Abstract
Defects in the XPD gene can result in several clinical phenotypes, including xeroderma pigmentosum (XP), trichothiodystrophy, and, less frequently, the combined phenotype of XP and Cockayne syndrome (XP-D/CS). We previously showed that in cells from two XP-D/CS patients, breaks were introduced into cellular DNA on exposure to UV damage, but these breaks were not at the sites of the damage. In the present work, we show that three further XP-D/CS patients show the same peculiar breakage phenomenon. We show that these breaks can be visualized inside the cells by immunofluorescence using antibodies to either γ-H2AX or poly-ADP-ribose and that they can be generated by the introduction of plasmids harboring methylation or oxidative damage as well as by UV photoproducts. Inhibition of RNA polymerase II transcription by four different inhibitors dramatically reduced the number of UV-induced breaks. Furthermore, the breaks were dependent on the nucleotide excision repair (NER) machinery. These data are consistent with our hypothesis that the NER machinery introduces the breaks at sites of transcription initiation. During transcription in UV-irradiated XP-D/CS cells, phosphorylation of the carboxy-terminal domain of RNA polymerase II occurred normally, but the elongating form of the polymerase remained blocked at lesions and was eventually degraded.
Xeroderma pigmentosum (XP), trichothiodystrophy (TTD), and Cockayne syndrome (CS) are genetic disorders, all associated with defects in nucleotide excision repair (NER) of DNA damage. Defects in XP have been assigned to eight complementation groups (XP-A through -G and variant) corresponding to proteins involved in different stages of the NER process. The XP-D complementation group is of particular interest and complexity, because mutations in the XPD gene can result in any of at least five different clinical phenotypes (23). XP and TTD are relatively frequent outcomes of XPD mutations, whereas the combined features of XP and CS, XP and TTD, or cranio-oculofacial-skeletal syndrome are rare outcomes (7, 16, 23). XPD protein is a subunit of transcription factor TFIIH, which is a multifunctional protein required for NER, basal transcription by RNA polymerase I (17) and II (10), and transcriptional activation (21, 27). It is likely that the complexity of the clinical outcomes arises because different mutations affect these various functions differentially. Thus, XP is thought to result if the mutation affects NER but has little effect on transcription. Conversely, TTD is thought to be the result of transcriptional deficiencies. In support of this hypothesis, each mutation site is specific for a particular disorder (37). Thus, for example, many XP patients with the XP clinical phenotype have a mutation at Arg683, whereas no TTD patients have this mutation. Conversely R112H and R722W are mutations found in several TTD patients but not in any XP patients (23). Furthermore, a mouse generated with the R722W mutation had many of the features of TTD (11).
At the start of this study, only two patients with the combined features of XP and CS were known in the XP-D group. Patient XP8BR was a severely affected boy who died at age 16 months and was a compound heterozygote with mutations G675R in one allele and del A in codon 669 in the other (8). The latter mutation is not expected to produce functional protein, and the phenotype can be largely attributed to the former mutation. Patient XPCS2 was a severely affected patient who died at age 13 with several tumors. The only expressed XPD allele had the mutation G602D (36). In our earlier work, we showed that fibroblasts from both of these patients were exquisitely sensitive to killing by UV irradiation (8) and that RNA synthesis failed to recover even after very low doses of UV (39). Curiously, unscheduled DNA synthesis was relatively high, and we discovered that breaks in these cells were generated following UV irradiation (2). Contrary to our expectations, however, we found that these breaks were not at the sites of DNA damage. They could be generated in the genomic DNA of undamaged cells following introduction into the cells of heavily UV-irradiated plasmid DNA (2). These results showed that the breaks were introduced in trans. We postulated that these breaks were introduced erroneously by the NER machinery at sites of transcription initiation, instead of at sites of damage, and that these erroneous breaks were the cause of the UV hypersensitivity.
In the present work, we have extended our previous study to include several further patients with the combined XP-D/CS phenotype. We show that the breaks are generated in genomic DNA on exposure of the cells to plasmids carrying several different types of DNA damage, and, consistent with our hypothesis, we show that the number of breaks is substantially diminished if the cells are treated with inhibitors of transcription following the UV treatment and that the breaks are abolished in the absence of the NER machinery.
MATERIALS AND METHODS
Cell culture.
Primary human fibroblasts from normal and repair-deficient individuals (Table 1) were cultured in Eagle's minimum essential medium (Gibco-BRL) containing 15% fetal calf serum (FCS) and kept at 37°C in a humidified atmosphere with 5% CO2. Primary mouse embryonic fibroblasts (MEFs) were cultured in Eagle's Alpha minimum essential medium (Gibco-BRL) with the oxygen tension reduced to 3%. Under these conditions, MEFs do not senesce (31). For transfection experiments, hTert-transformed fibroblasts were cultured under the same conditions as primary cells. For immunofluorescence experiments, cells were seeded onto sterile coverslips, allowed to grow to confluence, and incubated for 4 to 5 days in Eagle's minimum essential medium containing 0.5% FCS (human cells) or 15% FCS (MEFs).
TABLE 1.
Human cell lines used in this study
| Cell type | Cell line | Phenotype | Origin | XP-D causative mutation | Reference |
|---|---|---|---|---|---|
| Primary human fibroblasts | XP8BR | XP-D/CS | United Kingdom | G675R | 8 |
| XPCS2 | XP-D/CS | France | G602D | 36 | |
| XP1JI | XP-D/CS | Japan | G47R | 14a | |
| XPCS1PV | XP-D/CS | Spain | R666W | This paper | |
| XPCS118LV | XP-D/CS | Belgium | R666W | This paper | |
| XP1NE | XP-D/CS (?) | United Kingdom | G47R | 37 | |
| CS4BR | CS | United Kingdom | |||
| XP1BR | XP-D | United Kingdom | R683W | 37 | |
| 1BR3 | Normal | United Kingdom | |||
| Immortalized human fibroblasts | XP8BR-hTert | XP-D/CS | United Kingdom | G675R | 8 |
| 1604-hTert | Normal | United Kingdom | |||
| Primary mouse fibroblasts | Normal | ||||
| TTD | R722W | 11 | |||
| XP-D/CS | G602D | Andressoo et al., submitted | |||
| XP-D/CS | G602D (xpa del exons 3 and 4) | ||||
| XP-A |
UV irradiation.
For UV irradiation, medium was removed, and cells were washed once with phosphate-buffered saline (PBS) and then irradiated with four Westinghouse FS20 sunlamps (UVB) or a Philips 6W germicidal lamp (UVC), which emit primarily in the UVB and UVC ranges, respectively.
Transcription inhibition.
Cells under normal cell culture conditions were treated with α-amanitin (Sigma) at a concentration of 50 μg/ml for 24 h, with 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (ICN) at a concentration of 100 μM for 2 h, with N-[2-(methylamino)ethyl]-5-isoquinolinesulfonamide dihydrobromide (H8) (Sigma) at a concentration of 200 μM for 2 h, or with actinomycin D (Sigma) at a concentration of 0.5 μg/ml for 2 h. These treatments were followed immediately by preparation of cells for the comet assay as described below. Transcription inhibitors at these concentrations were also added during the postirradiation incubation period in the presence of 100 μM cytosine-β-arabinofuranoside (araC) and 10 mM hydroxyurea (HU).
Single-cell gel electrophoresis “comet” assay.
The comet assay was carried out as described previously (2). In short, cells were trypsinized and prepared in a thin layer of 0.6% low-melting-point agarose on fully frosted microscope slides at 2 × 104 cells/slide. Slides were maintained on ice and coverslips removed for UV irradiation. Immediately following irradiation, cells were incubated at 37°C in the dark with 100 μM araC and 10 mM HU to allow incisions to accumulate. Slides were subsequently immersed in a lysis solution (2.5 M NaCl, 0.2 M NaOH, 100 mM EDTA-Na2, 10 mM Tris base, 10% dimethyl sulfoxide, and 1% Triton-X, pH 10) for at least 1 hour at 4°C. Slides were then electrophoresed under alkaline conditions (0.3 M NaOH, 1 mM EDTA-Na2) at 20 V for 24 min. Following electrophoresis, slides were washed, at room temperature, with neutralizing buffer (0.4 M Tris-HCl, pH 7.5) and stained by the addition of 35 μl of 20-μg/ml ethidium bromide solution to the gel. Comet lengths were measured using the Casys system (Synoptics, Cambridge, United Kingdom), and comet tail moments were measured using the Comet Assay III software (Perceptive Instruments).
Plasmid preparation.
For UVB treatment, pCDNA3.1 plasmid was irradiated in a 3-cm dish in 0.5 ml H2O with 15,000 J/m2 of UVB. For methylene blue (MB) treatment, pCDNA3.1 was diluted at 1 μg/ml in 300 μl Tris-EDTA (TE) to which 300 μl MB solution (6 μM) was added in the dark. Photoactivation of MB was done under visible light for 54 min. The MB was then extracted with butanol, and the plasmid ethanol-precipitated. For N-methyl-N′-nitro-N-nitrosoguanidine (MNNG) and methyl methanesulfonate (MMS) treatments, 50 μg plasmid was diluted in 100 μl TE containing 1 mM MNNG or 50 mM MMS, respectively. Incubation was at room temperature for 1 hour. For OsO4 treatment, 50 μg plasmid was dissolved in 300 μl TE. OsO4 was added to 300 μg/ml (low dose) or 1.5 mg/ml (high dose). Incubation was at 70°C for 20 min. For the generation of double-strand breaks, the plasmid was digested with HinFI and MseI. For transfection, treated or untreated plasmid was ethanol precipitated together with pHook-1 plasmid in order to increase cotransfection efficiency.
Quantification of plasmid damage.
UVB-treated plasmid was digested by T4 endonuclease (diluted in 10 mM Tris-HCl, pH 8, 80 mM NaCl, 10 mM EDTA), specific for pyrimidine dimers; OsO4-treated plasmid was digested by Escherichia coli endonuclease III enzyme (Trevigen), specific for thymine glycol residues; and MB-treated plasmid was digested by E. coli formamidopyrimidine-DNA glycosylase (Trevigen), specific for 8-oxo-guanine residues (diluted in 40 mM HEPES-KOH, pH 8, 0.1 M KCl, 0.5 mM EDTA, 0.2 mg/ml bovine serum albumin [BSA]). MNNG- and MMS-treated plasmids were digested for 1 h at 37°C by E. coli 3-methyladenine DNA glycosylase type 1 (Tag) or yeast 3-methyladenine DNA glycosylase (Mag) enzyme (1 μg/ml in 70 mM HEPES-KOH pH 7.5, 0.5 mM EDTA, 5 mM mercaptoethanol, 5% glycerol), generously supplied by E. Seeberg, Oslo. Incision by damage-specific enzymes was analyzed by electrophoresis in 0.6% alkaline agarose, followed by Southern transfer and hybridization with pCDNA3.1.
Cotransfection.
The Capture-Tec system (Invitrogen) was used as described previously (2). In short, hTert-transformed fibroblasts were plated at 2.5 × 105 cells per 3-cm dish and transfected 24 h later with FuGENE6 (Roche) according to the manufacturer's protocol with a mixture of pCDNA3.1 (treated or untreated at 1 μg per transfection) and pHook-1 plasmids (0.25 μg per transfection). After 12 h of incubation, cells were washed together with phOx hapten-linked magnetic beads, bound to the beads, and collected according to the manufacturer's protocol. This was followed by the standard comet assay procedure.
Immunofluorescence microscopy.
For the detection of phosphorylated H2AX (γ-H2AX) and XRCC1 proteins, cells grown on coverslips were rinsed in PBS and fixed in 2 to 3.7% paraformaldehyde-2% sucrose for 10 min at room temperature. After being rinsed twice in PBS, cells were lysed in 0.2% Triton X-100 in PBS for 2 min at room temperature. This was followed by another wash in PBS. Cells were then incubated at room temperature for 25 min. with 3% BSA-PBS, followed by incubation for 30 min at room temperature or 16 h at 4°C with 0.1 ml mouse monoclonal anti-γ-H2AX (Upstate or AbCam), diluted 1:800. For the detection of poly-ADP-ribosylation, cells grown on coverslips were fixed in methanol-acetone (1:1) for 10 min at 4°C and washed three times with PBS supplemented with 0.1% Tween 20. Cells were then incubated for 30 min at 37°C with a monoclonal anti-poly-ADP-ribose (anti-PAR) antibody (10H) (Apotech), diluted 1:200 in 3% BSA-PBS. For detection of each protein, the primary antibody incubations were followed by incubation with a tetramethyl rhodamine isocyanate-conjugated anti-mouse (Jackson Immunoresearch Laboratories) or Alexafluor Cy555-conjugated anti-mouse (Molecular Probes) secondary antibody for 1 hour at room temperature. Coverslips were then washed three times in PBS and mounted onto slides in Vectashield mounting medium containing 0.05 μg/ml DAPI (4′,6′-diamidino-2-phenylindole). For the inhibition of poly-ADP-ribose-polymerase (PARP-1) activity, cells grown on coverslips were treated with 2 mM 3-aminobenzamide (Sigma) for 2 h prior to irradiation and also during the appropriate postirradiation incubation times.
Transcription elongation.
Cells grown on coverslips were UV irradiated, incubated for 1 h, and then washed twice with cold physiological buffer 1 (PB1) (130 mM KCl, 10 mM KPO4, pH 7.4, 1 mM Na2ATP grade II [Sigma-Aldrich], 2.5 mM MgCl2, 1 mM dithiothreitol [DTT]). Subsequently, streptolysin O (Murex Diagnostics, Dartford, United Kingdom) in PB1 was added to the cells at a final concentration of 0.1 unit/ml, and the cells were kept on ice for 30 min. Following a single wash with cold PB1, permeabilization of the cells was achieved by shifting the temperature to 37°C and by maintaining the cells at this temperature for 5 min as described previously (5, 18). Permeabilized cells were washed once with cold PB1, and run-on RNA synthesis was carried out by incubating the cells in PB1 containing ATP, CTP, and GTP (Gibco BRL) and BrUTP (Boehringer Mannheim) at 1 mM each, 50 mM KPO4, pH 7.4, and 4 mM MgCl2 (equimolar to the nucleotides) for 30 min at 37°C. Subsequently, cells were washed three times with cold PB1, and fluorescence labeling was performed as described above, using rat antibromodeoxyuridine (Harlan Seralabs) at a 1:100 dilution or mouse anti-cyclobutane pyrimidine dimer (immunoglobulin G2a), TD-M2, generously supplied by T. Mori, at a 1:1,000 dilution. Secondary antibodies were goat anti-mouse antibody-Texas red (used at 1:200) and donkey anti-rat antibody-fluorescein isothiocyanate (1:200), both from Jackson Laboratories.
Local UV irradiation.
Cells grown on coverslips were washed in PBS and covered with Isopore polycarbonate filters (Millipore) with a diameter of 0.8 μm. The cells were UVC irradiated with a Philips 6W germicidal lamp at a dose rate of approximately 1 J/m2/s. The filter was removed, and cells were incubated under normal culture conditions for appropriate postirradiation incubation times.
Cell fractionation and analysis of RNA polymerase II.
Cells were either UV irradiated or mock treated and subsequently incubated at 37°C for different time periods. They were then washed twice with ice-cold PBS, collected by scraping, and centrifuged (5 min, 1,000 rpm, 4°C). Subsequently, the cell pellet was processed at 4°C by two different protocols to generate soluble cellular extracts. In the first protocol the cell pellet was washed once with physiological buffer 2 (130 mM KCl, 10 mM Na2HPO4, 1 mM Na2ATP, 2.5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM DTT, pH 7.4), centrifuged, and resuspended in three cell pellet volumes of physiological buffer 2 containing 0.25% Triton X-100, 5 μg/ml proteinase inhibitors (proteinase inhibitor mixture from Roche), and 10 mM sodium pyrophosphate. Cells were rotated on a rollerbank for 10 min at 4°C and subsequently centrifuged at 2,000 rpm for 10 min to separate supernatant soluble and pellet fractions. The protein concentration was estimated, and the soluble fraction was analyzed directly with sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting or aliquoted and stored at −80°C.
In the second protocol, the cell pellet was resuspended in two cell pellet volumes of buffer A (25 mM HEPES, pH 7.6, 0.45 M NaCl, 1 mM EDTA, 0.5 mM EGTA, 12 mM NaF, 25% glycerol, 1 mM phenylmethylsulfonyl fluoride, 5 μg/ml proteinases inhibitors, 10 mM sodium pyrophosphate, and 0.5 mM DTT) and snap frozen in liquid nitrogen. Cells were then thawed in a 30°C water bath and snap frozen again. This freeze-thawing was repeated three times. After the last thawing step, the cell suspension was incubated on ice for 10 min and centrifuged at 13,000 rpm for 10 min at 4°C to pellet cell debris. The supernatant fraction, designated 0.45 M whole-cell extract, was aliquoted and stored at −80°C.
Antibodies used for Western blot analysis were mouse monoclonal 8WG16 and H5 (immunoglobulin M subclass) antibodies (Babco) against the nonphosphorylated C-terminal domain of the large subunit of RNA polymerase II (RNAPIIa) and the hyperphosphorylated RNAPIIo, respectively, and the rabbit polyclonal p89 (Santa Cruz) against the p89 subunit of TFIIH.
Western blot analysis of both the soluble and 0.45 M whole-cell extract was performed as described previously (38) except that the transfer was done overnight at 4°C at 30 V and prior to immunostaining, the membrane was blocked for 1 to 3 h at room temperature with 5% dry nonfat milk powder (Marvel; Premier Brands UK, Ltd.). Protein bands were visualized via chemiluminescence (ECL-Plus; Amersham Biosciences) using horseradish peroxidase-conjugated secondary antibodies and exposure to X-ray ECL-Hyperfilm (Amersham Biosciences).
RESULTS
UV-induced breaks in XP-D/CS cells.
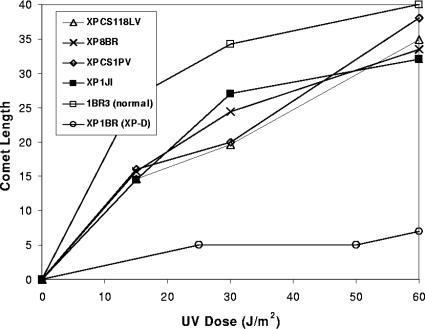
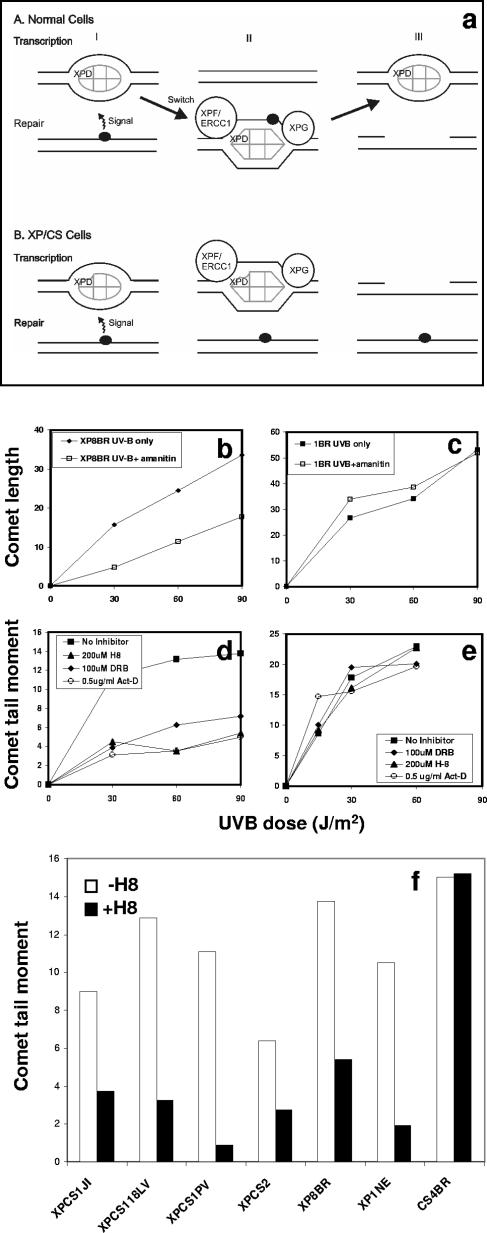
Following UV irradiation, breaks generated by NER accumulate in normal cells if the repair synthesis step is inhibited with a combination of araC and HU. Such breaks are not detected in XP-D cells defective in NER. However, we previously showed that XP-D/CS cells generated breaks in their DNA in response to UV damage, but these breaks were not located at the sites of DNA damage (2). Only two cases with the combined features of XP-D and CS have so far been described in the literature. However, we became aware of three further cases during the course of this work. Details of these patients will be presented in separate publications and are summarized in Table 1. In brief, XP1JI was a Japanese child with features of both CS and XP, including a squamous cell carcinoma. He died at age 24 months (14a). XPCS1PV was the first child of clinically normal and unrelated parents. She was delivered in the 41st week of gestation with weight (1,820 g) below the third percentile. Over the following months, she showed severe retardation in physical and mental development, bilateral cataract, and progressive worsening of general conditions. She had photosensitivity, pigmentary alterations on the face, and hypopigmented macules on the body. She died at age 15 months. XPCS118LV was diagnosed in childhood as having XP with neurological abnormalities, but neurological examination when he was in his 20s led to a diagnosis of CS. His unscheduled DNA synthesis level was rather high (75% of normal). The only expressed allele in the Japanese patient was the mutation G47R, whereas the other two both contained the same mutation, R666W, in one allele. In XPCS1PV this was the only allele expressed, whereas XPCS118LV was a compound heterozygote, with the second mutation being the combined L461V and del 716-730, shown previously to be a null allele (37). We measured breaks in cellular DNA by using the comet assay. Figure 1 shows that cells from these three individuals generate breaks in response to UVB damage exactly like those that we observed previously in XP8BR and XPCS2 (2). The number of breaks observed in the presence of araC and HU approached that in the normal cells, whereas in XP-D patient XP1BR, very few breaks were detected under these conditions, consistent with the defect in NER. These results confirm the association of the UV breakage phenotype with the clinical features of XP-D/CS.
FIG. 1.
UV-induced breaks in different XP-D/CS cells. Different cell strains were exposed to the indicated doses of UVB irradiation and then incubated for 1 h in the presence of araC and HU, followed by analysis of DNA breaks by using the comet assay.
Proteins at the break sites.
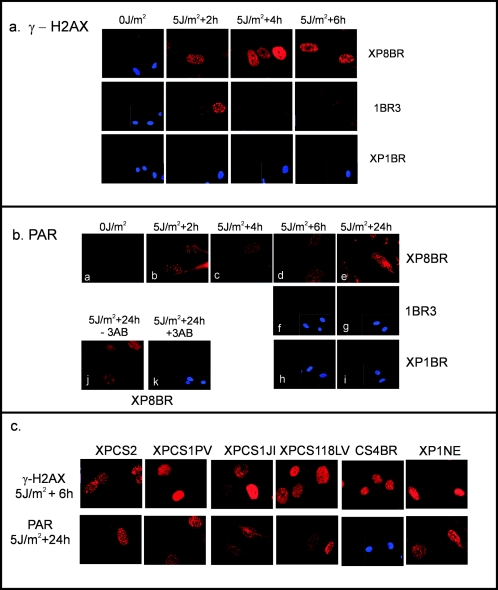
In order to gain further information as to the nature of the UV-induced breaks, we UV irradiated quiescent normal and XP8BR cells, and after various times, the cells were fixed and analyzed by immunofluorescence for proteins known to accumulate at sites of breaks produced by other agents such as ionizing radiation. (For these experiments it was more convenient to use UVC rather than UVB irradiation; we have previously shown that the both types of irradiation generate the same type of breaks in XP-D/CS cells.) The phosphorylated form of the histone variant H2AX (γ-H2AX) is one of the first proteins to appear at the sites of breaks induced by ionizing radiation (14, 34). Following low doses of UVC irradiation, γ-H2AX foci were detected in XP8BR cells 2 h after irradiation and persisted for many hours (Fig. 2a, top panels). In contrast, in normal 1BR3 cells, γ-H2AX foci could be seen 2 h after irradiation, but they had disappeared by 4 h (Fig. 2a, middle panels). In the XP-D cell line XP1BR, from a patient without the features of CS, no γ-H2AX foci were detected within the 6-hour period after irradiation (Fig. 2a, bottom panels).
FIG. 2.
Cellular detection of breaks following UV irradiation. (a) XP8BR, 1BR, or XP1BR cells were mock-irradiated or irradiated with 5 J/m2 UVC and incubated for the indicated times. The cells were then stained with antibody specific for γ-H2AX. In cases where no signal was visible, a section of the coverslip stained with DAPI is shown to demonstrate that cells were indeed present on the coverslips. (b) The conditions were similar to those for panel a except that the staining was with anti-PAR antibody. 3AB, 3-aminobenzamide. (c) Different cell strains were UV-irradiated and incubated as indicated, followed by staining with either anti-γ-H2AX or anti-PAR antibody. Four different XP-D/CS cell strains are compared with CS strain CS4BR and strain XP1NE.
Following treatment of cells with many types of DNA-damaging agents, breaks generated in cellular DNA result in the activation of PARP-1 and the synthesis of PAR (35). Using an anti-PAR antibody, we detected PAR by immunofluorescence, persisting for at least 24 h in UV-irradiated XP8BR cells (Fig. 2b, panels a to e). Two hours after irradiation, PAR staining could be detected in normal cells (not shown), but, importantly, no such signal was observed at later times (Fig. 2b, panels f and g). No significant PAR staining was detected in XP1BR cells at any time (Fig. 2b, panels h and i, and data not shown). No PAR signal was observed if the irradiated cells were incubated with 3-aminobenzamide, a well-documented inhibitor of PARP-1 (35) (Fig. 2b, panels j and k). This shows that the PAR was synthesized by PARP-1 in response to the DNA breaks. For both γH2AX and PAR, the staining patterns of XPCS2, XPCS1PV, XP1JI, and XPCS118LV were similar to that of XP8BR (Fig. 2c). Furthermore, cells from CS-B patient CS4BR showed γ-H2AX staining at 6 h that was similar to that of the XP-CS cells, but no staining was detected with the anti-PAR antibody. Cell strain XP1NE has the same G47R mutation as XP1JI, but the patient was originally diagnosed as having XP (24). Recent reevaluation of this patient showed some features of CS, and the cells show the breakage characteristic of XP-D/CS cells (14a). These cells showed the same pattern of γ-H2AX and PAR staining as the other XP-D/CS cells (Fig. 2c, right panels).
Taken together, these results show that the breaks that we previously detected by the comet assay are recognized by the cell as bona fide DNA breaks.
Damage spectrum.
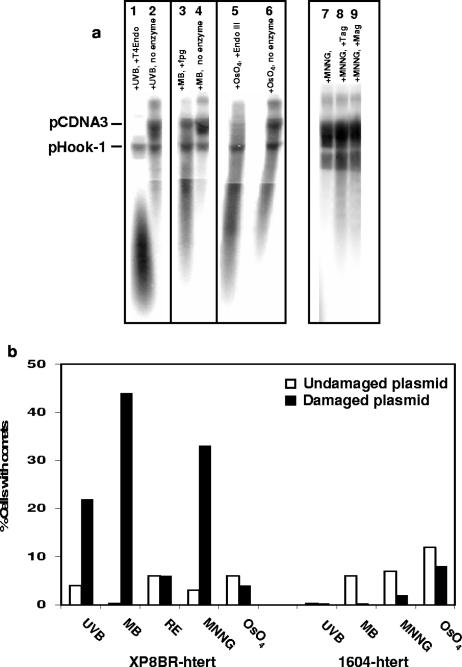
Our previous studies showed that the breaks could be generated in undamaged genomic DNA in response to ectopic UV damage introduced into the cell by means of a heavily irradiated plasmid. In our next series of experiments, we determined the types of damage that could elicit this break response in the XP-D/CS cells. Plasmid pCDNA3 was treated with different types of damaging agents. For each damaging treatment, a sample of the damaged plasmid, together with undamaged pHook-1 (see below), was treated with a glycosylase-AP-lyase specific for the type of damage introduced by the agent and analyzed on alkaline agarose gels to check that the plasmid had sustained substantial damage. As shown in Fig. 3a, CPD-specific T4 endonuclease V cleaved the UVB-irradiated plasmid (lane 1), formamidopyrimidine-DNA glycosylase (specific for 8-oxoguanine) cleaved the plasmid treated with methylene blue plus visible light (lane 3), and endonuclease III cleaved the OsO4-treated plasmid (lane 5). In all cases, the enzyme treatment resulted in a large reduction in the molecular weight of the plasmid, confirming that the treatments had generated substantial amounts of damage in the plasmids. MNNG generates a variety of methylation products in DNA. One of these, 3-methyladenine, is a substrate for E. coli Tag1 (3) or Saccharomyces cerevisiae Mag1 (4). Digestion of the MNNG-treated DNA (Fig. 3a, lane 7) with either of these enzymes (lanes 8 and 9) reduced the molecular weight of the plasmid, confirming the presence of methylation damage.
FIG. 3.
Breaks induced by plasmids damaged in different ways. pCDNA3 damaged with different agents was mixed with undamaged pHook-1. (a) pCDNA3 treated with the indicated agents was incubated with or without the appropriate damage-specific glycosylase. The samples were run on alkaline agarose gels and analyzed by Southern blotting and hybridization with pCDNA3. (b) The plasmid DNA, damaged with the indicated agents (but without enzyme treatment), was transfected into normal 1604-hTert or XP8BR-hTert cells, and 12 h later, breaks in the genomic DNA were analyzed using the comet assay. RE, pCDNA3 was digested with HinFI and MseI.
Having ascertained that all plasmids had sustained a substantial amount of damage, transformed XP8BR cells were cotransfected with the damaged pCDNA3 plasmids together with undamaged plasmid pHook-1. The latter contains the gene for the antibody phOx sFv, which is expressed on the surface of the cell. The transfected cells expressing this antibody together with the cotransfected damaged plasmid were separated by binding to phOx antigen attached to magnetic beads. The transfected cells separated by this procedure were then analyzed for breaks by using the comet assay. The results of the comet assays are presented in Fig. 3b. As shown previously, UVB-damaged plasmids generated breaks in a significant fraction of the cells. The methylating agents MNNG and MMS methylate DNA at several different positions. MMS damage is largely confined to methylation of nitrogen atoms, whereas MNNG also methylates on oxygen atoms. Plasmids damaged with MNNG generated breaks in recipient XP8BR cells (Fig. 3b). Similar results were obtained with MMS (data not shown).
Exposure of DNA to visible light in the presence of methylene blue is a well-documented method for producing oxidative damage in DNA, particularly 8-oxoguanine (1). DNA treated with this agent also generated breaks in the XP8BR cells (Fig. 3b). Osmium tetroxide also produces oxidative damage in DNA, but this is largely confined to oxidized pyrimidines, such as thymine glycols. Surprisingly, DNA damaged in this way did not generate breaks in the recipient cells (Fig. 3b). Finally, we treated the plasmid with restriction enzymes HinFI and MseI, which generate 40 fragments from pCDNA3.1. Transfection with restricted DNA did not result in breaks in the XP-CS cells (Fig. 3b). In no case were breaks generated in normal cells following transfection with any of the damaged plasmids. We conclude that the generation of breaks in genomic DNA in XP-D/CS cells in response to ectopic damage is not confined to UV photoproducts but is also seen in response to methylation and some types of oxidative damage.
Effect of local irradiation.
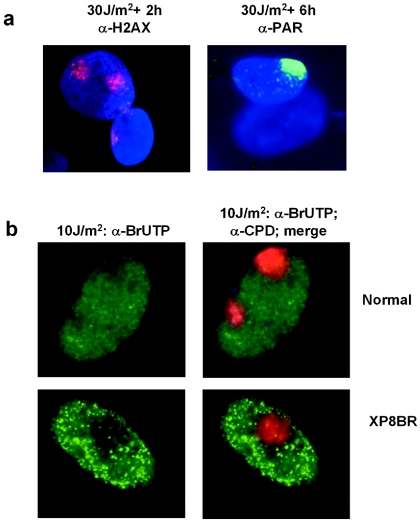
In order to gain further insight into the breakage in trans manifested in response to ectopic damage, we generated localized UV damage inside cells by irradiation through a membrane filter. This technique has been used previously by our laboratories to analyze the effects of local UV damage on RNA synthesis (28), NER (40), and DNA replication (20). We irradiated XP8BR cells through the filter and then looked at the localization of γ-H2AX and PAR. Based on our results with irradiated plasmid, we expected that breaks might be generated throughout the nucleus and that γ-H2AX and PAR would therefore be found distributed throughout the nucleus. Figure 4a shows that this is not the case. In XP8BR cells, γ-H2AX and PAR are found only at clearly defined regions of the nucleus where the damage has been inflicted. This result shows that, although the effects of damage can be mediated in trans, the signal can only operate within a localized area and is not transmitted through the whole nucleus.
FIG. 4.
Local UV irradiation. (a) XP8BR cells were UV irradiated (30 J m−2) through a membrane with 8-μm pores and incubated for 2 or 6 h, followed by staining with anti-γ-H2AX or anti-PAR antibodies, respectively. (b) Normal or XP8BR cells were UV irradiated (10 J m−2) through the filter, and after 1 h, the cells were permeabilized and incubated with ribonucleotides, including BrUTP. Cells were fixed and stained with antibromodeoxyuridine antibody (left panels) or with antibromodeoxyuridine, DAPI, and anti-CPD antibodies (right panels).
Moné et al. showed that in normal cells, local irradiation results in a decreased rate of transcription that is confined to the area of irradiation (28). This is confirmed in Fig. 4b, in which RNA synthesis was measured by the incorporation of BrUTP into permeabilized cells (green stain). There is a decrease in green staining at the sites of UV damage (red stain). Similar results were obtained with XP8BR cells. For this response, too, effects are observed only in the area of irradiation (Fig. 4b).
Breaks and transcription.
To explain the generation of breaks in response to ectopic damage, we previously proposed a model, reproduced in Fig. 5a, in which we suggested that the presence of damage changes the conformation of TFIIH and that in normal cells the TFIIH is recruited from sites of transcription initiation to the sites of the breaks. In the XP-D/CS cells, the conformational change still occurs, but the mutated XPD component prevents the TFIIH from being recruited to the damage. The TFIIH in its “repair conformation” remains at the site of transcription initiation, and the NER nucleases cut the DNA at sites of transcription initiation (2).
FIG. 5.
UV-induced breaks are dependent on transcription. (a) Model for origin of breaks, as proposed earlier (2). In normal cells, it is proposed that TFIIH changes conformation when it shuttles between transcription and NER modes. In XP-D/CS cells, the conformational change occurs in response to damage, but the recruitment to the site of damage cannot take place. Consequently the NER nucleasesXPG and XPF-ERCC1 cleave the DNA at sites of transcription initiation rather than at damaged sites. (b to e) Inhibition of UV-induced breaks in XP8BR cells by α-amanitin (b) and by H8, DRB, or actinomycin D (d) but not at NER sites in normal 1BR cells (c and e). (f) The indicated cell lines were exposed to 90 J/m2 and incubated with araC and HU for 1 h. Breaks in genomic DNA were analyzed using the comet assay.
A key point of the model in Fig. 5a is that the breaks are associated with transcription. Thus, the model predicts that if transcription is inhibited, the number of breaks should be reduced. Figure 5b to e show the effects of four different inhibitors of transcription. α-Amanitin prevents transcription initiation and results in the degradation of RNAPII (29). The breaks generated after UV treatment of XP8BR were indeed substantially reduced in the presence of α-amanitin (Fig. 5b), whereas α-amanitin had no significant effect on induction of breaks during NER in normal cells (Fig. 5c). DRB and H8 prevent transcription elongation, by inhibiting the phosphorylation of the C-terminal domain of RNAPII, which converts the initiating hypophosphorylated form RNAPIIa to the hyperphosphorylated form RNAPIIo (41, 42). Actinomycin D intercalates into the DNA and prevents progression of the transcriptional apparatus (22). As with α-amanitin, these three inhibitors all resulted in a much-reduced number of UV-induced breaks in XP8BR cells (Fig. 5d). In marked contrast, the breaks generated during NER in normal cells were unaffected by any of these inhibitors (Fig. 5e). Finally, Fig. 5f shows that breaks induced in the XP-D/CS cells XP1JI, XPCS118LV, XPCS1PV, XPCS2, and XP1NE are also reduced by treatment with H8. In contrast, breaks generated as intermediates in global NER in CS4BR, a CS cell strain with normal GGR, were, as in normal cells, unaffected by treatment with H8. These data provide strong evidence that the breaks in XP-D/CS cells are associated with transcription.
Breaks and NER.
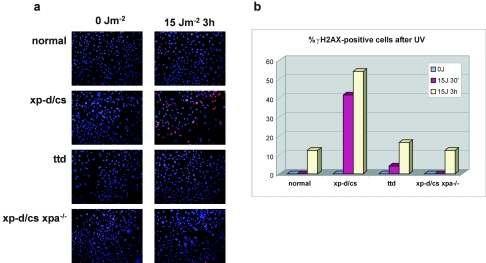
A further postulate of the model is that the NER machinery generates the breaks in the XP-D/CS cells. In order to investigate this, we have used embryonic fibroblasts derived from mice in which the xpd gene contains a homozygous mutation at the same site as the functional mutation in XPCS2, namely, G602D (J.-O. Andressoo et al., submitted for publication). We measured breaks induced by UV irradiation in these cells by staining for γ-H2AX 3 h after irradiation with 15 J m−2. As with the human XP-D/CS cells, breaks were generated in the xp-d/cs mouse cells, whereas very few were detected under these conditions in MEFs from a normal mouse or from the “TTD mouse” carrying the R722W mutation (Fig. 6a). However, when we compared cells from the xp-d/cs mouse with those from a double mutant mouse with both the xp-d/cs mutation and a knockout mutation in the xpa gene (12), the γ-H2AX staining was greatly reduced in the latter. The results are quantitated in Fig. 6b. They demonstrate that the breaks induced in XP-D/CS cells are indeed dependent on intact NER machinery.
FIG. 6.
UV-induced breaks are dependent on XPA protein. MEFs from animals with the genotypes indicated were irradiated with 15 J m−2 and incubated for different times. (a) The cells were fixed and then stained with anti-γH2AX. (b) an estimate of the proportion of cells staining positive for γ-H2AX was made using Adobe Photoshop software. Ellipses were drawn around randomly selected nuclei by using the elliptical marquee tool. The signal in the red channel was measured by the software, and the fraction of nuclei with signal greater than a value of 45 units was calculated.
Effect of UV irradiation on RNA polymerase II.
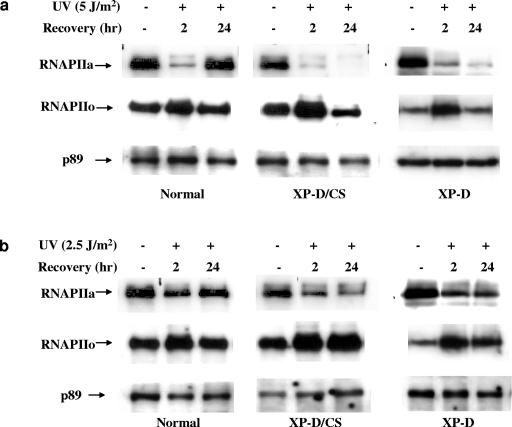
Normal, XP-D (XP1DU) and XP-D/CS (XP8BR) cells were UV irradiated, and at different times after irradiation cells were lysed and extracted either in the presence of low concentrations of nonionic detergents under physiological buffer conditions mimicking the ionic salt concentrations in the cell (19) or in the presence of higher salt concentrations (0.45 M NaCl). The extracts were then analyzed by immunoblotting, using antibodies specific for the initiating form of RNAPII that contains a hypophosphorylated C-terminal domain of the large subunit of RNAPII and is designated RNAPIIa (Fig. 7a and b, top panels). RNAPIIa is converted into a hyperphosphorylated elongating form, designated RNAPIIo, by phosphorylation of the C-terminal domain. This was detected by a different antibody (Fig. 7a and b, middle panels). In the absence of DNA damage, most of the RNAPIIa can be recovered from the soluble cellular and nuclear fraction (also shown previously [6, 33]). This situation was drastically altered after UV irradiation. The amount of RNAPIIa in the soluble extracts was severely reduced 2 h after UV irradiation, with this effect being more marked after 5 J m−2 (Fig. 7a) than after 2.5 J m−2 (Fig. 7b). In normal cells, this was followed by a restoration 24 h later (see also reference 33). Conversely, the amount of RNAPIIo increased after 2 h, returning to normal after 24 h. These results reflect the conversion of RNAPIIa to RNAPIIo after initiation and the RNAPIIo molecules becoming blocked at lesions, where they remain attached to the chromatin until either the lesions are repaired or the polymerase molecules are prematurely released into the nucleoplasm. This explains the depletion of RNAPIIa from the soluble fraction, followed by restoration after most of the blocking lesions have been repaired.
FIG. 7.
Effect of UV irradiation on RNAPII in normal, XPD, and XP-D/CS cells. Normal, XP-D/CS (XP8BR), and XP-D (XP1DU) cells were UV or mock irradiated with either 5 J/m2 (a) or 2.5 J/m2 (b) and incubated at 37°C for the indicated times before lysis and extraction of the soluble fraction. Western blotting was carried out using 8WG16 and H5 antibodies for the detection of RNAPIIa and RNAPIIo, respectively. The detection of the p89 subunit of TFIIH was used as a loading control, since we have never detected any UV-induced changes in p89 levels under these extraction conditions.
In the XP-D and XP-D/CS cell strains, after a UV dose of 5 J/m2, the initial rapid depletion of RNAPIIa was observed, as in normal cells, but it was not restored after 24 h, presumably because the blocking lesions were not repaired (Fig. 7a) (there was more pronounced depletion in XP-D/CS cells). Furthermore, the amount of RNAPIIo increased after 1 h but then decreased dramatically, even though no RNAPIIa reappeared. This suggests that in the defective cells the RNAPII was degraded. After a lower dose of 2.5 J/m2, the situation was similar in the normal cells (Fig. 7b), although the initial depletion of RNAPIIa was not as marked as with the higher dose. Interestingly, the extent of RNAPIIa depletion in XP-D cells after the low UV dose was less marked than that in the XP-D/CS cells. This correlates with the recovery of RNA synthesis after this dose of irradiation in XP-D cells compared to XP-D/CS cells (39). After this dose, RNAPII was not degraded but remained in the RNAPIIo form and was not restored to RNAPIIa (Fig. 7b, right panels).
DISCUSSION
In our earlier work, we showed that cells from the two XP-D/CS patients described in the literature generated breaks in their DNA following UV irradiation (2). We have now identified three further XP-D/CS patients, and they all show the same breakage phenomenon. These UV-induced breaks, which are transcription dependent, can be regarded as diagnostic for XP-D/CS. Indeed, they may provide a prognostic marker for development of later CS symptoms in XP-D patients. We detected similar breaks in cell strain XP1NE, from a patient previously diagnosed with XP but carrying the same causative mutation as one of the new XP-D/CS patients. As described in detail elsewhere, XP1NE also has some features of CS (14a).
In our earlier study, we used the comet assay to analyze the breaks. This assay includes a prolonged treatment of the agarose-embedded cells in strong alkali. It was therefore possible that the breaks visualized in the assay were actually alkali-labile lesions, which were converted into breaks during the alkaline treatment. The results in Fig. 2 suggest that this is not the case. All of the XP-D/CS cell strains show persisting PAR and γ-H2AX staining. γ-H2AX (Fig. 2a) and PAR staining are seen in normal cells after 2 h, presumably as a consequence of breaks introduced by NER of 6-4 photoproducts. However no significant staining is visible in normal cells at later times. PAR is the product of PARP-1 activity, which is stimulated by DNA nicks and breaks (35). γ-H2AX accumulates at sites of DNA breaks. It is normally considered to be a marker of double-strand breaks. However, O'Driscoll et al. have recently shown that cells from a patient with Seckel syndrome, defective in the ATR gene, are unable to phosphorylate H2AX in response to replication blocks, whereas there was no defect in response to ionizing-radiation-induced double-strand breaks (30). Since the ATR gene is known to be activated by single-stranded regions of DNA rather than double-strand breaks, these observations suggest that H2AX can be phosphorylated in response to the accumulation of single-stranded regions as well as double-strand breaks. We have no reason to suggest that the breaks that we observe in genomic DNA in XP-D/CS cells are double-strand breaks, and we consider it more likely that they are single-stranded regions (see also reference 32).
XRCC1 is a scaffold protein involved in base excision repair, as well as in the repair of single-strand breaks (9), and it can be observed by immunofluorescence following treatment of cells with H2O2 (13). However, we did not detect any XRCC1 foci following treatment of XP8BR with UV irradiation (results not shown). This indicates that base excision repair is probably not involved in generation of the breaks.
We previously demonstrated that the breaks were not localized at the sites of DNA damage, since they could be generated in trans by introducing UV-irradiated plasmid DNA into cells (2). However, we have now shown that localized irradiation in the nucleus generates nicks (as measured by PAR and γ-H2AX staining) only within the radiation field. Furthermore, inhibition of transcription is also localized to the damaged area. This suggests that the trans effect of the damage can only act on neighboring DNA molecules, rather than generating a signal that is transmitted throughout the nucleus.
We discovered that the breaks could be induced in response not only to UV damage but also to 8-oxoguanine and to methylation damage, although not in response to oxidized pyrimidines or to broken DNA. Several reports have indicated a deficiency in the repair of 8-oxoguanine in CS and XP-D/CS cells (25, 26), whereas reports of similar deficiencies in repair of oxidized pyrimidines have been retracted (15). However, there are no reports of deficiencies in repair of methylation damage in NER-defective syndromes and no suggestions that TFIIH might be involved in repair of alkylation damage. The repair of methylated bases is mediated by base excision repair, alkyltransferases, or oxidative demethylation, none of which involves TFIIH. Our finding of breaks induced in XP-D/CS cells in response to methylation damage is therefore quite unexpected.
We previously speculated that the breaks induced in XP-D/CS cells were at sites of transcription initiation. We have not been able to detect such breaks directly in the promoter regions of the ICAM-1 and β-actin genes (data not shown), but we consider that these negative findings are inconclusive, as only a very small proportion of promoter sites would be involved in transcription initiation at any one time. However, our results shown in Fig. 5 and 7 provide strong evidence for a link with transcription. Four transcription inhibitors all reduced the UV-induced breaks in XP-D/CS cells by approximately 70%, while having no effect on NER-induced incisions in normal cells. DNA damage is linked with transcription in at least three different ways: (i) it results in inhibition of transcription elongation, (ii) damage in the transcribed strands of active genes is preferentially repaired by transcription-coupled repair, and (iii) TFIIH is required for both NER and transcription by RNA polymerase I and II. It is difficult to understand how inhibition of transcription or transcription-coupled repair might be involved in generation of breaks, since the breaks can be induced in undamaged genomic DNA. We therefore consider that our previously proposed model of aberrant induction of breaks by the NER nucleases close to sites of transcription initiation remains the most attractive hypothesis and is consistent with the data obtained in this study. It is intriguing that although transcription inhibitors reduced the number of breaks in the XP-D/CS cells substantially, they did not abolish them, suggesting that there might be a transcription-independent process also taking place. In further support of our hypothesis, we have shown that the breaks are dependent on active XPA protein. This suggests that introduction of the breaks requires the NER machinery and that they are most likely introduced by the NER nucleases, XPG and XPF-ERCC1.
Our results on the effects of UV on RNAPII showed a depletion of RNAPIIa (Fig. 7) and accumulation of RNAPIIo within the first 2 hours after irradiation in normal, XP-D, and XP-D/CS cells. Thus, there does not appear to be any defect in XP-D/CS cells in the ability of the TFIIH-associated Cdk7 to phosphorylate the C-terminal domain of RNAPII. According to current models of transcription-coupled repair and unpublished data (M. I. Fousteri and L. H. Mullenders), when RNAPII is stalled at a UV lesion, it recruits the CSA and CSB proteins. These in turn recruit TFIIH to the stalled polymerase, and subsequent steps of NER remove the lesion. In normal cells, RNAPII can then proceed, and after completion of transcription, the RNAPIIo is dephosphorylated back to RNAPIIa. In CS cells, the blocked polymerase remains stalled at the lesion and RNAPIIo remains for long periods ((33); Fousteri and Mullenders, unpublished data). However, in the case of XP-D/CS and, to a lesser extent, XP-D cells, we speculate that the mutated TFIIH does not bind properly to the RNAPIIo-CS protein complex. As a consequence, RNAPIIo is not dephosphorylated and there is no reappearance of RNAPIIa. The relationship of these effects on transcription elongation to the proposed breaks at the sites of transcription initiation remains to be established in future work.
Acknowledgments
We are grateful to E. Seeberg for the Tag and Mag1 enzymes, to M. Berneburg for help with early experiments, and to R. Wood for helpful suggestions.
This work was supported by EC contracts QLG1-CT-1999-00181 and MRTN-CT-2003-503618 and an MRC programme grant to A.R.L. and by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC), the Italian Ministry of Health (Ricerca Finalizzata), and the MIUR (FIRB grant RBNE01RNN7) to M.S.
REFERENCES
- 1.Adam, W., C. R. Saha-Moller, A. Schonberger, M. Berger, and J. Cadet. 1995. Formation of 7,8-dihydro-8-oxoguanine in the 1,2-dioxetane-induced oxidation of calf thymus DNA: evidence for photosensitized DNA damage by thermally generated triplet ketones in the dark. Photochem. Photobiol. 62:231-238. [DOI] [PubMed] [Google Scholar]
- 2.Berneburg, M., J. E. Lowe, T. Nardo, S. Araujo, M. I. Fousteri, M. H. Green, J. Krutmann, R. D. Wood, M. Stefanini, and A. R. Lehmann. 2000. UV damage causes uncontrolled DNA breakage in cells from patients with combined features of XP-D and Cockayne syndrome. EMBO J. 19:1157-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjelland, S., and E. Seeberg. 1987. Purification and characterization of 3-methyladenine DNA glycosylase I from Escherichia coli. Nucleic Acids Res. 15:2787-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjoras, M., A. Klungland, R. F. Johansen, and E. Seeberg. 1995. Purification and properties of the alkylation repair DNA glycosylase encoded the MAG gene from Saccharomyces cerevisiae. Biochemistry 34:4577-4582. [DOI] [PubMed] [Google Scholar]
- 5.Bouayadi, K., A. van der Leer-van Hoffen, A. S. Balajee, A. T. Natarajan, A. A. van Zeeland, and L. H. Mullenders. 1997. Enzymatic activities involved in the DNA resynthesis step of nucleotide excision repair are firmly attached to chromatin. Nucleic Acids Res. 25:1056-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bregman, D. B., L. Du, S. van der Zee, and S. L. Warren. 1995. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129:287-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broughton, B. C., M. Berneburg, H. Fawcett, E. M. Taylor, C. F. Arlett, T. Nardo, M. Stefanini, E. Menefee, V. H. Price, S. Queille, A. Sarasin, E. Bohnert, J. Krutmann, R. Davidson, K. H. Kraemer, and A. R. Lehmann. 2001. Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum. Mol. Genet. 10:2539-2547. [DOI] [PubMed] [Google Scholar]
- 8.Broughton, B. C., A. F. Thompson, S. A. Harcourt, W. Vermeulen, J. H. J. Hoeijmakers, E. Botta, M. Stefanini, M. King, C. Weber, J. Cole, C. F. Arlett, and A. R. Lehmann. 1995. Molecular and cellular analysis of the DNA repair defect in a patient in xeroderma pigmentosum complementation group D with the clinical features of xeroderma pigmentosum and Cockayne syndrome. Am. J. Hum. Genet. 56:167-174. [PMC free article] [PubMed] [Google Scholar]
- 9.Caldecott, K. W. 2001. Mammalian DNA single-strand break repair: an X-ra(y)ted affair. Bioessays 23:447-455. [DOI] [PubMed] [Google Scholar]
- 10.Coin, F., E. Bergmann, A. Tremeau-Bravard, and J. M. Egly. 1999. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 18:1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer, J., J. de Wit, H. van Steeg, R. J. W. Berg, H. Morreau, P. Visser, A. R. Lehmann, M. Duran, J. H. J. Hoeijmakers, and G. Weeda. 1998. A mouse model for the basal transcription/ DNA repair syndrome trichothiodystrophy. Mol. Cell 1:981-990. [DOI] [PubMed] [Google Scholar]
- 12.de Vries, A., C. T. van Oostrom, F. M. Hofhuis, P. M. Dortant, R. J. Berg, F. R. de Gruijl, P. W. Wester, C. F. van Kreijl, P. J. Capel, H. van Steeg, and S. J. Verbeek. 1995. Increased susceptibility to ultraviolet-B and carcinogens of mice lacking the DNA excision repair gene XPA. Nature 377:169-173. [DOI] [PubMed] [Google Scholar]
- 13.El-Khamisy, S. F., M. Masutani, H. Suzuki, and K. W. Caldecott. 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31:5526-5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandez-Capetillo, O., C. D. Allis, and A. Nussenzweig. 2004. Phosphorylation of histone H2B at DNA double-strand breaks. J Exp. Med. 199:1671-1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14a.Fujimoto, M., S. N. Leech, T. Theron, M. Mori, H. Fawcett, E. Botta, Y. Nozaki, T. Yamagata, S. Moriwaki, M. Stefanini, M. Y. Momoi, H. Nakagawa, S. Schuster, C. Moss, and A. R. Lehmann. 2005. Two new XPD patients compound heterozygous for the same mutation demonstrate diverse clinical features. J. Invest. Dermatol. 125:86-92. [DOI] [PubMed]
- 15.Gowen, L. C., A. V. Avrutskaya, A. M. Latour, B. H. Koller, and S. A. Leadon. 1998. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science 281:1009-1012. (Retraction, 300:1657, 2003.) [DOI] [PubMed] [Google Scholar]
- 16.Graham, J. M., K. Anyane-Yeboa, A. Raams, E. Appeldoorn, W. J. Kleijer, V. H. Garritsen, D. Busch, T. G. Edersheim, and N. G. Jaspers. 2001. Cerebro-oculo-facio-skeletal syndrome with a nucleotide excision-repair defect and a mutated XPD gene, with prenatal diagnosis in a triplet pregnancy. Am. J. Hum Genet. 69:291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iben, S., H. Tschochner, M. Bier, D. Hoogstraten, P. Hozak, J. M. Egly, and I. Grummt. 2002. TFIIH plays an essential role in RNA polymerase I transcription. Cell 109:297-306. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, D. A., A. S. Balajee, L. Mullenders, and P. R. Cook. 1994. Sites in human nuclei where DNA damaged by ultraviolet light is repaired: visualization and localization relative to the nucleoskeleton. J. Cell Sci. 107:1745-1752. [DOI] [PubMed] [Google Scholar]
- 19.Jackson, D. A., J. Yuan, and P. R. Cook. 1988. A gentle method of preparing cyto- and nucleo-skeletons and associated chromatin. J. Cell Sci. 90:365-378. [DOI] [PubMed] [Google Scholar]
- 20.Kannouche, P., B. C. Broughton, M. Volker, F. Hanaoka, L. H. F. Mullenders, and A. R. Lehmann. 2001. Domain structure, localization and function of DNA polymerase η, defective in xeroderma pigmentosum variant cells. Genes Dev. 15:158-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keriel, A., A. Stary, A. Sarasin, C. Rochette-Egly, and J. M. Egly. 2002. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell 109:125-135. [DOI] [PubMed] [Google Scholar]
- 22.Kimura, H., K. Sugaya, and P. R. Cook. 2002. The transcription cycle of RNA polymerase II in living cells. J. Cell Biol. 159:777-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehmann, A. R. 2001. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15:15-23. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann, A. R., and S. Stevens. 1980. A rapid procedure for measurement of DNA repair in human fibroblasts and for complementation analysis of xeroderma pigmentosum cells. Mutat. Res. 69:177-190. [DOI] [PubMed] [Google Scholar]
- 25.Le Page, F., E. E. Kwoh, A. Avrutskaya, A. Gentil, S. A. Leadon, A. Sarasin, and P. K. Cooper. 2000. Transcription-coupled repair of 8-oxoguanine: requirement for XPG, TFIIH, and CSB and implications for Cockayne syndrome. Cell 101:159-171. [DOI] [PubMed] [Google Scholar]
- 26.Licht, C. L., T. Stevnsner, and V. A. Bohr. 2003. Cockayne syndrome group B cellular and biochemical functions. Am. J. Hum Genet. 73:1217-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, J., S. Akoulitchev, A. Weber, H. Ge, S. Chuikov, D. Libutti, X. W. Wang, J. W. Conaway, C. C. Harris, R. C. Conaway, D. Reinberg, and D. Levens. 2001. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell 104:353-363. [DOI] [PubMed] [Google Scholar]
- 28.Moné, M. J., M. Volker, O. Nikaido, L. H. Mullenders, A. A. van Zeeland, P. J. Verschure, E. M. Manders, and R. van Driel. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2:1013-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, V. T., F. Giannoni, M. F. Dubois, S. J. Seo, M. Vigneron, C. Kedinger, and O. Bensaude. 1996. In vivo degradation of RNA polymerase II largest subunit triggered by alpha-amanitin. Nucleic Acids Res. 24:2924-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Driscoll, M., V. L. Ruiz-Perez, C. G. Woods, P. A. Jeggo, and J. A. Goodship. 2003. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nature Genet. 33:497-501. [DOI] [PubMed] [Google Scholar]
- 31.Parrinello, S., E. Samper, A. Krtolica, J. Goldstein, S. Melov, and J. Campisi. 2003. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat. Cell Biol. 5:741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrini, J. H., and T. H. Stracker. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458-462. [DOI] [PubMed] [Google Scholar]
- 33.Rockx, D. A., R. Mason, A. van Hoffen, M. C. Barton, E. Citterio, D. B. Bregman, A. A. van Zeeland, H. Vrieling, and L. H. Mullenders. 2000. UV-induced inhibition of transcription involves repression of transcription initiation and phosphorylation of RNA polymerase II. Proc. Natl. Acad. Sci. USA 97:10503-10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogakou, E. P., D. R. Pilch, A. H. Orr, V. S. Ivanova, and W. M. Bonner. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858-5868. [DOI] [PubMed] [Google Scholar]
- 35.Shall, S., and G. de Murcia. 2000. Poly(ADP-ribose) polymerase-1: what have we learned from the deficient mouse model? Mutat. Res. 460:1-15. [DOI] [PubMed] [Google Scholar]
- 36.Takayama, K., E. P. Salazar, A. R. Lehmann, M. Stefanini, L. H. Thompson, and C. A. Weber. 1995. Defects in the repair and transcription gene ERCC2 in the cancer-prone disorder xeroderma pigmentosum. Cancer Res. 55:5656-5663. [PubMed] [Google Scholar]
- 37.Taylor, E. M., B. C. Broughton, E. Botta, M. Stefanini, A. Sarasin, N. G. J. Jaspers, H. Fawcett, S. A. Harcourt, C. F. Arlett, and A. R. Lehmann. 1997. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc. Natl. Acad. Sci. USA 94:8658-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Gool, A. J., E. Citterio, S. Rademakers, R. van Os, W. Vermeulen, A. Constantinou, J.-M. Egly, D. Bootsma, and H. J. Hoeijmakers. 1997. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in a RNA polymerase II containing complex. EMBO J. 16:5955-5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Hoffen, A., W. H. Kalle, A. Jong-Versteeg, A. R. Lehmann, A. A. Zeeland, and L. H. Mullenders. 1999. Cells from XP-D and XP-D-CS patients exhibit equally inefficient repair of UV-induced damage in transcribed genes but different capacity to recover UV-inhibited transcription. Nucleic Acids Res. 27:2898-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8:213-224. [DOI] [PubMed] [Google Scholar]
- 41.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yankulov, K., K. Yamashita, R. Roy, J. M. Egly, and D. L. Bentley. 1995. The transcriptional elongation inhibitor 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole inhibits transcription factor IIH-associated protein kinase. J. Biol. Chem. 270:23922-23925. [DOI] [PubMed] [Google Scholar]