Abstract
Replication protein A (RPA) is a single-stranded-DNA-binding protein composed of three subunits with molecular masses of 70, 32, and 14 kDa. The protein is involved in multiple processes of eukaryotic DNA metabolism, including DNA replication, repair, and recombination. In Xenopus, Xenopus RPA-interacting protein α has been identified as a carrier molecule of RPA into the nucleus. In this study, human RPA-interacting protein α (hRIPα) and five novel splice isoforms (named hRIPα, hRIPβ, hRIPγ, hRIPδ1, hRIPδ2, and hRIPδ3 according to the lengths of their encoding peptides) were cloned. Among hRIP isoforms, hRIPα and hRIPβ were found to be the major splice isoforms and to show different subcellular localizations. While hRIPα localized to the cytoplasm, hRIPβ was found in the PML nuclear body. Modification of hRIPβ by sumoylation was found to be required for localization to the PML nuclear body. The results of the present work demonstrate that hRIPβ transports RPA into the PML nuclear body and releases RPA upon UV irradiation. hRIPβ thus plays an important role in RPA deposition in PML nuclear bodies and thereby supplements RPA for DNA metabolism.
Replication protein A (RPA) (also known as replication factor A) is a single-stranded-DNA-binding protein that is composed of three subunits of 70, 32, and 14 kDa and involved in multiple processes of eukaryotic DNA metabolism, including DNA replication, repair, and recombination (10, 39, 40). RPA binds tightly to single-stranded DNA but also interacts with double-stranded DNA with much lower affinity (21, 41). This binding activity of RPA has been localized to the 70-kDa subunit (20, 25, 41).
Several groups have reported the 32-kDa subunit of RPA to be phosphorylated in a cell cycle-dependent manner (8, 11) or in response to DNA-damaging reagents such as ionizing radiation, UV radiation, and camptothecin (5, 23, 26, 29, 33, 37, 38). The cyclin-dependent kinase family plays a central role in phosphorylating RPA p32 for the regulation of DNA replication and for progression through the cell cycle (9, 11, 12). Several protein kinases, including DNA-dependent protein kinase and ataxia-telanogiectasia mutated, have been implicated in the DNA damage-induced phosphorylation of RPA p32 (3, 4, 13, 24, 26, 29).
Since RPA localizes primarily to the nucleus, RPA should be transported into the nucleus by the cellular transport machinery. Yeast two-hybrid screening identified Xenopus RPA-interacting protein α (xRIPα) as a carrier molecule for the import of RPA into the nucleus (19). Specifically, xRIPα was found to serve as an adapter molecule, linking RPA to importin β just as importin α links nuclear localization signal-containing proteins to importin β.
The xRIPα protein can be divided into three subdomains (19): the N-terminal domain (amino acids 1 to 45) is rich in basic residues, the central domain (amino acids 46 to 140) is acidic, and the C terminus contains a putative zinc finger domain (amino acids 141 to 226). While the N-terminal domain is responsible for interaction with importin β, the central region interacts with RPA (19).
Because xRIPα is conserved among higher eukaryotes but not in yeast, higher eukaryotes and yeast employ divergent pathways for the import of RPA. In yeast, Msn5p protein was identified as a carrier molecule that imports RPA into the nucleus but is also involved in the export of other cargo molecules (42).
The nucleus is compartmentalized into highly organized structures, with numerous subnuclear structures identified by their biochemical function (44). The PML (promyelocytic leukemia) nuclear body is one of these subnuclear structures and has a diameter of between 0.2 and 1 μm (44). Cells generally contain 10 to 30 PML bodies per nucleus. The formation of PML nuclear bodies is dependent on the PML gene, with inactivation of the PML gene resulting in complete disruption of PML nuclear bodies (18, 43). Several PML nuclear body components have recently been identified.
While PML nuclear bodies are present in many cells, they are not essential for cell survival. The nonessential nature of the PML nuclear bodies suggests that they are regulated depots in the nucleus (28). Negorev and Maul postulated a nuclear defense mechanism whereby PML nuclear bodies recruit specific proteins and release them upon an external stress such as heat shock or UV irradiation (28). Supporting this hypothesis of a defensive role for PML nuclear bodies, Bischof et al. (2) found them to contain several proteins associated with DNA metabolism and, specifically, with DNA repair and recombination.
The PML nuclear body is considered to be a nuclear organelle, although it is more accurately described as an aggregate of PML-associated proteins and its formation is dependent on the specific interactions of these aggregated proteins. Sumoylation plays an important role in PML nuclear body formation, as demonstrated by a PML-3K mutant lacking SUMO modification sites (43). The PML-3K mutant aberrantly localizes to the nucleus and fails to recruit nuclear body proteins such as Sp100. Sumoylation of PML bodies is further associated with release of various proteins from the PML nuclear body. Such external stressors as heat shock induce the desumoylation of PML nuclear body proteins and lead to their release (27). A SUMO isopeptidase, SENP-1, has been identified as one component of PML-associated proteins (14). SENP-1 removes SUMO-1 conjugates from PML body-associated proteins and releases them from the PML nuclear body (27, 28).
Recently, it was reported that a fraction of RPA localizes to the PML nuclear body with other components of the DNA repair machinery (1, 2, 31). Additionally, DNA-damaging agents such as ionizing radiation and UV were shown to lead to the translocation of RPA to the site of DNA damage (1, 45). The present study demonstrated that the hRIPα gene encodes human RIPα (hRIPα) and that alternative splicing leads to multiple isoforms of this protein. Further, the splice isoforms of hRIPα were found to deposit RPA into PML nuclear bodies and then release RPA upon UV irradiation.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa, HEK293T, U2OS, and COS cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum; Jurkat and BJAB cells were grown in RPMI medium also supplemented with 10% fetal bovine serum. Transfections of HEK293T and COS cells were carried out using the standard calcium phosphate method (15). Leptomycin B and actinomycin D were purchased from Sigma (St. Louis, MO) and used at final concentrations of 20 nM and 5 μg/ml, respectively. Jurkat cell synchronization was performed by the thymidine double block as shown previously (17). For UV treatment, cells were irradiated with 254-nm UV light from a Spectrolinker XL-1000 (Spectroline, Westbury, NY). Because UV light of this wavelength penetrates poorly into water, the medium was replaced with a thin layer of phosphate-buffered saline (PBS) during UV irradiation.
Immunofluorescence and confocal microscopy.
Cells were grown on sterilized glass coverslips, fixed with 4% paraformaldehyde, and blocked with 0.1% bovine serum albumin in PBS. Cells were stained with 1:500-diluted primary antibody in PBS and then reacted with 1:5,000-diluted Alexa 488-conjugated or Alexa 568-conjugated secondary antibody (Vector, Burlingame, CA). Finally, slides were washed three times with PBS and mounted in mounting medium (Vector). Images were captured with a Zeiss confocal microscope (Oberkochen, Germany). For counterstaining, slides were incubated in 100 μg/ml DNase-free RNase for 30 min at room temperature and then stained with 500 nM propidium iodide (Sigma) in PBS.
cDNA cloning and plasmid construction.
Reverse transcription-PCR (RT-PCR) was used to amplify the entire coding region of hRIPα and its splicing variants from Jurkat-T RNA, using the 5′ primer GGGCTCGAGATGGCGGAGTCGTTGAGGTCTC and the 3′ primer GGGTCTAGACTAGAGGATCACAGCCCAAG. The amplified cDNA fragments were ligated into pcDNA3/HA and completely sequenced. After sequencing, cDNA for hRIPα or hRIPβ was subcloned into pGEX4T-1 and pEBG plasmids. Point mutants of hRIPα and hRIPβ were generated using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA), and each mutant was completely sequenced to verify the presence of the intended mutation and the absence of any others.
Human RPA p70 cDNA was kindly provided by M. S. Wold (16). To generate FLAG-tagged RPA p70, RPA p70 cDNA was amplified by PCR and subcloned into pME18S plasmids (34). The plasmid encoding His-tagged SENP1 was provided by C. Y. Choi (SKKU, Korea), the importin β cDNA was provided by D. Gorlich (22), and SENP1 small interfering RNA (siRNA) was provided by E. T. Yeh (6, 7).
Antibody production, immunoprecipitation, and immunoblotting.
Glutathione S-transferase (GST)-hRIPα fusion protein was used as a source antigen for anti-hRIP antibody production. The GST fusion protein was prepared as described previously (35) and used to immunize rabbits with mixed Freund's adjuvant (Sigma). Nonspecific antibodies were removed from anti-hRIP antibody to produce a purified mechanism for the detection of the endogenous protein. Anti-hRIP antibody was mixed with GST protein-bound beads for 10 min at 4°C. After brief centrifugation, the supernatant was transferred and used for immunostaining. Anti-hRIPAS (alpha-specific) antibody was generated using an GST-hRIPα deletion mutant (amino acids 107 to 219), and the antibody, which recognized GST-hRIPβ recombinant protein, was removed to obtain specificity to hRIPα as described above. Antibodies for RPA p70 and p32 were generously provided by B. Stillman (8), while antibodies for PML and lamin were purchased from Santa Cruz (Santa Cruz, CA) and antibody for SENP1 was purchased from IMGENEX (San Diego, CA).
For immunoprecipitation, cells were harvested and resuspended in lysis buffer (150 mM NaCl, 50 mM HEPES [pH 8.0], 0.5% NP-40) containing a protease inhibitor cocktail (Roche, Germany). Immunoprecipitated proteins from precleared cell lysates were used for immunoblotting. Immunoblot detection was performed with a 1:1,000 or 1:2,000 dilution of primary antibody and an enhanced chemiluminescence system (ECL; Amersham, Chicago, IL). To detect the sumoylated protein, cells were washed with PBS and directly lysed with sodium dodecyl sulfate (SDS) loading buffer (100 mM Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 0.001% bromophenol blue) supplemented with 20 μM N-ethylmaleimide (Sigma). Finally, cell lysates were boiled for 5 min, spun in a centrifuge for 10 min at 13,000 rpm, and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Subcellular fractionation.
Cells were harvested with 500 μl CLB buffer (10 mM Tris-HCl [pH 7.5], 10 mM NaCl, 1 mM KH2PO4, 5 mM NaHCO3, 1 mM CaCl2, 0.5 mM MgCl2) with a protease inhibitor cocktail. Cells were allowed to swell for 5 min and were then homogenized 50 times using a Dounce homogenizer (Wheaton, Millville, IL). Cell lysates were spun for 10 min at 7,500 rpm in a centrifuge at 4°C, and the cytoplasmic fraction was transferred to a clean tube. The nuclear pellet was washed once with CLB buffer before being lysed by addition of SDS loading buffer.
Nucleotide sequence accession numbers.
The nucleotide and deduced amino acid sequences of the seven novel cDNAs identified in this study have been registered in GenBank (accession numbers AY680654 to AY680660).
RESULTS
Cloning of hRIPα and its isoforms.
Searches of the expressed sequence tag database identified the hRIPα protein as a human homologue of xRIPα, with the two proteins displaying similar domain structures (19). hRIPα cDNA was obtained by RT-PCR using RNA extracted from Jurkat cells as a template and primers complementary to the 5′ and 3′ ends of the hRIPα open reading frame, which encodes the full-length hRIPα protein (Fig. 1A).
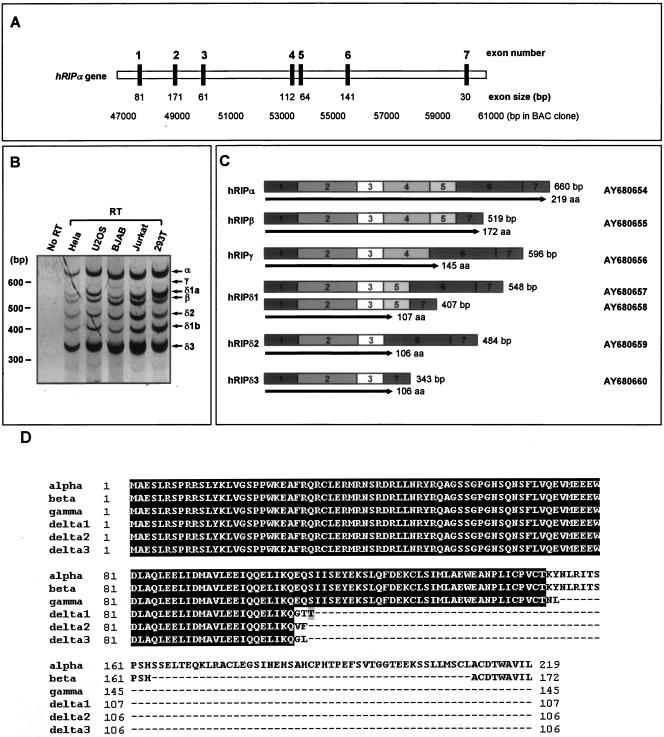
FIG. 1.
hRIPα gene structure and alternative splicing. (A) Schematic diagram of the hRIPα gene and the alternative splicing that leads to its multiple isoforms. Coding exons are indicated by black boxes, as mapped on bacterial artificial chromosome (BAC) clone HCIT524C5. (B) Alternative splicing of the hRIPα gene. The mRNA transcripts of hRIPα were analyzed by reverse transcription-PCR using total RNA from the indicated human cell lines. (C) Structure of alternative splicing products of the hRIPα gene. Exons are shown as boxes, while introns have been omitted. The protein products are shown as arrows under each isoform. Seven novel cDNAs were identified, and their GenBank accession numbers are indicated. aa, amino acids. (D) Alignment of hRIPα and its isoforms. hRIPα, hRIPβ, hRIPγ, hRIPδ1, hRIPδ2, and hRIPδ3 were aligned using the CLUSTALW multiple-alignment program. Identical sequences are shaded. N-terminal sequences (residues 1 to 104) are conserved among all isoforms.
RT-PCR analysis revealed that hRIPα yields seven splice isoforms, and various cell lines, including HeLa, U2OS, BJAB, Jurkat, and HEK293T, were employed to verify expression of the alternatively spliced mRNAs (Fig. 1B). Characterization of the splice isoforms of hRIPα was accomplished by cloning the various PCR products into a hemagglutinin (HA)-tagged eukaryotic expression vector (pcDNA3/HA) and obtaining their nucleotide sequences by standard methods. Seven isoforms were cloned, and nucleotide sequencing indicated that each was derived from a unique combination of hRIPα exons 1 through 7 (Fig. 1B and C).
Any one of the seven clones can potentially encode the polypeptide, and further analysis was necessary. The present work yielded seven different cDNA clones, but two of these (hRIPδ1a and hRIPδ1b) were found to encode the same polypeptide (hRIPδ1). Thus, a total of six polypeptides were produced through alternative splicing of the hRIPα gene (Fig. 1B and C). The longest gene product was the hRIPα protein, and the smaller polypeptides were named hRIPβ, hRIPγ, hRIPδ1, hRIPδ2, and hRIPδ3 in order of decreasing length. While hRIPα and hRIPβ are terminated by the same codon, the termination codons of the remaining clones are produced by a frameshift within their cDNA sequences. hRIPδ1, hRIPδ2, and hRIPδ3 have been grouped into the hRIPδ family, as they differ by only 2 or 3 amino acids while sharing 104 amino acids (Fig. 1D).
hRIPβ localizes to the PML nuclear body.
In order to determine which of the splice variants indeed encode the protein of interest, hRIPα and its various splice isoforms were expressed in rabbit reticulocytes in the presence of [35S]methionine. PAGE revealed that hRIPα and all of its isoforms could be translated into a protein of the appropriate size in vitro (Fig. 2A). Their expression in the eukaryotic cell was verified by transfection of plasmids encoding hRIPα and its isoforms into HEK293T cells and subsequent immunoblot analysis with anti-HA antibody. While hRIPα and hRIPβ proteins were readily detected, the expression of other isoforms was low, indicating that hRIPα and hRIPβ are the major splice isoforms in vivo (Fig. 2B). Frequently throughout the experiment reported here, hRIPβ expression resulted in a shifted band at around 45 kDa that will be discussed further in the next section.
FIG. 2.
Expression analysis of hRIPα and its isoforms and differential localization of hRIPα and hRIPβ. (A) In vitro translation of hRIPα and its isoforms. hRIPα and its isoforms were translated in vitro using [35S]methionine and analyzed by 15% SDS-PAGE. (B) Transient expression of hRIPα and its isoforms. Plasmids encoding hRIPα and its isoforms were transiently transfected into HEK293T cells. Equal amounts of protein were subjected to SDS-PAGE, and hRIPα and its isoforms were identified by immunoblotting (IB) with anti-HA antibody. Nontransfected cell lysate was also probed with anti-hRIP antibody and anti-hRIPAS antibody to show expression of the endogenous proteins. The star indicates nonspecific bands. (C) Differential localization of hRIPα and hRIPβ. HEK293T cells or COS cells were transiently transfected with plasmids encoding hRIPα or hRIPβ. The transfected cells were fixed and then immunostained with anti-hRIP or anti-HA antibody. DNA was visualized using propidium iodide staining. Bars, 10 μm. (D) Subcellular fractionation of hRIPα- and hRIPβ-transfected cells. Extracts from transfected cells were fractionated to give cytoplasmic (C) and nuclear (N) fractions as described in Materials and Methods. Aliquots of 5% of each fraction were immunoblotted with anti-HA (upper panel) and antilamin (bottom panel) antibodies. (E) hRIPβ localizes to the PML nuclear body. Cells were prepared as for panel A and immunostained with anti-hRIP antibody (green) and anti-PML antibody (red).
Endogenous expression of hRIPα and hRIPβ was demonstrated by generating a rabbit polyclonal antibody against bacterially expressed GST-hRIPα recombinant protein that contained full-length hRIPα and was named hRIP antibody. Additionally, an hRIPα-specific antibody was generated using hRIPα-specific exon 6 domain to recognize the endogenous hRIPα expression and was named hRIPAS (alpha-specific) antibody. A GST-hRIPα deletion mutant (amino acid 107 to 219) was used to immunize rabbits, and the antibody, which recognized GST-hRIPβ recombinant protein, was removed to obtain specificity to hRIPα as described in Materials and Methods. Immunoblotting with the hRIP antibody revealed that it could recognize the double bands near 30 kDa as well as the 45-kDa band. The larger band is consistent with the shifted band of HA-hRIPβ, while the two smaller bands approximate HA-hRIPα and HA-hRIPβ. Finally, immunoblotting with the hRIPAS antibody demonstrated that the upper of the smaller two bands is hRIPα. These findings strongly support hRIPα and hRIPβ as the major translation products of the hRIPα gene (Fig. 2B).
Since hRIPα and hRIPβ are predominantly expressed in eukaryotic cells, the initial focus here was on the subcellular localization of these two isoforms. Plasmids encoding HA-hRIPα and HA-hRIPβ were transiently expressed in HEK293T cells, which were then immunostained with anti-hRIP antibody. Confocal microscopy revealed hRIPα to be localized primarily in the cytoplasm but also detectable in the nucleus. By contrast, the majority of hRIPβ was localized to the nucleus with a dot-like staining pattern (Fig. 2C). These results were confirmed in an analogous experiment with COS cells and anti-HA antibody (Fig. 2C). The distinct subcellular localizations of hRIPα and hRIPβ were corroborated by transient expression of hRIPα and hRIPβ in cells that were subjected to cellular fractionation to give cytoplasmic and nuclear fractions. While the majority of hRIPα proteins were detected in the cytoplasmic fraction, hRIPβ proteins were mostly found in the nuclear fraction (Fig. 2D). Taken together, these results indicate that hRIPα and hRIPβ indeed display distinct subcellular localization patterns.
After observing the dot-like localization of hRIPβ in the nucleus, investigation shifted to further characterizing these dot-like structures. Previous studies had documented that RPA, the binding partner of hRIPα, was often found in PML nuclear bodies (1, 2, 31), and because of this finding it was postulated here that the observed nuclear spots may have indeed been PML nuclear bodies. This hypothesis was tested by transfecting plasmids encoding hRIPα and hRIPβ into HEK293T cells and then double immunostaining with anti-hRIP antibody (rabbit, green) and anti-PML antibody (mouse, red). Confocal microscopy revealed that more than 90% of hRIPβ foci were completely associated with PML spots in the transiently transfected cells, indicating that hRIPβ localized to the PML nuclear body (Fig. 2E).
Sumoylation of hRIPβ.
When hRIPβ was transiently expressed in HEK293T cells, there was repeated detection of a more slowly migrating form of hRIPβ (slower by ∼20 kDa). This band was also detected by anti-hRIP blotting using endogenous proteins (Fig. 2B). Because many proteins in the PML nuclear body are modified by sumoylation (28, 44), it was assumed that the larger band was the sumoylated form of hRIPβ. This hypothesis was investigated by determining whether hRIPβ was in fact sumoylated in vivo.
Transfections were performed that paired plasmids encoding HA-hRIPα and HA-hRIPβ with plasmids for FLAG-SUMO-1 and thus revealed if conjugation was taking place. Figure 3A shows that the more slowly migrating form of hRIPβ was detected by both anti-HA and anti-FLAG antibodies but was not detected with anti-FLAG antibody in the absence of FLAG-SUMO-1 expression. These findings indicate that the slower-migrating hRIPβ band is the sumoylation product of the protein.
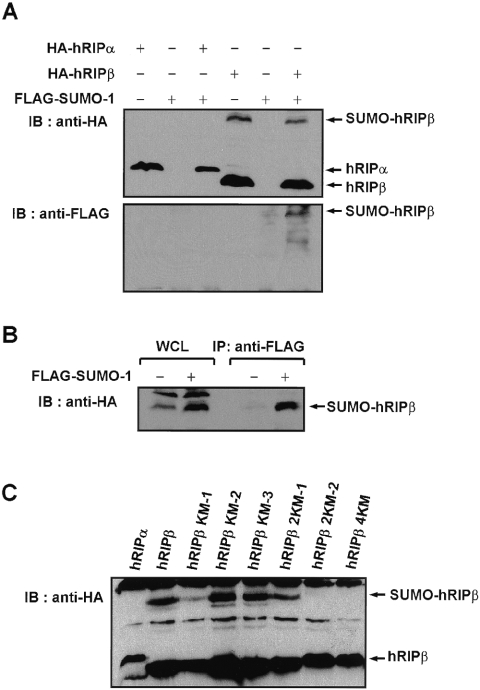
FIG. 3.
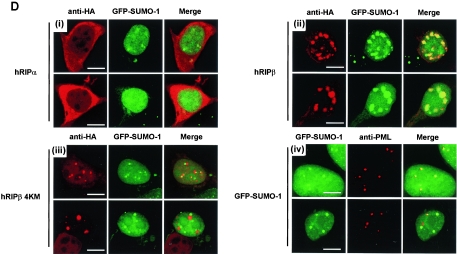
Sumoylation of hRIPβ and generation of SUMO-deficient hRIPβ mutants. (A) Sumoylation of hRIPβ. HEK293T cells were transiently transfected with plasmids encoding hRIPα or hRIPβ in combination with a plasmid encoding FLAG-SUMO-1. Cell lysates were probed with anti-HA antibody (upper panel) and anti-FLAG antibody (lower panel). IB, immunoblotting. (B) Immunoprecipitation (IP) of FLAG-SUMO-1-conjugated HA-hRIPβ protein. Cells were transfected with hRIPβ in combination with a plasmid encoding FLAG-SUMO-1, and the lysates were boiled and immunopurified with anti-FLAG antibody. FLAG-SUMO-1-conjugated HA-hRIPβ protein was probed with anti-HA antibody. WCL, whole-cell lysate. (C) Generation of SUMO-deficient hRIPβ mutants. Cells were transfected with plasmids encoding hRIPα, hRIPβ, hRIPβ KM-1 (K103N), hRIPβ KM-2 (K114R), hRIPβ KM-3 (K121R), hRIPβ 2KM-1 (K103N and K114R), hRIPβ 2KM-2 (K103N and K121R), and hRIPβ 4KM (K103N, K114R, K121R, and K142R). Cell lysates were immunoblotted with anti-HA antibody. (D) hRIPβ colocalizes with GFP-SUMO-1. Panels i to iii, cells were transfected with hRIPα, hRIPβ, and hRIPβ 4KM in combination with GFP-SUMO-1 (green) and immunostained with anti-HA antibody (red). Panel iv, colocalization of GFP-SUMO-1 with PML body was also shown. Bars, 10 μm.
So as to provide further evidence for the sumoylation of hRIPβ in vivo, sumoylated hRIPβ was immunopurified. SUMO protease is capable of readily cleaving the isopeptide bond between SUMO and the target protein, so the cell lysate was boiled to inactivate SUMO protease (32). The denatured cell lysates were incubated with anti-FLAG antibody in order to purify proteins conjugated to FLAG-SUMO-1, and the resulting precipitates were probed with anti-HA antibody for detection of HA-hRIPβ. The band corresponding to FLAG-SUMO-1-conjugated HA-hRIPβ was purified and positively identified by anti-HA antibody (Fig. 3B). hRIPβ was thus shown to be a substrate for sumoylation.
The obvious extension of showing that hRIPβ can be sumoylated was to determine the significance of such a modification to the protein. To this end, a SUMO-deficient mutant of hRIPβ was generated: the lysine residue of the sumoylation consensus sequence (IKQE) was replaced with asparagine (30). Yet sumoylation of hRIPβ was not completely diminished by the K103N mutation (hRIPβ KM-1) (Fig. 3C), and the other lysine residues were replaced by arginine so as to generate additional mutants. As shown in Fig. 3C, sumoylation of hRIPβ 2KM-2 (carrying both the K103N and K121R mutations) was significantly reduced, and the sumoylated form was completely lost for hRIPβ 4KM, modified by the K103N and K114,121,142R mutations. This strongly implicates the multiple lysine residues as the acceptor site of sumoylation (Fig. 3C).
Establishment of the sumoylation-deficient mutants of hRIPβ led to an examination of the role of hRIPβ sumoylation. Plasmids encoding hRIPα, hRIPβ, and hRIPβ 4KM were transiently transfected with GFP-SUMO-1, whose expression showed dense spots, which indicate PML nuclear bodies (Fig. 3D, panel iv). More than 90% of hRIPβ foci showed complete association with GFP-SUMO-1 spots, while none of the hRIPα was colocalized with GFP-SUMO-1 (Fig. 3D, panels i and ii). In contrast, 70% of hRIPβ 4KM exhibited spots with no association or only adjacent localization with GFP-SUMO-1 (Fig. 3D, panel iii). The remaining hRIPβ 4KM was diffusely dispersed in the nucleus. These results suggest that hRIPβ must be sumoylated in order to localize to the PML nuclear body.
Interaction between RPA and hRIPα splice isoforms.
Jullien et al. (19) reported that RPA p70 is the binding partner of hRIPα and that the acidic-residue-rich domain of hRIPα is the binding region for RPA p70. Because both hRIPα and hRIPβ contain this acidic domain, we examined whether hRIPβ could interact with the RPA p70 protein. The interaction between RPA and hRIPβ was probed by transfecting HEK293T cells with plasmids encoding FLAG-tagged RPA p70 (FLAG-RPA) in the presence or absence of GST-hRIPα and GST-hRIPβ. Forty-eight hours after transfection, GST, GST-hRIPα, and GST-hRIPβ were precipitated with glutathione beads. Bead-bound FLAG-RPA proteins were probed with anti-FLAG antibodies. A binding assay revealed that both hRIPα and hRIPβ interacted with the RPA protein in vivo (Fig. 4A). Because RPA p70 is one subunit of the RPA complex, it was demonstrated that the transiently expressed FLAG-RPA p70 was associated with endogenous RPA p32 in vivo (Fig. 4B). A coimmunoprecipitation assay with endogenous proteins provided yet more evidence for the interaction between RPA and hRIPα/hRIPβ. This coimmunoprecipitation assay with HEK293T cells also showed interactions of RPA with endogenous hRIPα and hRIPβ (Fig. 4C).
FIG. 4.
Interaction between RPA and hRIPβ. (A) hRIPα and hRIPβ interact with RPA in HEK293T cells. Cells were transiently transfected with a plasmid encoding FLAG-RPA in combination with GST, GST-hRIPα, or GST-hRIPβ. Whole-cell lysates (WCL) were precipitated with glutathione-Sepharose beads. The bead-bound proteins were eluted and immunoblotted (IB) with anti-FLAG antibody (upper panel) or anti-GST antibody (lower panel). (B) Interaction between FLAG-RPA p70 and endogenous (Endo.) RPA p32. Cells were transfected with either blank vector (lane 1) or a plasmid encoding FLAG-RPA p70 (lane 2). FLAG-RPA p70 was immunopurified (IP) with anti-FLAG antibody. To roughly compare the amount of bound RPA p32 protein with that of FLAG-RPA p70, the blotted membrane was simultaneously probed with anti-FLAG and anti-RPA p32 antibodies. IgG, immunoglobulin G. (C) Interaction of RPA with endogenous hRIPα and hRIPβ. HEK293T cells were immunoprecipitated with either anti-HA antibody (control) or anti-RPA p70 antibody, followed by immunoblotting with anti-hRIP antibody (upper panel), anti-RPA p32 antibody (middle panel), and anti-RPA p70 antibody (lower panel). (D) Identification of the regions of hRIPα required for RPA interaction. GST and GST-RPA protein were used for GST pull-down assays with 35S-labeled, in vitro-translated hRIP constructs. Bead-bound proteins were eluted and visualized by autoradiography. GST and GST-RPA proteins were visualized by Coomassie brilliant blue staining.
In order to elucidate the interactions between RPA and the many splice isoforms of hRIPα, a GST pull-down assay was performed. GST-RPA p70 recombinant protein was produced in bacteria (Fig. 4D), and the in vitro binding assays used in vitro-translated forms of hRIPα, hRIPβ, and hRIPδ3. While the last of these, hRIPδ3, interacted with the RPA protein, the hRIPα mutant (amino acids 1 to 84) devoid of the acidic domain lacked binding affinity to RPA (Fig. 4D), as shown previously by (19). An hRIPδ3 mutant (amino acids 45 to 106) lacking the basic domain required for interaction with importin β was also constructed. While the hRIPδ3(45-106) mutant did not interact with importin β in vitro (data not shown), an interaction with RPA was identified that was slightly weaker than that of normal hRIPα and its splicing variants (Fig. 4D). These results indicate that the splice isoforms of hRIPα, as well as hRIPα, interact with RPA.
Nuclear export signal sequence of hRIPα.
Based on an in vitro transport assay, Jullien et al. (19) reported that xRIPα transports RPA into the nucleus, but the detailed mechanism of the process was not known at the time. Because hRIPα localizes primarily to the cytoplasm, it is supposed that hRIPα shuttles between the nucleus and the cytoplasm and thereby transports RPA into the nucleus. This hypothesis was supported by identifying the region of hRIPα required for nuclear export. Because hRIPα and hRIPβ share the majority of their peptide sequences yet display different subcellular localizations, it was supposed that the additional hRIPα-specific domain (amino acids 164 to 210) induced nuclear export of that protein (Fig. 5A). To verify this hypothesis, several deletion mutants that displayed differential localization were generated (Fig. 5A).
FIG. 5.
hRIPα contains the specific domain for cytoplasmic localization. (A) Schematic diagram of hRIPα, its isoforms, and deletion mutants. Numbers correspond to the amino acid sequence. Note the presence or absence of the indicated region (amino acids 163 to 180) in each construct. (B) hRIPα contains the specific domain (amino acids 164 to 180) required for cytoplasmic retention. The constructs shown in panel A were transiently expressed in HEK293T cells and stained with anti-hRIP antibody (green).
Confocal microscopy data revealed that hRIPβ, hRIPγ, and hRIPδ3 generally localized to the nucleus in a dot-like pattern (Fig. 5B). Because hRIPβ, hRIPγ, and hRIPδ3 lack the hRIPα-specific domain (amino acids 164 to 210), however, one can conclude that the hRIPα-specific domain is required for nuclear export. Moreover, two hRIPα deletion mutants (amino acids 1 to 180 and 1 to 200) localized to the cytoplasm (Fig. 5C), indicating that the 17-amino-acid region from amino acid 164 to 180 was sufficient for nuclear export of hRIPα.
Interaction with importin β is also important for nuclear import, as illustrated by an hRIPδ3 mutant that does not interact with importin β and localizes to the cytoplasm. On the other hand, an hRIPα(1-84) mutant that does not interact with RPA localizes to both the nucleus and cytoplasm. The above results indicate that hRIPα indeed harbors the specific domain (amino acids 164 to 180) required for its nuclear export.
hRIPβ targets RPA to the PML nuclear body.
Previous studies documented that RPA protein was mainly or partially localized to the PML nuclear body and that colocalization with the PML nuclear body was dependent on cell type (1, 2, 31). Little is known, however, about the transport of RPA into the PML nuclear body. As hRIPβ is associated with RPA and also localizes in the PML nuclear body, we postulated that hRIPβ was the carrier molecule for the targeting of RPA to the PML nuclear body.
To assess whether hRIPβ indeed targets RPA to the PML nuclear body, RPA was transiently expressed in HEK293T cells with either blank vector or a plasmid encoding hRIP isoforms and mutants. After 24 h, the transfected cells were fixed and double immunostained with anti-hRIP antibody (rabbit, green) and anti-FLAG antibody (mouse, red). While ectopic expression of FLAG-RPA alone induced accumulation of FLAG-RPA protein in the cytoplasm, expression of hRIPα, hRIPβ, and hRIPδ3 led to the transport of RPA protein into the nucleus (Fig. 6).
FIG. 6.
hRIPβ transports RPA into the PML nuclear body. HEK293T cells were transfected with a plasmid encoding FLAG-RPA in combination with either blank vector or hRIP constructs. After 24 h, the cells were fixed and stained with anti-hRIP antibody (green) and anti-FLAG antibody (red). (B) Colocalization of RPA with hRIP. HEK293T cells were stained with anti-hRIP antibody (green) and anti-RPA p32 antibody. To remove the background, anti-hRIP antibody was purified as described in Materials and Methods. Bars, 10 μm.
The nuclear distribution pattern of RPA with hRIPβ differs from the pattern of RPA with hRIPα. While RPA localizes throughout the entire nucleus when hRIPα is expressed, hRIPβ expression leads to a dot-like staining pattern of RPA. Previous microscopy data suggest that these dots are PML nuclear bodies. Moreover, hRIPβ was tightly associated with RPA, and hRIPα did not colocalize with RPA. While a fraction of the hRIPβ 4KM mutant colocalized with RPA, the level of this colocalization was weaker than that for wild-type hRIPβ. In the case of the hRIPα(1-84) mutant, which was devoid of the RPA binding region, localization was limited to the cytoplasm, indicating that the interaction between RPA and hRIPα or its isoforms is required for the transport of RPA into the nucleus.
hRIPβ is desumoylated upon UV irradiation and during S phase of the cell cycle.
Negorev and Maul (28) postulated that the PML nuclear body acts as a nuclear depot and can recruit specific proteins and release them upon external stress. Furthermore, regulated recruitment into the PML nuclear body, as well as controlled release, is closely related to sumoylation of the proteins. Because hRIPβ can transport RPA into the PML nuclear body and because this process is dependent on the sumoylation of hRIPβ, it is supposed that hRIPβ recruits RPA into the PML nuclear body and releases RPA upon external stress. UV irradiation was used to determine if the hRIPβ protein would be desumoylated by an external stressor.
HA-hRIPβ was transiently expressed in HEK293T cells, and the cells were either left untreated or irradiated with 50 J/m2 of UV light. After UV irradiation, the cells were harvested and subjected to direct lysis. The sumoylation status of hRIPβ was examined by immunoblot analysis with anti-HA antibody. As shown in Fig. 7A, the sumoylated hRIPβ band was reduced to less than half that for untreated cells at 1 h postirradiation. By 2 h postirradiation, sumoylated hRIPβ was hardly detected, although the amount of unmodified hRIPβ had not been significantly altered (Fig. 7A, panel i). A similar experiment with the endogenous proteins was performed to provide more evidence, and the results with the endogenous hRIPβ were identical to those with the transiently expressed hRIPβ (Fig. 7B, panel ii). These results indicate that UV light desumoylates hRIPβ in vivo.
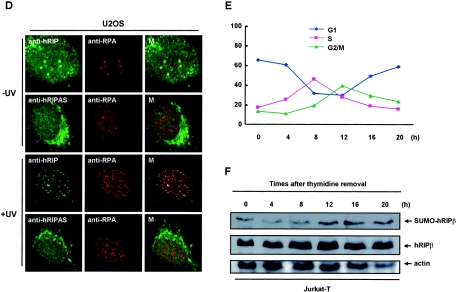
FIG. 7.
Sumoylation of hRIPβ is regulated by UV irradiation and the normal cell cycle. (A) Panel i, UV irradiation desumoylates hRIPβ. HA-hRIPβ was transiently expressed in HEK293T cells. After 24 h, the cells were either left untreated (lane 1) or irradiated with 50 J/m2 of UV (lanes 2 and 3). The irradiated cells were harvested at the indicated times and the cell lysates blotted with anti-HA antibody. Panel ii, the sumoylation of endogenous (endo.) hRIPβ upon UV irradiation was measured to confirm the results. (B) SENP1 desumoylates hRIPβ. Panel i, cells were transfected with a plasmid encoding HA-hRIPβ in the presence or absence of a plasmid encoding His-SENP1. After 48 h, the cells were collected and the cell lysates blotted with anti-HA antibody (upper panel) or anti-His antibody (lower panel). Panel ii, the transiently expressed SENP1 desumoylated the endogenous hRIPβ. Panel iii, SENP1 siRNA was used to inhibit SENP1-mediated desumoylation of hRIPβ. Panel iv, SENP1 siRNA treatment decreased the endogenous SENP1 protein. (C) UV irradiation disrupts the interaction between hRIPβ and RPA. hRIPβ and RPA were expressed in HEK293T cells grown on glass coverslips. After 24 h, the cells were either left untreated or irradiated with 50 J/m2 of UV. Two hours after UV irradiation, the cells were fixed and stained with anti-hRIP antibody (green) and anti-FLAG antibody (red). The second and fourth rows show higher magnifications of the areas indicated by the white dotted boxes in the respective panels of the first and third rows. Bars, 10 μm. UV irradiation also disrupts the association of hRIPβ with PML protein. (D) U2OS cells were used to show the colocalization of endogenous hRIPβ with RPA. Cells were either left untreated or irradiated with 50 J/m2 of UV and were stained with anti-hRIP antibody or anti-hRIPAS antibody together with anti-RPA antibody. M, merge. (E) Cell cycle analysis of Jurkat cell synchronization. Jurkat cells were synchronized by the thymidine double-block method. After removal of thymidine, the cells were harvested at the indicated times and subjected to cell cycle analysis. The cell cycle index was calculated by cell numbers in G1, S, and G2/M phases. (F) Sumoylation of hRIPβ is downregulated during S phase. The synchronized cells were harvested, and equal amounts of the cell lysates were blotted with anti-hRIP and antiactin antibodies.
Recently, the SUMO isopeptidase SENP1 was identified (14). SENP1 is responsible for the desumoylation of the proteins in the PML nuclear body upon external stress (27, 28). Because hRIPβ is desumoylated by UV irradiation, it is supposed that SENP1 removes SUMO from hRIPβ. This hypothesis was tested by expressing HA-hRIPβ in HEK293T cells in the presence or absence of His-SENP1. Figure 7B, panel i, shows that transient expression of SENP1 results in a dramatic reduction of sumoylated hRIPβ. The same results were obtained with the endogenous hRIPβ (Fig. 7B, panel ii). To further confirm the effect of SENP1 on regulation of hRIPβ desumoylation, a SENP1 siRNA plasmid was used to silence endogenous SENP1 in HEK293T cells. Cheng et al. (6, 7) reported that SENP1 siRNA treatment decreases the endogenous SENP1 mRNA. As expected, the desumoylation of hRIPβ upon UV irradiation was decreased in SENP1 siRNA-treated cells (Fig. 7B, panels iii and iv). Consequently, SENP1 is responsible for the desumoylation of hRIPβ.
Next, the localization of hRIPβ and RPA upon UV irradiation was examined. hRIPβ and FLAG-RPA were expressed in HEK293T cells, and these cells were either left untreated or irradiated with 50 J/m2 of UV light. At 2 h postirradiation, the cells were fixed and reacted with anti-hRIP antibody (green) and anti-FLAG antibody (red). While hRIPβ showed obvious colocalization with RPA without UV treatment (Fig. 7C, upper panels), UV irradiation led to the dissociation of RPA from hRIPβ (Fig. 7C, lower panels). hRIPβ still showed dot-like patterns in the nucleus, but the spots were enlarged and the level of colocalization with RPA was much weaker than it was without UV treatment. RPA also exhibited smaller nuclear foci, which were believed to be the sites of DNA damage and repair as had been previously reported (1). These results indicate that UV light disrupts the interaction between hRIPβ and RPA, with this process accompanied by the desumoylation of hRIPβ.
Further demonstration of the localization change of RPA with hRIPβ upon treatment with UV light was accomplished through immunohistochemistry with endogenous RPA and hRIPβ protein in U2OS cells. Background interference was reduced by using the more sensitive anti-RPA p32 antibody and the purified anti-hRIP antibody to detect the endogenous protein. It should be recognized that all of the hRIPβ amino acid sequences are also included in hRIPα sequences. This redundancy contributed to difficulties when attempting to immunostain hRIPβ with the endogenous proteins. Purified anti-hRIP antibody detects the splice isoforms of hRIPα as well as hRIPβ. For this reason, anti-hRIPAS antibody was used to stain endogenous hRIPα as a control. As shown in Fig. 7D, anti-hRIP antibody reacts with the spots in the nucleus, and anti-RPA also reacts with the same spots. As a control, anti-hRIPAS antibody was used to detect the endogenous hRIPα, and anti-hRIPAS antibody does not react with the spots whereas anti-RPA antibody does. These data strongly support the supposition that hRIPβ transports RPA into the PML nuclear body.
In the same way, the localization of endogenous hRIPβ and RPA upon UV irradiation was examined. After UV irradiation, endogenous RPA was translocated to small and abundant spots (Fig. 7D). While a fraction of RPA spots were colocalized with hRIP spots, the rest of them were not colocalized with hRIP spots, indicating that the level of colocalization of RPA with hRIPβ was reduced upon UV irradiation. Taken together with the transient-expression data, these results indicate that UV light induces the dissociation of RPA from hRIPβ.
Finally, changes in hRIPβ sumoylation status were tracked throughout the cell cycle. Jurkat cells were synchronized using the thymidine double-block method (17). After thymidine removal, the cells were harvested at the indicated times and subjected to cell cycle analysis (Fig. 7D). Immunoblotting with anti-hRIP antibody showed that sumoylation of endogenous hRIPβ was lowered during early and full S phase (4 h and 8 h after thymidine removal, respectively) and that sumoylation was increased during G2/M phase (Fig. 7E). These findings show that sumoylation of hRIPβ varies during the cell cycle.
DISCUSSION
hRIPα and hRIPβ are expressed primarily among six splice isoforms of hRIP.
hRIPα has been identified as a human analog of xRIPα, the nuclear transporter of RPA in Xenopus (19). Here, the splice isoforms of hRIPα were cloned and designated hRIPα, hRIPβ, hRIPγ, hRIPδ1, hRIPδ2, and hRIPδ3 according to the sizes of their encoding polypeptides. Among six splice isoforms, hRIPα and hRIPβ appear to be the major splice products; several lines of evidence support this conclusion.
First, hRIPα and hRIPβ are easily detected by anti-hRIP immunoblotting, while hRIPγ and hRIPδ1 to -3 are hardly detected. Because they are easily detected after in vitro translation, it is likely that low levels of hRIPγ and hRIPδ1 to -3 are due to low protein stability. Second, hRIPα and hRIPβ share the same termination codon, while other splice isoforms use a termination codon formed by a frameshift in the middle of the combined exon sequences. This frameshift implies that hRIPγ and hRIPδ1 to -3 are by-products as opposed to the major products of this particular pathway. In contrast to hRIPα and hRIPβ, the hRIPγ and hRIPδ1 to -3 cDNAs contain the untranslated exons 5, 6, and 7. The low-level expression of hRIPγ and hRIPδ is likely due to the lack of translation of exons 5 through 7. Despite their minimal expression, hRIPγ and hRIPδ1 to -3 may later be found to have as-yet-unknown functions. The main focus of the present study was on the characterization of hRIPβ. Further study will be required for elucidation of the exact roles of these other isoforms.
Sumoylation of hRIPβ is required for localization in the PML nuclear body.
In the present report, it has been demonstrated that hRIPβ is a novel substrate for SUMO modification. hRIPβ has a sumoylation consensus sequence (IKQE/ψKXE) (30), and the replacement of four lysines, including the consensus lysine, resulted in the complete loss of hRIPβ sumoylation. The 103rd (consensus) and 121st lysines in hRIPβ were found to be the major sites for sumoylation: the expression of hRIPβ 2KM-2, carrying K103N and K121R mutations, revealed a dramatic reduction in sumoylated protein compared to the sumoylation level of wild-type protein. The data collected so far suggest that hRIPβ is sumoylated at one of the four lysine residues in hRIPβ, as a shifted band was repeatedly detected in hRIPβ immunoblotting. It is also possible, however, that multiply sumoylated hRIPβ was not detected owing to a low level of multiple sumoylation.
Sumoylation has recently emerged as an important regulatory mechanism for protein localization, and several studies have demonstrated the close relation between the sumoylation of PML-associated proteins and their localization to the PML nuclear body (44). In one such study, the transcription repressor Bach2 was sumoylated and targeted to the PML nuclear body by oxidative stress, while a SUMO-deficient Bach2 mutant showed no association or only adjacent localization with PML nuclear bodies (36). Similarly, it was demonstrated here that hRIPβ localizes to the PML nuclear body and that a SUMO-deficient mutant, hRIPβ 4KM, exhibits a localization pattern similar to that of the Bach2 mutant. These findings are not completely analogous, however, because Bach2 is the functional protein itself, while hRIPβ has only a regulatory role in RPA localization to the PML nuclear body. To the best of our knowledge, hRIPβ is the first regulatory protein whose sumoylation has been found to control the PML localization of the target protein.
hRIPα and hRIPβ transport RPA to distinct locations.
It has been reported, based on an in vitro nuclear import assay, that xRIPα transports RPA into the nucleus (19), and the present study yielded results in agreement with this finding. Transient expression of FLAG-RPA in HEK293T cells induced cytoplasmic accumulation of FLAG-RPA. In contrast, expression of hRIPα or hRIPβ clearly shifted the localization of FLAG-RPA into the nucleus, indicating that both hRIPα and hRIPβ can transport RPA into the nucleus. Because the RPA protein was localized predominantly to the cytoplasm when transporter proteins were not expressed, the level of endogenous RPA nuclear transporter is likely insufficient for transporting the transiently expressed RPA protein into the nucleus, It is of particular interest that the localization patterns of transported RPA are different for hRIPα and hRIPβ. While hRIPα transports RPA close to the nuclear membrane, hRIPβ targets RPA to the PML nuclear body. Moreover, hRIPα returns to the cytoplasm due to an hRIPα-specific domain required for efficient nuclear export, while hRIPβ is colocalized with RPA in the PML nuclear body. These results led us to propose that hRIPα and hRIPβ have different roles in the nuclear transport of RPA; that is, hRIPα simply transports RPA into the nucleus, but hRIPβ deposits RPA in the PML nuclear body. Because RPA is involved in DNA repair, recombination, and replication, which require DNA machinery including RPA, it is possible that hRIPβ deposits RPA into PML nuclear bodies because of RPA insufficiency.
RPA is stored in PML nuclear bodies as a nuclear defense against external insults.
Several studies suggest that RPA is transported to DNA damage sites and is involved in DNA repair and recombination (1, 45, 46). In this report, it was demonstrated that RPA is stored in the PML nuclear body by its interaction with hRIPβ and that RPA is released from hRIPβ upon UV irradiation. It is postulated that sumoylation of hRIPβ is involved in RPA transport from the PML nuclear body to DNA damage sites. In addition to the sumoylation of hRIPβ, ATR protein kinase activity plays an important role in the translocation of RPA upon exposure to a DNA-damaging agent (1), and many other DNA repair proteins are required for the full response to DNA damage. For this reason, it is possible that the deposition of RPA in PML nuclear bodies contributes to the coordinated action of DNA repair processes with other repair machinery in PML nuclear bodies. If this is true, the deposition of RPA in PML nuclear bodies by hRIPβ sumoylation is a novel nuclear defense mechanism for the protection of cellular DNA from external stresses.
In summary, hRIPβ was cloned as a novel splice isoform of hRIPα, and hRIPβ was found to be modified by sumoylation. This sumoylation was found to be required for localization to the PML nuclear body. Furthermore, hRIPβ was shown to transport RPA into the PML nuclear body and to release RPA upon UV irradiation. Taken together, the findings of this study indicate that hRIPβ regulates RPA storage in the PML nuclear body and supplements RPA in cases of RPA insufficiency.
Acknowledgments
We especially thank B. Stillman for providing anti-RPA antibody and M. S. Wold for providing RPA p70 clones.
This work was supported in part by grants from the National Research Laboratory Program of the Korea Institute of Science and Technology Evaluation and Planning (KISTEP) and from the Korea Science and Engineering Foundation (KOSEF) through the Protein Network Research Center at Yonsei University.
REFERENCES
- 1.Barr, S. M., C. G. Leung, E. E. Chang, and K. A. Cimprich. 2003. ATR kinase activity regulates the intranuclear translocation of ATR and RPA following ionizing radiation. Curr. Biol. 13:1047-1051. [DOI] [PubMed] [Google Scholar]
- 2.Bischof, O., S. H. Kim, J. Irving, S. Beresten, N. A. Ellis, and J. Campisi. 2001. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 153:367-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boubnov, N. V., and D. T. Weaver. 1995. scid cells are deficient in Ku and replication protein A phosphorylation by the DNA-dependent protein kinase. Mol. Cell. Biol. 15:5700-5706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brush, G. S., C. W. Anderson, and T. J. Kelly. 1994. The DNA-activated protein kinase is required for the phosphorylation of replication protein A during simian virus 40 DNA replication. Proc. Natl. Acad. Sci. USA 91:12520-12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brush, G. S., D. M. Morrow, P. Hieter, and T. J. Kelly. 1996. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc. Natl. Acad. Sci. USA 93:15075-15080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J., N. D. Perkins, and E. T. Yeh. 2005. Differential regulation of c-Jun-dependent transcription by SUMO-specific proteases. J. Biol. Chem. 280:14492-14498. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, J., D. Wang, Z. Wang, and E. T. Yeh. 2004. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 24:6021-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Din, S., S. J. Brill, M. P. Fairman, and B. Stillman. 1990. Cell-cycle-regulated phosphorylation of DNA replication factor A from human and yeast cells. Genes Dev. 4:968-977. [DOI] [PubMed] [Google Scholar]
- 9.Dutta, A., and B. Stillman. 1992. cdc2 family kinases phosphorylate a human cell DNA replication factor, RPA, and activate DNA replication. EMBO J. 11:2189-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fairman, M. P., and B. Stillman. 1988. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 7:1211-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, F., and J. W. Newport. 1993. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J. Cell Sci. 106:983-994. [DOI] [PubMed] [Google Scholar]
- 12.Fotedar, R., and J. M. Roberts. 1992. Cell cycle regulated phosphorylation of RPA-32 occurs within the replication initiation complex. EMBO J. 11:2177-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gately, D. P., J. C. Hittle, G. K. Chan, and T. J. Yen. 1998. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol. Biol. Cell 9:2361-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong, L., S. Millas, G. G. Maul, and E. T. Yeh. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355-3359. [DOI] [PubMed] [Google Scholar]
- 15.Graham, F. L., and A. J. van der Eb. 1973. Transformation of rat cells by DNA of human adenovirus 5. Virology 54:536-539. [DOI] [PubMed] [Google Scholar]
- 16.Henricksen, L. A., C. B. Umbricht, and M. S. Wold. 1994. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 269:11121-11132. [PubMed] [Google Scholar]
- 17.Hiroi, N., H. Maruta, and S. Tanuma. 1999. Fas-mediated apoptosis in Jurkat cells is suppressed in the pre-G2/M phase. Apoptosis 4:255-261. [DOI] [PubMed] [Google Scholar]
- 18.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jullien, D., D. Gorlich, U. K. Laemmli, and Y. Adachi. 1999. Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J. 18:4348-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kenny, M. K., U. Schlegel, H. Furneaux, and J. Hurwitz. 1990. The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem. 265:7693-7700. [PubMed] [Google Scholar]
- 21.Kim, C., R. O. Snyder, and M. S. Wold. 1992. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 12:3050-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kutay, U., E. Izaurralde, F. R. Bischoff, I. W. Mattaj, and D. Gorlich. 1997. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 16:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, J. S., S. R. Kuo, M. M. McHugh, T. A. Beerman, and T. Melendy. 2000. Adozelesin triggers DNA damage response pathways and arrests SV40 DNA replication through replication protein A inactivation. J. Biol. Chem. 275:1391-1397. [DOI] [PubMed] [Google Scholar]
- 24.Liu, V. F., and D. T. Weaver. 1993. The ionizing radiation-induced replication protein A phosphorylation response differs between ataxia telangiectasia and normal human cells. Mol. Cell. Biol. 13:7222-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marton, R. F., P. Thommes, and S. Cotterill. 1994. Purification and characterisation of dRP-A: a single-stranded DNA binding protein from Drosophila melanogaster. FEBS Lett. 342:139-144. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, S. E., and M. B. Kastan. 1997. Dissociation of radiation-induced phosphorylation of replication protein A from the S-phase checkpoint. Cancer Res. 57:3386-3389. [PubMed] [Google Scholar]
- 27.Nefkens, I., D. G. Negorev, A. M. Ishov, J. S. Michaelson, E. T. Yeh, R. M. Tanguay, W. E. Muller, and G. G. Maul. 2003. Heat shock and Cd2+ exposure regulate PML and Daxx release from ND10 by independent mechanisms that modify the induction of heat-shock proteins 70 and 25 differently. J. Cell Sci. 116:513-524. [DOI] [PubMed] [Google Scholar]
- 28.Negorev, D., and G. G. Maul. 2001. Cellular proteins localized at and interacting within ND10/PML nuclear bodies/PODs suggest functions of a nuclear depot. Oncogene 20:7234-7242. [DOI] [PubMed] [Google Scholar]
- 29.Oakley, G. G., L. I. Loberg, J. Yao, M. A. Risinger, R. L. Yunker, M. Zernik-Kobak, K. K. Khanna, M. F. Lavin, M. P. Carty, and K. Dixon. 2001. UV-induced hyperphosphorylation of replication protein A depends on DNA replication and expression of ATM protein. Mol. Biol. Cell 12:1199-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654-12659. [DOI] [PubMed] [Google Scholar]
- 31.Sanz, M. M., M. Proytcheva, N. A. Ellis, W. K. Holloman, and J. German. 2000. BLM, the Bloom's syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet. Cell Genet. 91:217-223. [DOI] [PubMed] [Google Scholar]
- 32.Sapetschnig, A., G. Rischitor, H. Braun, A. Doll, M. Schergaut, F. Melchior, and G. Suske. 2002. Transcription factor Sp3 is silenced through SUMO modification by PIAS1. EMBO J. 21:5206-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao, R. G., C. X. Cao, H. Zhang, K. W. Kohn, M. S. Wold, and Y. Pommier. 1999. Replication-mediated DNA damage by camptothecin induces phosphorylation of RPA by DNA-dependent protein kinase and dissociates RPA:DNA-PK complexes. EMBO J. 18:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiio, Y., T. Yamamoto, and N. Yamaguchi. 1992. Negative regulation of Rb expression by the p53 gene product. Proc. Natl. Acad. Sci. USA 89:5206-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 36.Tashiro, S., A. Muto, K. Tanimoto, H. Tsuchiya, H. Suzuki, H. Hoshino, M. Yoshida, J. Walter, and K. Igarashi. 2004. Repression of PML nuclear body-associated transcription by oxidative stress-activated Bach2. Mol. Cell. Biol. 24:3473-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, H., J. Guan, A. R. Perrault, Y. Wang, and G. Iliakis. 2001. Replication protein A2 phosphorylation after DNA damage by the coordinated action of ataxia telangiectasia-mutated and DNA-dependent protein kinase. Cancer Res. 61:8554-8563. [PubMed] [Google Scholar]
- 38.Wang, Y., X. Y. Zhou, H. Wang, M. S. Huq, and G. Iliakis. 1999. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J. Biol. Chem. 274:22060-22064. [DOI] [PubMed] [Google Scholar]
- 39.Wold, M. S. 1997. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 40.Wold, M. S., and T. Kelly. 1988. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 85:2523-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wold, M. S., D. H. Weinberg, D. M. Virshup, J. J. Li, and T. J. Kelly. 1989. Identification of cellular proteins required for simian virus 40 DNA replication. J. Biol. Chem. 264:2801-2809. [PubMed] [Google Scholar]
- 42.Yoshida, K., and G. Blobel. 2001. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J. Cell Biol. 152:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]
- 44.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:85-90. [DOI] [PubMed] [Google Scholar]
- 45.Zou, L., and S. J. Elledge. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542-1548. [DOI] [PubMed] [Google Scholar]
- 46.Zou, L., D. Liu, and S. J. Elledge. 2003. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc. Natl. Acad. Sci. USA 100:13827-13832. [DOI] [PMC free article] [PubMed] [Google Scholar]