Jensen L. H. Structure of rubredoxin from Desulfovibrio vulgaris at 1.5 A resolution. J Mol Biol. 1991 Jan 20;217(2):337–352. doi: 10.1016/0022-2836(91)90547-j. [DOI] [PubMed] [Google Scholar]
- Adman E., Watenpaugh K. D., Jensen L. H. NH---S hydrogen bonds in Peptococcus aerogenes ferredoxin, Clostridium pasteurianum rubredoxin, and Chromatium high potential iron protein. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4854–4858. doi: 10.1073/pnas.72.12.4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning M. M., Meyer T. E., Rayment I., Holden H. M. Molecular structure of the oxidized high-potential iron-sulfur protein isolated from Ectothiorhodospira vacuolata. Biochemistry. 1994 Mar 8;33(9):2476–2483. doi: 10.1021/bi00175a016. [DOI] [PubMed] [Google Scholar]
- Breiter D. R., Meyer T. E., Rayment I., Holden H. M. The molecular structure of the high potential iron-sulfur protein isolated from Ectothiorhodospira halophila determined at 2.5-A resolution. J Biol Chem. 1991 Oct 5;266(28):18660–18667. doi: 10.2210/pdb2hip/pdb. [DOI] [PubMed] [Google Scholar]
- Carter C. W., Jr, Kraut J., Freer S. T., Nguyen-Huu-Xuong, Alden R. A., Bartsch R. G. Two-Angstrom crystal structure of oxidized Chromatium high potential iron protein. J Biol Chem. 1974 Jul 10;249(13):4212–4225. [PubMed] [Google Scholar]
- Churg A. K., Warshel A. Control of the redox potential of cytochrome c and microscopic dielectric effects in proteins. Biochemistry. 1986 Apr 8;25(7):1675–1681. doi: 10.1021/bi00355a035. [DOI] [PubMed] [Google Scholar]
- Day M. W., Hsu B. T., Joshua-Tor L., Park J. B., Zhou Z. H., Adams M. W., Rees D. C. X-ray crystal structures of the oxidized and reduced forms of the rubredoxin from the marine hyperthermophilic archaebacterium Pyrococcus furiosus. Protein Sci. 1992 Nov;1(11):1494–1507. doi: 10.1002/pro.5560011111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey M., Sieker L., Payan F., Haser R., Bruschi M., Pepe G., LeGall J. Rubredoxin from Desulfovibrio gigas. A molecular model of the oxidized form at 1.4 A resolution. J Mol Biol. 1987 Oct 5;197(3):525–541. doi: 10.1016/0022-2836(87)90562-6. [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Gleason F. K. Mutation of conserved residues in Escherichia coli thioredoxin: effects on stability and function. Protein Sci. 1992 May;1(5):609–616. doi: 10.1002/pro.5560010507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunner M. R., Honig B. Electrostatic control of midpoint potentials in the cytochrome subunit of the Rhodopseudomonas viridis reaction center. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9151–9155. doi: 10.1073/pnas.88.20.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen G. M., Warshel A., Stephens P. J. Calculation of the redox potentials of iron-sulfur proteins: the 2-/3-couple of [Fe4S*4Cys4] clusters in Peptococcus aerogenes ferredoxin, Azotobacter vinelandii ferredoxin I, and Chromatium vinosum high-potential iron protein. Biochemistry. 1994 Sep 13;33(36):10911–10924. doi: 10.1021/bi00202a010. [DOI] [PubMed] [Google Scholar]
- Langen R., Brayer G. D., Berghuis A. M., McLendon G., Sherman F., Warshel A. Effect of the Asn52----Ile mutation on the redox potential of yeast cytochrome c. Theory and experiment. J Mol Biol. 1992 Apr 5;224(3):589–600. doi: 10.1016/0022-2836(92)90546-v. [DOI] [PubMed] [Google Scholar]
- Langen R., Jensen G. M., Jacob U., Stephens P. J., Warshel A. Protein control of iron-sulfur cluster redox potentials. J Biol Chem. 1992 Dec 25;267(36):25625–25627. [PubMed] [Google Scholar]
- LeGall J., Prickril B. C., Moura I., Xavier A. V., Moura J. J., Huynh B. H. Isolation and characterization of rubrerythrin, a non-heme iron protein from Desulfovibrio vulgaris that contains rubredoxin centers and a hemerythrin-like binuclear iron cluster. Biochemistry. 1988 Mar 8;27(5):1636–1642. doi: 10.1021/bi00405a037. [DOI] [PubMed] [Google Scholar]
- Lee W. Y., Brune D. C., LoBrutto R., Blankenship R. E. Isolation, characterization, and primary structure of rubredoxin from the photosynthetic bacterium, Heliobacillus mobilis. Arch Biochem Biophys. 1995 Apr 1;318(1):80–88. doi: 10.1006/abbi.1995.1207. [DOI] [PubMed] [Google Scholar]
- Lovenberg W., Sobel B. E. Rubredoxin: a new electron transfer protein from Clostridium pasteurianum. Proc Natl Acad Sci U S A. 1965 Jul;54(1):193–199. doi: 10.1073/pnas.54.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt E. A., Murphy M. E. Raster3D Version 2.0. A program for photorealistic molecular graphics. Acta Crystallogr D Biol Crystallogr. 1994 Nov 1;50(Pt 6):869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- Meyer T. E., Przysiecki C. T., Watkins J. A., Bhattacharyya A., Simondsen R. P., Cusanovich M. A., Tollin G. Correlation between rate constant for reduction and redox potential as a basis for systematic investigation of reaction mechanisms of electron transfer proteins. Proc Natl Acad Sci U S A. 1983 Nov;80(22):6740–6744. doi: 10.1073/pnas.80.22.6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moura I., Moura J. J., Santos M. H., Xavier A. V., Le Gall J. Redox studies on rubredoxins from sulphate and sulphur reducing bacteria. FEBS Lett. 1979 Nov 15;107(2):419–421. doi: 10.1016/0014-5793(79)80421-4. [DOI] [PubMed] [Google Scholar]
- Parson W. W., Chu Z. T., Warshel A. Electrostatic control of charge separation in bacterial photosynthesis. Biochim Biophys Acta. 1990 Jun 26;1017(3):251–272. doi: 10.1016/0005-2728(90)90192-7. [DOI] [PubMed] [Google Scholar]
- Rayment I., Wesenberg G., Meyer T. E., Cusanovich M. A., Holden H. M. Three-dimensional structure of the high-potential iron-sulfur protein isolated from the purple phototrophic bacterium Rhodocyclus tenuis determined and refined at 1.5 A resolution. J Mol Biol. 1992 Nov 20;228(2):672–686. doi: 10.1016/0022-2836(92)90849-f. [DOI] [PubMed] [Google Scholar]
- Shen B., Jollie D. R., Stout C. D., Diller T. C., Armstrong F. A., Gorst C. M., La Mar G. N., Stephens P. J., Burgess B. K. Azotobacter vinelandii ferredoxin I. Alteration of individual surface charges and the [4FE-4S]2+/+ cluster reduction potential. J Biol Chem. 1994 Mar 18;269(11):8564–8575. [PubMed] [Google Scholar]
- Shenoy V. S., Ichiye T. Influence of protein flexibility on the redox potential of rubredoxin: energy minimization studies. Proteins. 1993 Oct;17(2):152–160. doi: 10.1002/prot.340170205. [DOI] [PubMed] [Google Scholar]
- Sieker L. C., Stenkamp R. E., Jensen L. H., Prickril B., LeGall J. Structure of rubredoxin from the bacterium Desulfovibrio desulfuricans. FEBS Lett. 1986 Nov 10;208(1):73–76. doi: 10.1016/0014-5793(86)81535-6. [DOI] [PubMed] [Google Scholar]
- Sweeney W. V., Rabinowitz J. C. Proteins containing 4Fe-4S clusters: an overview. Annu Rev Biochem. 1980;49:139–161. doi: 10.1146/annurev.bi.49.070180.001035. [DOI] [PubMed] [Google Scholar]
- Watenpaugh K. D., Sieker L. C., Jensen L. H. Crystallographic refinement of rubredoxin at 1 x 2 A degrees resolution. J Mol Biol. 1980 Apr 15;138(3):615–633. doi: 10.1016/s0022-2836(80)80020-9. [DOI] [PubMed] [Google Scholar]
- Yelle R. B., Park N. S., Ichiye T. Molecular dynamics simulations of rubredoxin from Clostridium pasteurianum: changes in structure and electrostatic potential during redox reactions. Proteins. 1995 Jun;22(2):154–167. doi: 10.1002/prot.340220208. [DOI] [PubMed] [Google Scholar]