Abstract
Biochemical and genetic studies have determined that retinoblastoma protein (pRB) tumor suppressor family members have overlapping functions. However, these studies have largely failed to distinguish functional differences between the highly related p107 and p130 proteins. Moreover, most studies pertaining to the pRB family and its principal target, the E2F transcription factor, have focused on cells that have reinitiated a cell cycle from quiescence, although recent studies suggest that cycling cells exhibit layers of regulation distinct from mitogenically stimulated cells. Using genome-wide chromatin immunoprecipitation, we show that there are distinct classes of genes directly regulated by unique combinations of E2F4, p107, and p130, including a group of genes specifically regulated in cycling cells. These groups exhibit both distinct histone acetylation signatures and patterns of mammalian Sin3B corepressor recruitment. Our findings suggest that cell cycle-dependent repression results from recruitment of an unexpected array of diverse complexes and reveals specific differences between transcriptional regulation in cycling and quiescent cells. In addition, factor location analyses have, for the first time, allowed the identification of novel and specific targets of the highly related transcriptional regulators p107 and p130, suggesting new and distinct regulatory networks engaged by each protein in continuously cycling cells.
E2F/retinoblastoma tumor suppressor (pRB) complexes play a critical role in the regulation of cell cycle progression. There are at least eight E2F transcription factors expressed in mammals (reviewed in references 3 and 29). pRB and related family members regulate the transcription of E2F-responsive genes by interacting with the E2Fs. Three pRB family members, pRB, p130, and p107 (termed pocket proteins), are expressed in mammalian cells. pRB family members in the hypophosphorylated state bind to E2Fs and inhibit transcription. This inhibition is relieved when the pocket proteins are released from E2F complexes following their phosphorylation by cyclin/CDK activity (15). There are functional and structural differences within the E2F family. E2F-1, E2F-2, and E2F-3 are transcriptional activators and interact with pRB. E2F-4 and E2F-5 are transcriptional repressors and preferentially bind p130 and p107 (17). Although E2F-6 apparently does not have a pocket protein interaction domain, it interacts with Polycomb proteins and thus represses transcription (7, 18, 32, 45). The recently identified E2F-7 and E2F-8 are also believed to repress specific promoters (14, 29).
Although p107, p130, and pRB are closely related members of the same family, they have different affinities for E2F family members, and they exhibit distinct temporal regulation during the cell cycle. While E2F4/p130 complexes are the most abundant in quiescent cells, E2F4/p107 and E2F4/pRB complexes accumulate in G1 cells (31). In addition, while p130 and p107 are bound to a number of promoters in asynchronously growing cells, only p130 is recruited to promoters in quiescent or serum-restimulated human cells (43).
These and other observations suggest unique properties among pRB and E2F family members. Indeed, a recent study revealed a distinct novel function for p107 as a negative regulator of stem cell development in the adult brain (47). Furthermore, knockout studies implicate both p130 and p107 in embryonic development and cell cycle regulation, though the mechanisms are likely different (23, 24, 47). Mice lacking either p107 or p130 in a mixed genetic background are viable and fertile, and exhibit no overt phenotypes, and mutant embryonic fibroblasts exhibit normal cell cycle kinetics (10, 21, 25). However, ablation of the p130 gene in BALB/cJ mice results in death between embryonic days E11 and E13, whereas in the same genetic background, p107-null mice display impaired growth but are viable and fertile (23, 24).
E2F cell cycle control mechanisms have been studied primarily by analyzing quiescent cells stimulated by mitogens. However, recent evidence suggests that cells reentering the cell cycle from quiescence exhibit differences in gene regulatory mechanisms compared to cells that are continuously cycling. Studies of cycling cells revealed that certain E2F-regulated genes are expressed without significant fluctuations following cell cycle reentry from mitosis (26). In addition, E2F4 and E2F5 are dispensable in quiescent cells, but necessary in cycling cells for pocket protein-mediated G1 arrest (17). Embryonic fibroblasts isolated from E2F4- and E2F5-null mice proliferate normally and reenter the cell cycle from G0 with kinetics similar to those of wild-type cells. However, they fail to arrest in G1 in response to p16INK4 (17). Moreover, it has been suggested that an E2F6 complex consisting of HP1, Polycomb group proteins, and a novel histone methyltransferase preferentially occupies target promoters in G0 rather than G1 cells (34), suggesting that chromatin modifiers contribute to silencing of E2F-responsive genes in quiescent but not G1 cells. More recent studies, however, suggest that E2F6 functions to repress certain genes in S phase, thereby limiting their expression to the G1/S transition (19).
It is becoming clear that elucidating the regulatory mechanisms that differentiate quiescent, mitogenically stimulated, and cycling cells will be essential, as it will allow the identification of specific targets that are regulated under growth-suppressive conditions and whose improper regulation could lead to cellular transformation. Thus, it has been shown that the disruption of normal regulatory networks that maintain adult stem cells in a stable, quiescent state may convert them to the malignant progenitors of cancer (2).
Several new experimental approaches, including combined use of chromatin immunoprecipitation (ChIP) and DNA microarrays (termed ChIP-on-chip or location analysis), have extended our understanding regarding cell cycle control by E2F and pocket protein complexes. This technique allows identification of novel targets of a given transcription factor on a genomic scale. These studies have revealed a diverse array of processes controlled by E2Fs, including DNA replication and repair, chromatin assembly and modification, mitosis, and cell cycle checkpoints (5, 39, 49). Although these data have immensely expanded our understanding of how E2F and pRB regulate transcription, these studies focused on transcriptional regulation of E2F-dependent genes under conditions of growth arrest or mitogenic stimulation.
Here, we have investigated how E2F-dependent genes are regulated in continuously cycling cells. Location analysis reveals differences between pocket proteins and shows that E2F4, p107, and p130 regulate common and distinct sets of genes: some genes appear to be regulated exclusively by one protein or diverse combinations of two or more of these proteins. Moreover, we have shown that these groups have distinct histone acetylation and mammalian Sin3B corepressor recruitment signatures. Interestingly, we have identified new target genes in cycling cells unique to E2F4 and p107 that are involved in stress and immune responses. Thus, we suggest that location analysis provides a unique advantage over genetic methods to discriminate between targets bound by highly related and functionally overlapping family members. We conclude that E2F4, p107, and p130 regulate distinct classes of targets, enabling them to play distinct roles in the regulation of cell cycle progression.
MATERIALS AND METHODS
Cell culture.
Human T98G cells were obtained from the American Type Culture Collection. Double and triple knockout mouse embryo fibroblasts (MEFs) derived from p107−/−, p130−/−, and RB−/− mice were described previously (12). p130- and p107-null MEFs were described previously (23, 24). T98G cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). MEFs derived from p107−/−, p130−/−, and RB−/− mice were grown in DMEM containing 10% calf serum. MEFs derived from p107−/− or p130−/− mice were grown in DMEM plus 10% FBS. Quiescent T98G cells and MEFs were synchronized by removal of serum for 3 days and stimulation with 10% FBS in the presence of 2 mM hydroxyurea for 22 to 24 h. Cells were released from hydroxyurea block for indicated lengths of time to acquire synchronized populations of cells. Propidium iodide staining and flow cytometry were used to monitor cell cycle stage as previously described (43). Only those populations exhibiting at least 80% enrichment in the early G1 fraction were used for subsequent experiments.
Western blot analysis.
Western blot analysis was performed with antibodies against pRB (G3-245; BD Biosciences), p107 (sc-318), p130 (sc-317) and cyclin B1 (sc-245) from Santa Cruz Biotechnologies, and Ser 10-phosphorylated histone H3 (clone RR002) purchased from Upstate Biotechnologies.
Construction of Hu13K promoter DNA microarrays.
The construction and use of the custom DNA microarray (Hu13K) containing 13,000 human gene promoter regions spanning ∼700 bp upstream and ∼200 bp downstream of the transcription start sites have been described previously (5, 33). The complete ChIP-on-chip data set and supplemental information can be found at http://www.med.nyu.edu/Research/dynlab01.html.
Chromatin immunoprecipitation.
Chromatin immunoprecipitation (ChIP) was performed as described previously (39, 43), with some modifications. Briefly, cells were cross-linked with 1% formaldehyde and sonicated to chromatin fragments of 500 to 1,000 bp in length. Crude chromatin was incubated with 2 μg of antibodies against p107 (sc-18), E2F4 (sc-1082), or p130 (sc-317) (all from Santa Cruz). Recovered immunoprecipitates were treated as described previously. The DNA was purified from resulting samples and assayed by semiquantitative PCR using corresponding primers for the indicated E2F-regulated promoter (sequences available upon request). The resultant PCR products were separated on either a polyacrylamide gel (if a radioactive nucleotide was used in PCRs) and visualized by exposure to X-ray film, or on 2% agarose gels (visualized by ethidium bromide staining). In some experiments, as indicated in figure legends, the intensities of the enrichment in ChIP assays were quantitated using Quantiti One and normalized to the input. The data are presented in graphs where the y axis shows input-normalized intensity (Int × mm2) of each ChIP band: (ChIP volume − mock volume)/(input volume − mock volume).
Location analysis.
Preparation of the hybridization probe was carried out essentially as described previously (38) with some modifications. Briefly, immunoprecipitated DNA was amplified by ligation-mediated PCR and 0.5 to 1 μg of amplification product was used for labeling. The absorbance of Cy3- and Cy5-labeled DNA was monitored at 550 nm for Cy3 and 650 nm for Cy5. Only the probes that contained at least 80 to 100 pmol of the dye were used for the hybridization on microarray slides. The hybridization was carried out in a hybridization buffer containing 3× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate, 20 μg human Cot-1 DNA, and 40 μg yeast tRNA at 60°C for 16 to 18 h. Following washing, promoter arrays were scanned with a Genomic Solutions LS IV or Axon 4000B scanner, and scanned images were analyzed with ImaGene and GenePix Pro software, respectively. A previously described error model (39) was used to assess the relative numbers of enriched targets. Data from three independent experiments were used to calculate average P value and enrichment ratios. The binding of a factor to DNA was deemed significant if the average P value is <0.005.
RNAi.
Small interfering RNA (siRNA) oligonucleotides targeting p130 and E2F4 were designed by and purchased from Dharmacon Inc. The sequences were as follows: p130 (5′-AAGAGCAGAGCTTAATCGAAT-3′ and 5′-AAGGACTTAGTTTATGGAAAT-3′), and E2F4 (5′-AAGGAGATTGCTGACAAACTG-3′). T98G cells were plated at the density of 6 × 105 per 100-mm tissue culture dish. At 24 h, cells were switched to serum-free Dulbecco's modified Eagle's medium. Following 36 h incubation in serum-free medium, cells were transfected with 400 pmol of siRNA oligonucleotides per 100-mm dish using sImporter transfection reagent (Upstate) per the manufacturer's instructions. At 16 h posttransfection, cells were switched to DMEM plus 10% FBS containing 2 mM final concentration of hydroxyurea. Following a 21-h hydroxyurea treatment, cells were released into DMEM plus 10% FBS and harvested at 3, 9, and 14 h for reverse transcription (RT)-PCR, Western blotting, and fluorescence-activated cell sorting (FACS) analysis.
RT-PCR.
Total RNA was extracted using TRIzol reagent (Invitrogen). First-strand cDNA synthesis was performed using SuperScript First-Strand synthesis system for RT-PCR (Invitrogen) as described by the manufacturer. The resulting cDNA was amplified using gene-specific primers. Linear amplification was assured for each gene, and three independent reactions were performed in all cases. Primer sequences are available upon request.
Affymetrix DNA microarrays.
Hybridization to human U133Av2.0 microarrays was carried out according to the manufacturer's instructions. Briefly, total RNA was isolated with TRIzol from synchronized T98G cells; 10 μg of total RNA was used for reverse transcription and subsequent in vitro cRNA synthesis. We designated a gene as derepressed if its relative signal in RNA interference (RNAi)- treated cells was ≥1.5-fold higher compared to nonspecific oligonucleotide-treated cells. Data extraction and analysis were performed with dChip (27). Each experiment was performed two times. The complete Affymetrix data set is available at http://www.med.nyu.edu/Research/dynlab01.html.
RESULTS
Analysis of cycling cells.
In order to obtain homogeneous populations of continuously proliferating cells, we tested several human cell lines, including T98G (a human glioblastoma cell line) and normal diploid IMR90 and WI38 fibroblasts for the ability to sustain synchronous cell cycle progression following release from a drug block. The most efficient and reproducible synchrony was achieved by treating T98G cells with hydroxyurea, which arrests cells in S phase, followed by a timed release into drug-free medium. A very large proportion of cells (85%) entered G1 phase 14 h after removal of drug, as verified by FACS analysis (Fig. 1A). T98G cells retain growth arrest mechanisms characteristic of normal cells, including density-mediated growth arrest and induction of quiescence in response to serum deprivation (42), and they represent a well-characterized cell line with normal expression of pocket proteins (43). Importantly, we have recently shown that T98G cells closely resemble other cell lines, including normal WI38 fibroblasts, with respect to the spectrum of target genes bound by E2F4 and p130 during cell cycle arrest (5).
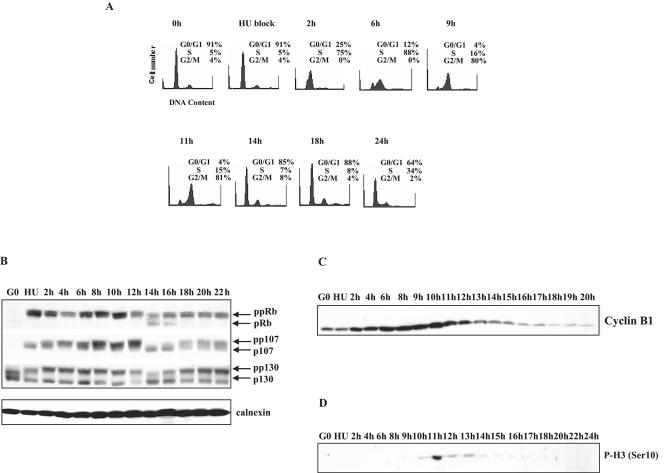
FIG. 1.
Synchronization of T98G cells. (A) Cell synchronization. Quiescent T98G cells were stimulated with medium containing 10% FBS in the presence of 2 mM hydroxyurea (HU). After 22 h, the G1/S-arrested cells were washed and released into DMEM plus 10% FBS. At the indicated times following release from hydroxyurea, cells were harvested, stained with propidium iodide, and processed for FACS analysis. (B to D) T98G cells were synchronized as for panel A. At the indicated times following release from hydroxyurea, cell lysates were harvested and subjected to SDS-PAGE and Western blot analysis with antibodies against (B) Rb, p107, and p130, (C) cyclin B1, and (D) phosphorylated histone H3 (Ser 10). A protein abundance control (calnexin) was included as indicated.
To examine the homogeneity of these cell populations, we monitored the phosphorylation of pRB family members. The hypophosphorylated form of pRB is present in quiescent (G0) and early G1 cells when transcription of E2F-responsive genes is inhibited (28, 44). As shown in Fig. 1B, pRB, p107, and p130 are hypophosphorylated in cells released from a hydroxyurea block for 14 to 16 h. These data confirm that cells released from drug for 14 to 16 h represent a population of cells in early G1 phase of the cell cycle (Fig. 1B). We also monitored levels of cyclin B1, which is degraded during mitosis (35). Figure 1C shows that cyclin B1 levels decreased 14 to 16 h after hydroxyurea release, further confirming that these cells are in early G1 phase. Finally, we assessed histone H3 serine 10 phosphorylation, which has been reported to be an excellent marker for chromosome condensation during mitotic prophase (48). Figure 1D shows that histone H3 phosphorylation is detected 11 h after hydroxyurea release and decreases to basal levels at later time points. These data demonstrate the utility of this system in isolating a highly synchronized population of cells in early G1 phase.
Promoter occupancy by E2F4, p107, and p130 throughout the cell cycle.
E2F4 represses transcription in quiescent cells through the recruitment of p130 to target promoters (5, 43). Interestingly, p107 is bound to a number of promoters in asynchronously growing cells, but not in quiescent or serum-restimulated human cells (5, 43). Figure 1B shows that p130, but not p107 or pRB, is expressed in G0 cells. However, all three pocket proteins are expressed in continuously cycling cells (Fig. 1B). This suggests that the occupancy of E2F-regulated promoters by transcriptional repressors in quiescent cells and cycling cells may not be equivalent. Therefore, we investigated the occupancy of E2F-responsive promoters in the S, G2, M, early G1, and G1/S phases of cycling cells.
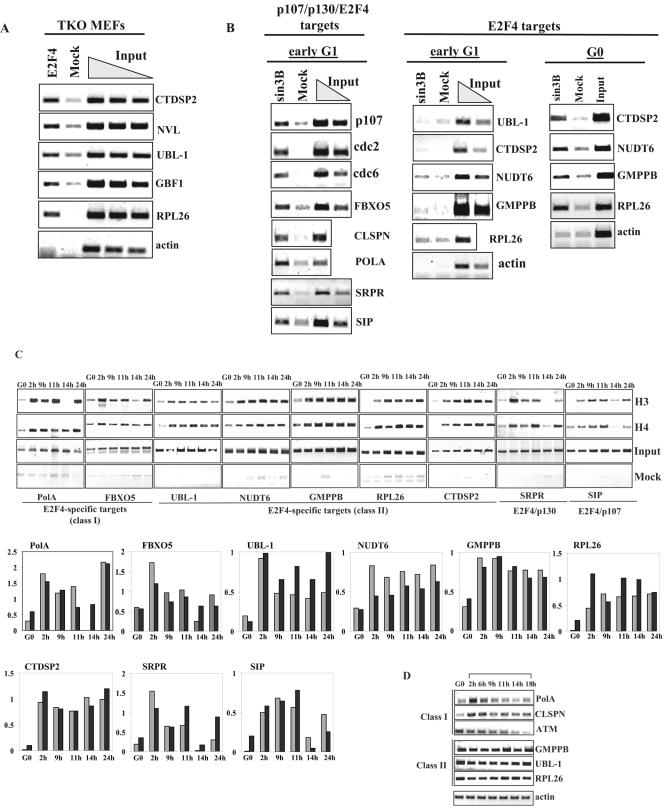
We used chromatin immunoprecipitation to show that p130 and E2F4, but not p107, are recruited to E2F-regulated promoters in quiescent cells (Fig. 2A), in agreement with previously published results (43). Interestingly, p107, p130, and E2F4 were bound to several E2F-regulated promoters in the early G1 phase of cycling cells (14 h after release from hydroxyurea; Fig. 2A). These data suggest that transcriptional repression in cycling cells is mediated by both p107 and p130. pRB was not detected on several E2F-regulated promoters in cycling cells, consistent with biochemical and genetic studies in human and mouse cells showing that pRB does not directly regulate these genes (data not shown).
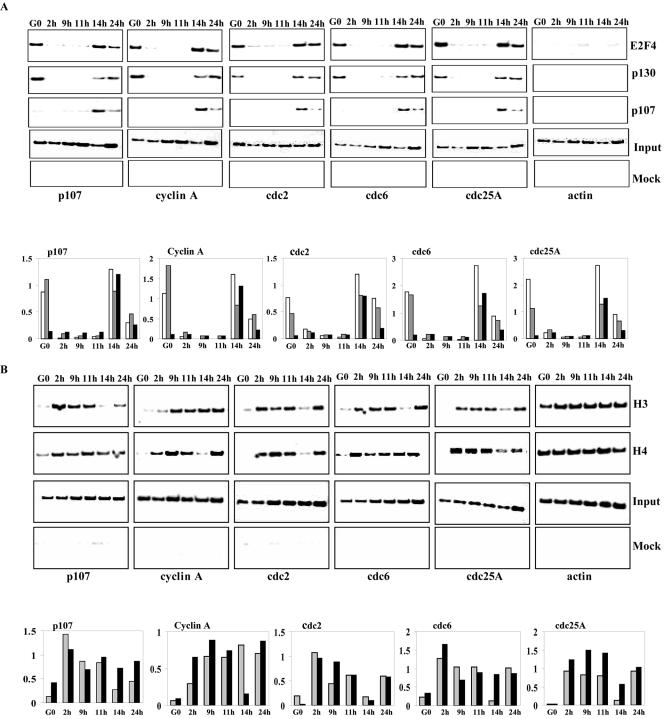
FIG. 2.
Promoter occupancy and acetylation of histones H3 and H4 throughout the cell cycle. (A) T98 G cells were synchronized by hydroxyurea (HU) block. Cells were released for 2, 9, 11, 14, and 24 h to obtain enriched populations of cells in S, G2, M, early G1, and G1/S phases, respectively. Chromatin was prepared and immunoprecipitated with antibodies against E2F4, p107, p130, or p57 (negative control). The resulting ChIPs were amplified with primers to the promoters of p107, cyclin A, cdc2, cdc6, cdc25A, and actin. The input reaction represents 0.5% of the total chromatin for each sample. (B) T98G cells were treated as above and ChIP analysis was performed using antibodies against the panacetylated forms of histone H3 and histone H4. Histograms for both panels A and B show the relative binding activity of each transcription factor or the extent of acetylation of histones as quantitated from data in the upper panel. In panel A, white, gray, and black bars represent binding of E2F4, p130, and p107, respectively. In panel B gray and black bars represent acetylated H3 and acetylated H4, respectively.
Deacetylation of histones at E2F-regulated promoters in cycling cells.
In quiescent cells, E2F4 recruits pocket proteins and histone deacetylases (HDACs) to target promoters, resulting in histone deacetylation and transcriptional repression. The HDACs then dissociate from promoters during progression from G0 to S (37), and dissociation of HDAC appears to result in a general increase in acetylation of histones H3 and H4 at E2F-responsive promoters (43). However, these studies did not address whether such changes in acetylation also occur in cycling cells upon entry into early G1 phase.
We used ChIP to investigate the cell cycle-dependent patterns of histone acetylation on a subset of E2F-responsive promoters. As shown in Fig. 2B, histones H3 and H4 are extensively acetylated at E2F-responsive promoters, in the S, G2 and M phases of the cell cycle. In contrast, both histones are deacetylated in early G1, corresponding to the period during which p107 and p130 are recruited to these promoters (Fig. 2A), although the extent of deacetylation varied among the promoters (Fig. 2B). In stark contrast with quiescent cells, either histone H3 or H4, but generally not both, was deacetylated on certain promoters. Interestingly, we could distinguish three different patterns of deacetylation: promoters that are deacetylated on histone H3 alone (p107 and CDC6), histone H4 alone (cyclin A), or a combination of both (CDC2 and CDC25A). These findings suggest that promoters may have specific histone acetylation signatures that must be reset with each cell cycle and that deacetylation of either histone may suffice in some cases to resilence each during early G1 of cycling cells. This could occur through cycles of recruitment of one or more HDAC with distinct specificities for a given histone.
ChIP-on-chip analysis reveals binding of E2F4 and pocket proteins to common and distinct sets of targets.
To further investigate the role of E2F4, p107, and p130 in cell cycle-dependent gene regulation, we performed genome-wide location analysis to identify promoters bound by each factor during early G1 in cycling cells. For this analysis, we made use of a custom DNA microarray (Hu13K) containing 13,000 human gene promoter regions spanning ∼700 bp upstream and ∼200 bp downstream of the transcription start sites (5, 33). Figure 3A shows representative scatter plots for the G1-phase location analyses of E2F4, p130, and p107 on the Hu13K promoter microarray. Using a P value cutoff of ≤ 0.005, we found that E2F4 occupied 301 promoters, while p130 and p107 were bound to 226 and 244 genes, respectively (supplemental data S1, located at http://www.med.nyu.edu/Research/dynlab01.html).A comparison of targets revealed that the majority (171) of targets were bound by all three factors (Fig. 3B). Remarkably, a substantial number of targets (74) identified in the E2F4 ChIP-on-chip experiment did not overlap with either p107 or p130 targets and were thus unique for E2F4 (Fig. 3B). Moreover, we identified another subset of genes (40) that were uniquely bound by p107. We estimated the false positive and false negative rates for all three antibodies in our genomewide location analysis experiments to be 12% and 18%, respectively, when a P value cutoff of ≤0.005 was used.
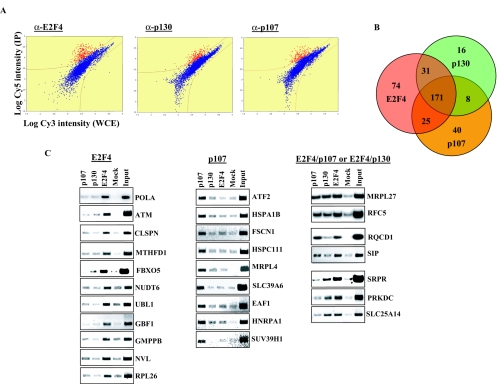
FIG. 3.
Location analysis of E2F4, p107 and p130 in early G1 of cycling T98G cells. (A). Representative scatter plots for location analyses on Hu13K promoter microarrays. Fluorescence intensities in the Cy5 channel (ChIP) are plotted against the Cy3 channel (input chromatin). Red spots represent enriched promoters and blue spots represent nonenriched promoters. (B). Venn diagram comparing E2F4, p130, and p107 targets. (C). Subset of targets identified by ChIP-on-Chip confirmed by conventional ChIP and gene-specific PCR. Chromatin immunoprecipitation was performed as described in Materials and Methods. The input lane indicates PCRs containing 0.25% of total chromatin for each sample.
This analysis suggested that genes bound exclusively by E2F4 or p107 are likely to be true targets and revealed subgroups of genes that are differentially regulated by these factors. Furthermore, scatter plots in which promoter occupancy was compared among these proteins showed that P values for a subset of targets bound by E2F4, p130, or p107 are significantly lower compared to the same targets in different immunoprecipitations (supplemental data S2), further supporting the notion that a majority of the targets exclusively bound by each transcription factor in our ChIP-on-chip experiment are likely to be true hits. In order to validate these results, we conducted extensive conventional ChIP analysis. Figure 3C shows a selected group of targets that are exclusively bound by E2F4 or p107, a combination of E2F4 and either p107 or p130, or all three factors. These results indicate that we have succeeded in identifying and confirming promoters that are selectively bound by E2F4 and different pocket proteins in the early G1 phase of the cell cycle.
Functional categories of targets.
E2F/pocket protein complexes are known to regulate the G1-S phase transition and DNA replication. Recent studies have shown that the role of E2F/pRB family complexes extends beyond these well-established functions to mitosis, chromatin assembly, chromatin modification, and apoptosis (5, 6, 22, 36, 39, 50). We used Gene Ontology to categorize the novel early G1 phase targets of E2F4, p130, and p107 into functional categories and found that E2F/p107/p130 complexes regulate genes within a diverse range of functional categories, including apoptosis, mitochondrial biogenesis, transcriptional regulation, protein ubiquitination and turnover, and organelle assembly and function (Fig. 4A and supplemental data S3). We compared the functional categories of targets found in cycling cells to those identified in growth-inhibited conditions (5). Interestingly, we found that genes in three functional categories-stress response, signal transduction, and immune response—were uniquely bound by E2F4 and/or p107 in the early G1 phase of cycling cells (Fig. 4A) (5).
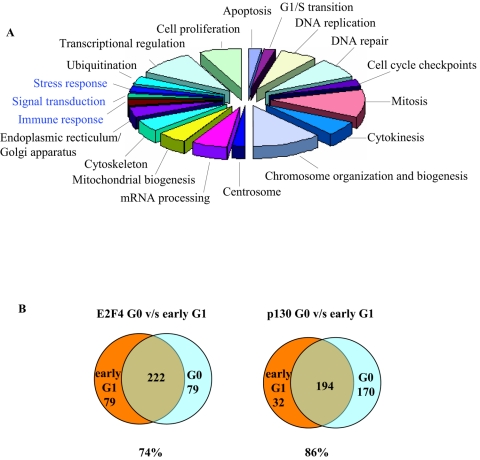
FIG. 4.
Functional categories of ChIP-on-chip targets in constantly cycling and growth inhibited cells. (A) Pie chart describing major functional gene ontology categories of E2F4, p107, and p130 targets enriched in the early G1 phase of cycling cells. Functional categories in blue represent targets identified in cycling cells but not growth-inhibited cells. (B) Venn diagrams comparing E2F4 and p130 targets in the early G1 phase of cycling cells to that of G0 phase in quiescent cells.
We compared the set of genes that are bound by p130 in G0 and early G1 of cycling cells and found that the overlap between these groups of targets is 86% (Fig. 4B and supplemental data S1) (5). Thus, the difference in overlap observed between p130 targets in these two conditions fell within or below the false positive and negative rates, suggesting that p130 likely binds a nearly identical (if not identical) set of genes in quiescent and early G1 cycling cells. To further elucidate differences between quiescence and the early G1 phase of proliferating cells, we compared the genes bound by E2F4 and p130 in early G1 with ChIP-on-chip data from G0 cells. Figure 4B shows that 74% of genes bound by E2F4 in G0 are also bound by E2F4 in early G1 of cycling cells. The number of targets that are specific for each condition exceeds what might be expected given false positive and negative rates, suggesting that E2F4 regulates distinct subgroups of targets in each condition. These conclusions are further supported by scatter plots of P values for enrichment of E2F4 targets in early G1-phase and G0 cells (supplemental data S2) wherein we identified a subset of targets whose P values were significantly lower in early G1 than in G0.
Functional analysis of targets. (i) Subsets of targets are differentially regulated in pocket protein-null cells.
In contrast with observations that E2F4 and p130 function together to repress transcription in growth-arrested cells (5), we identified a number of novel genes that were bound by E2F4, but not by p107 or p130, in early G1 cells (Fig. 3B and 3C). These findings suggest that certain genes can be regulated by E2F4 in a p107/p130-independent manner in early G1. To confirm this finding, we analyzed expression of E2F target genes in primary mouse embryo fibroblasts that are wild type, deficient for p107 and p130 (double knockout), or all three pRB family members (triple knockout). Cells were synchronized as before by hydroxyurea treatment and released for 14 h to obtain early G1 cells prior to performing semiquantitative RT-PCR.
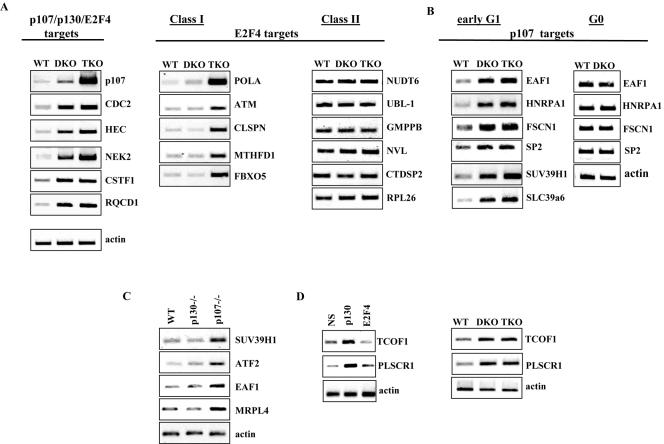
As expected, loss of p107 and p130 resulted in derepression of genes bound by both p107 and p130 (Fig. 5A; p107/p130/E2F4 targets). Interestingly, a subset of genes, including POLA, ATM, CLSPN, MTHFD1, and FBXO5, that are bound exclusively by E2F4 were derepressed in triple-knockout cells, but not in double-knockout cells, suggesting that these genes may be direct or indirect targets of pRB (Fig. 5A; E2F4 targets, class I). We have performed ChIP experiments to address whether pRB binds these promoters directly. However, we have failed to detect pRB on several class I promoters that we tested (data not shown). We note that we have only examined proximal promoters using this assay, and we cannot rule out the possibility that RB may regulate these genes by binding to another region beyond the boundaries of our assay or that it may indirectly regulate this class of promoters.
FIG. 5.
Regulation of E2F4-specific targets in early G1 cells deficient for p107/p130, and all pocket proteins. (A to D) RT-PCR analysis of targets identified by location analysis. (A) Expression of a subset of p107/p130/E2F4- and E2F4-specific targets in p107- and p130-null MEFs (DKO), and p107-, p130-, and pRB-null MEFs (TKO). Prior to RT-PCR analysis, MEFs were synchronized in early G1 by hydroxyurea block and release for 14 h. Class I E2F4 targets are those that show deregulation in triple-knockout but not double-knockout MEFs. Class II E2F4 targets are those that are not deregulated in either double-knockout or triple-knockout MEFs. (B) Expression of a subset of p107 targets in wild-type (WT), double-knockout (DKO), and triple-knockout (TKO) MEFs. Prior to RT-PCR analysis, MEFs were synchronized in either G0 or G1 as described in Materials and Methods. (C) Expression of p107 targets in p107- or p130-null MEFs. Cells were treated as for panel A. (D) Left panel: RT-PCR analysis of p130 targets in T98G (human) cells treated with nonspecific siRNA or siRNA specific for p130. T98G cell were synchronized by hydroxyurea (HU) treatment and released for 3 h from hydroxyurea block. Right panel: RT-PCR analysis of two p130 targets in early G1 double-knockout MEFs and triple-knockout MEFs.
Remarkably, we identified yet another set of E2F4-specific targets whose expression is unaffected by loss of all three pocket proteins (e.g., NUDT6 and others; Fig. 5, E2F4 targets, class II). We considered the possibility that these observations stemmed from altered cell cycle kinetics in the triple-knockout cells. Importantly, although it has been previously reported that loss of all three pocket proteins abolishes growth arrest by serum deprivation (12, 40), we were able to demonstrate that the kinetics of cyclin B1 expression in triple-knockout cells following release from an hydroxyurea block are similar to those observed in wild-type MEFs and T98G cells (data not shown). Thus, the failure of triple-knockout cells to cycle synchronously following hydroxyurea release is not a likely explanation for our results. In summary, our data support the notion that the regulation of a significant subset of E2F4-bound genes is not dependent on pocket proteins. On other promoters, E2F4 may cooperate with either p107/p130 or pRB.
We investigated whether the ablation of E2F4 by specific siRNAs would deregulate E2F4-only targets. We observed no dramatic changes in the expression of these targets, most likely as a result of functional compensation by either E2F5 or E2F6 (17, 19, 38). When we examined E2F6 binding to a subset of these E2F4-only promoters (CTDSP2, NVL1, GBF1, and RPL26) in wild-type and triple-knockout MEFs, we found that E2F6 is recruited to most of the targets that we tested, consistent with this possibility (data not shown).
We also examined the expression levels of genes identified as p107-specific by comparing wild-type, double-knockout, and triple-knockout cells. As expected and as shown in Fig. 5B (p107 targets), expression of certain p107 target genes (e.g., SUV39H1 and others) increases in cells lacking p107 and p130 and in cells devoid of all three pocket proteins. Interestingly, we found that the observed derepression of p107-specific genes in double-knockout and triple-knockout MEFs is specific to cycling cells, and derepression of the same genes is not seen in quiescent cells (Fig. 5B, compare early G1 and G0). These data further support the existence of a cadre of genes specifically regulated by p107 in cycling cells.
Next, we investigated the regulation of unique p107 targets in single-knockout MEFs that are deficient for p107 or p130. Strain-dependent defects have been observed in mice lacking either the p107 or p130 gene (23, 24). Interestingly, as shown in Fig. 5B, several genes bound exclusively by p107 (ATF2, SUV39H1, MRPL4, and EAF1) are up-regulated in p107−/− cells compared to wild-type and p130−/− cells. These data provide the first evidence for novel regulation of p107-selective targets in both human and mouse cells. Of particular interest is SUV39H1, a histone methyltransferase whose activity is required for heterochromatin formation and whose overexpression leads to growth retardation (11). Taken together with our observation that heterochromatin protein HP1 is also a target of pocket protein-E2F4 repression (5), our data indicate that expression of the SUV39H1-HP1 module may be entrained to the cell cycle via pocket protein-mediated regulation.
In an effort to uncover p130-specific targets, we performed genome-wide expression profiling of cells treated with p130 siRNAs using Affymetrix microarrays. Two genes that were bound exclusively by p130 in our ChIP-on-chip experiments were significantly up-regulated (4.1- and 1.7-fold) in cells treated with two different p130 siRNAs compared with cells treated with control siRNA (data not shown). These genes encode phospholipid scramblase 1 (PLSCR1) and Treacher-Collins syndrome factor 1 (TCOF1). We confirmed the effect of p130 knockdown by both p130 siRNAs using RT-PCR, and the expression of these genes did not change in cells treated with control or E2F4-specific siRNAs, in agreement with our previous ChIP-on-chip results (Fig. 5D and data not shown). Finally, we tested the expression levels of these genes in double-knockout and triple-knockout MEFs. Importantly, the RNA levels of both genes were increased in double-knockout and triple-knockout cells compared to wild-type cells (Fig. 5D).
(ii) E2F4 binding to promoters in triple-knockout MEFs.
Our previous findings indicated that p130 and p107 play a role in the nuclear localization of E2F4 in G0, and that binding of E2F4 to certain promoters is undetectable in quiescent cells lacking p107 and p130 (37). This suggests that regulation of a subset of genes bound exclusively by E2F4 in cycling cells may be driven by a novel mechanism. Thus, we investigated whether the binding of E2F4 to these promoters is affected in MEFs lacking all three pocket proteins. Interestingly, ChIP analysis revealed that E2F4 binding persists in cells lacking pocket proteins on the promoters that were bound exclusively by E2F4 in our ChIP-on-chip experiments (Fig. 6A). These data offer further support that E2F4 is indeed capable of binding to certain promoters independently of pocket proteins.
FIG. 6.
Analysis of E2F-specific promoters. (A) ChIP of E2F4 in triple-knockout (TKO) MEFs. Various amounts of total chromatin (0.5, 0.25, and 0.125%) were used to estimate the relative binding of E2F4 to the promoters. (B) ChIP of Sin3B in T98G cells. Cells were synchronized by hydroxyurea (HU) block and released for 14 h to enrich for cells in early G1 phase or serum starved for 72 h (G0). (C) Acetylation of E2F4-specific promoters. T98G cells were treated as described in the legend to Fig. 2B. Gene names and classifications are indicated. Graphs for both panels show the relative extent of acetylation of histones as quantitated from data in the figure. Gray and black bars represent acetylated H3 and acetylated H4, respectively. (D) Expression analysis of E2F4-only class I and class II genes. T98G cells were treated with hydroxyurea and released from this block for the indicated times as described in the legend to Fig. 2B. The brackets indicate times after release from hydroxyurea.
Recruitment of corepressor mSin3B and acetylation.
Our previous studies showed that the transcriptional corepressor complex mSin3B/HDAC1 is specifically recruited to endogenous E2F-regulated promoters in quiescent cells (37). Thus, we investigated whether mSin3B is recruited to promoters commonly bound by p107/130/E2F and promoters bound specifically by E2F4 in cycling cells. We found that mSin3B binds to the common E2F-regulated promoters p107, CDC2, and CDC6 and to promoters that are regulated by a combination of p107/E2F4 or p130/E2F4 (Fig. 6B). In striking contrast, we were not able to detect binding of mSin3B to the set of E2F4-specific promoters we previously identified as pocket protein-independent targets of E2F4 (class II targets) in early G1 of continuously cycling cells. Remarkably, however, we could show that mSin3B binding was detected on the same set of promoters in quiescent cells (Fig. 6B and 5A), indicating that Sin3B recruitment may be one key mechanistic discriminator for this set of genes (class II targets) that distinguishes between their regulation in G0 and early G1.
Sin3 can be recruited to promoters as a Sin3/HDAC complex by several transcription factors, altering their histone acetylation state and repressing gene expression (1, 37, 41). Our previous studies showed that the decrease in histone acetylation levels coincides with transcriptional repression of E2F-responsive genes (43). Thus, we compared the histone acetylation state of class I and class II E2F4-specific targets, as well as E2F4 targets that also recruit p107/p130. Interestingly, in cycling early G1 cells, we observed persistent acetylation at the promoters of class II E2F4-bound genes that do not recruit Sin3B (Fig. 6B and C), but not at class I targets, or E2F4 targets that are also bound by p107/p130 as well as Sin3B (Fig. 2B and Fig. 6B and C).
Finally, in order to further characterize the set of genes we identified as E2F4-specific, we examined previously published cDNA microarray experiments that monitored the expression of ∼29,600 human genes during cell cycle progression to determine whether E2F4-specific genes show cell cycle periodicity (51). Interestingly, we found that the vast majority of genes (85%) identified as E2F4-specific in our ChIP-on-chip analysis are not cell cycle regulated (supplemental data S1). To confirm this experimentally, we examined the cell cycle periodicity of expression of class I and class II E2F4-specific target genes. Strikingly, we found that the expression of class I E2F4-only targets fluctuates throughout the cell cycle, while class II E2F4 targets do not (Fig. 6D).
That class I targets are deregulated in triple-knockout but not double-knockout MEFs (suggesting a role for pRB) is particularly noteworthy in light of these observations (Fig. 5). Furthermore, the data in Fig. 6D augment the other differences we see in these two classes of genes: class I targets are bound by mSin3B and exhibit diminished histone acetylation at their promoters during early G1, while class II targets do not appear to recruit mSin3B and the levels of promoter histone acetylation remain nearly constant through early G1 (Fig. 6). This suggests that E2F-dependent cell cycle oscillations in gene expression may be driven by the periodic recruitment of an HDAC/mSin3B complex in early G1 phase in cycling cells.
DISCUSSION
Here we investigated the role of highly related pocket proteins and E2F4 on gene transcription in growing cells. We have shown that E2F4, p130, and p107 are major transcriptional regulators in early G1 phase in cycling cells. Genomewide analysis of transcription factor occupancy revealed that E2F4 and pocket proteins regulate common and distinct sets of targets in cycling cells. The comparison of targets that were identified by ChIP-on-chip analysis in quiescent cells (5) and continuously cycling cells revealed that E2F4 and pocket proteins bind to a cadre of genes that are specific to the latter population. Our studies reveal unanticipated complexity in the recruitment of E2F4 and pocket proteins to distinct classes of promoters, where they have differential effects on corepressor mSin3B recruitment, acetylation, and transcriptional repression.
Numerous molecular events contribute to transcriptional repression. One such event is chromatin condensation, which limits the accessibility of the DNA template to transcription factors and RNA polymerases. However, recent studies have shown that certain basal transcription factors remain associated with cell cycle-regulated promoters in asynchronous and mitotic human cell populations, arguing that such factor-promoter complexes withstand condensation of chromatin into transcriptionally silent chromosomes (8, 20, 30). Unlike these basal transcription factors, E2F4, p130, and p107 are not detected on E2F-regulated promoters in the S, G2, or M phase of cycling cells, but only in the G0 and early G1 phases. These data suggest that the binding of these pocket proteins and E2F4 to the promoter is restricted to cell cycle stages in which transcription is repressed.
E2F/pocket protein complexes repress transcription through the recruitment of histone-modifying enzymes, in particular HDACs, to target promoters. Recruitment of histone deacetylases results in histone H3 and H4 deacetylation and transcriptional silencing. We have found that, although histones associated with E2F-regulated promoters are generally acetylated throughout the cell cycle in growing cells, acetylation is significantly decreased in early G1 phase. Interestingly, our data show that histone acetylation profiles differ from promoter to promoter. These data are in agreement with previously published studies in Saccharomyces cerevisiae (13) showing that specific activators confer distinct patterns of histone acetylation on target promoters, and transcriptional activation is not necessarily associated with increased acetylation.
Our data show that promoters are acetylated in the S, G2 and M phases of the cell cycle. However, we detect activator E2F (E2F1 and E2F2) binding to promoters only in S phase, not in G2 or M phase (data not shown), in agreement with recently published data (19). This suggests that transcription might be silenced in the G2 and M phases of cell cycle, despite the acetylation of H3 and H4 that persists during these phases. Furthermore, promoter-selective deacetylation of histones H3 and H4 in early G1 suggests that distinct histone deacetylases may preferentially bind specific E2F targets and could imply the existence of an additional layer of specificity at E2F-responsive promoters. Alternatively, these promoters could recruit a single HDAC that acquires new specificities for histone H3 or H4 in a promoter-dependent context.
ChIP-on-chip and functional analysis of targets.
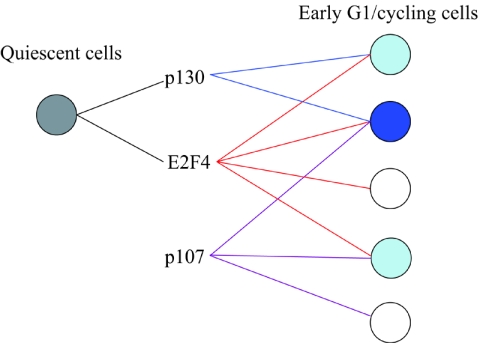
Here we investigated the differences between highly related pocket protein family members in cycling human cells. Previous studies revealed that p130 and E2F4 share highly overlapping functions in growth-arrested cells. In contrast, our findings suggest that E2F4, p130 and p107 regulate common and unique sets of targets in proliferating cells (Fig. 7). In addition, we have found evidence that E2F4 and p107 may regulate gene transcription independently of pRB family members and E2F4, respectively. These findings indicate that there may be at least five distinct groups of targets regulated by combinations of E2F4, p107, and p130. Additional classes may exist, given evidence suggesting promoter-specific patterns of histone acetylation.
FIG. 7.
Model summarizing different transcriptional networks regulated by E2F4 and/or pocket proteins in quiescent and early G1 cells. Color intensity indicates the number of factors that regulate a given class of target genes.
Our data are in agreement with previous studies showing that pRB family members p107 and p130 have overlapping functions as well as seemingly opposing functions (9). In addition, this notion is supported by the observation that overexpression of pRB in wild-type cells promotes differentiation, whereas overexpression of p107 antagonizes differentiation in the same setting (9). Moreover, our findings highlight potential differences in mechanisms of transcriptional suppression in continuously proliferating and growth-inhibited cells. In this regard, we provide strong evidence that the p107 repressor functions primarily in early G1 phase, since p107-specific targets are derepressed in early G1 cells deficient for this protein, but not in the same cells rendered quiescent through mitogen deprivation (Fig. 5B).
Furthermore, our data provide new insights into the transcriptional regulatory mechanisms of subgroups of E2F-regulated genes and suggest distinctive patterns of mSin3B corepressor recruitment to their promoters, resulting in differential acetylation: Differential recruitment of Sin3B (and HDACs) in quiescent or early G1 cells (or both) could establish a mark that mechanistically distinguishes certain E2F4 target genes and ensures that genes will be deacetylated and silenced under both conditions or exclusively during cell cycle arrest.
Our genomewide location analysis identified new classes of genes that are differentially regulated in continuously cycling cells. In particular, our examination of E2F4-specific genes raises the possibility that the recruitment of E2F4 to promoters does not necessarily correlate with gene regulation in normal conditions. Thus, E2F4 may be bound to specific promoters in an inactive state, awaiting activation of its repressive function through recruitment of additional transcriptional regulators in response to currently unknown cues. Such a poised state could allow rapid responses to changing extracellular cues (4, 5).
We have also found that certain genes (TCOF1 and PLSCR1) are derepressed in cells in which p130 expression has been suppressed. These genes were also identified as p130-specific targets. PLSCR1 has been shown to play a role in cell differentiation, apoptosis, and proliferation (16, 52). The TCOF1 gene product is involved in ribosomal RNA gene transcription by interacting with an upstream binding factor (46). Mutations in the Treacher-Collins syndrome gene, TCOF1, result in craniofacial defects, and Tcof+/− mouse embryos exhibit craniofacial defects and growth retardation (46). It will be interesting in future studies to test a role for regulation of PLSCR1 and/or TCOF1 transcription in cell cycle control and differentiation. This may offer further insights into the biological functions of p130 and the contribution of these functions to the phenotypes of knockout animals. We surmise that in the future, similar genome-scale analyses will uncover distinctions among targets of other fully or partially redundant regulatory proteins and reveal novel mechanisms through which they function.
Finally, our observations regarding gene expression of the two subsets of E2F4-only target genes (class I and class II, Fig. 5 and 6) demonstrate that these are truly distinct classes of E2F4 target genes, regulated by separate but not yet fully understood mechanisms. This observation highlights how careful studies of gene regulation during the early G1 phase of cycling cells reveal important phenomena that may not be disclosed by studies with quiescent and serum-stimulated cell populations.
Acknowledgments
We thank I. Sanchez, L. Gardner, A. Blais, M. Tsikitis, and members of the Dynlacht laboratory for critical input and suggestions. We are grateful to Michael S. Huh for the help with MEFs. We are grateful to the NYU Cancer Institute Genomics Facility for providing necessary instrumentation and expertise.
N.H.L. was initially supported by an NRSA postdoctoral fellowship (1F32-AG025617-01) and is now supported by a Susan G. Komen postdoctoral fellowship (PDF0504345). This work was initiated and completed using funds from the American Cancer Society (#RSG-0034204CCG) and NIH (2R01 CA77245-07), respectively, to B.D.D.
REFERENCES
- 1.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 2.Bissell, M. J., and M. A. Labarge. 2005. Context, tissue plasticity, and cancer: are tumor stem cells also regulated by the microenvironment? Cancer Cell 7:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blais, A., and B. D. Dynlacht. 2004. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr. Opin. Genet. Dev. 14:527-532. [DOI] [PubMed] [Google Scholar]
- 4.Blais, A., M. Tsikitis, D. Acosta-Alvear, R. Sharan, Y. Kluger, and B. D. Dynlacht. 2005. An initial blueprint for myogenic differentiation. Genes Dev. 19:533-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 6.Cam, H., and B. D. Dynlacht. 2003. Emerging roles for E2F: beyond the G1/S transition and DNA replication. Cancer Cell 3:311-316. [DOI] [PubMed] [Google Scholar]
- 7.Cartwright, P., H. Muller, C. Wagener, K. Holm, and K. Helin. 1998. E2F-6: a novel member of the E2F family is an inhibitor of E2F-dependent transcription. Oncogene 17:611-623. [DOI] [PubMed] [Google Scholar]
- 8.Christova, R., and T. Oelgeschlager. 2002. Association of human TFIID-promoter complexes with silenced mitotic chromatin in vivo. Nat. Cell Biol. 4:79-82. [DOI] [PubMed] [Google Scholar]
- 9.Classon, M., B. K. Kennedy, R. Mulloy, and E. Harlow. 2000. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 97:10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobrinik, D., M. H. Lee, G. Hannon, G. Mulligan, R. T. Bronson, N. Dyson, E. Harlow, D. Beach, R. A. Weinberg, and T. Jacks. 1996. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 10:1633-1644. [DOI] [PubMed] [Google Scholar]
- 11.Czvitkovich, S., S. Sauer, A. H. Peters, E. Deiner, A. Wolf, G. Laible, S. Opravil, H. Beug, and T. Jenuwein. 2001. Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech. Dev. 107:141-153. [DOI] [PubMed] [Google Scholar]
- 12.Dannenberg, J. H., A. van Rossum, L. Schuijff, and H. te Riele. 2000. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 14:3051-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deckert, J., and K. Struhl. 2001. Histone acetylation at promoters is differentially affected by specific activators and repressors. Mol. Cell. Biol. 21:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiStefano, L., M. R. Jensen, and K. Helin. 2003. E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J. 22:6289-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyson, N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 16.Frasch, S. C., P. M. Henson, J. M. Kailey, D. A. Richter, M. S. Janes, V. A. Fadok, and D. L. Bratton. 2000. Regulation of phospholipid scramblase activity during apoptosis and cell activation by protein kinase Cdelta. J. Biol. Chem. 275:23065-23073. [DOI] [PubMed] [Google Scholar]
- 17.Gaubatz, S., G. J. Lindeman, S. Ishida, L. Jakoi, J. R. Nevins, D. M. Livingston, and R. E. Rempel. 2000. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol. Cell 6:729-735. [DOI] [PubMed] [Google Scholar]
- 18.Gaubatz, S., J. G. Wood, and D. M. Livingston. 1998. Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. USA 95:9190-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giangrande, P. H., W. Zhu, S. Schlisio, X. Sun, S. Mori, S. Gaubatz, and J. R. Nevins. 2004. A role for E2F6 in distinguishing G1/S- and G2/M-specific transcription. Genes Dev. 18:2941-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottesfeld, J. M., and D. J. Forbes. 1997. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 22:197-202. [DOI] [PubMed] [Google Scholar]
- 21.Hurford, R. K., Jr., D. Cobrinik, M. H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 22.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LeCouter, J. E., B. Kablar, W. R. Hardy, C. Ying, L. A. Megeney, L. L. May, and M. A. Rudnicki. 1998. Strain-dependent myeloid hyperplasia, growth deficiency, and accelerated cell cycle in mice lacking the Rb-related p107 gene. Mol. Cell. Biol. 18:7455-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LeCouter, J. E., B. Kablar, P. F. Whyte, C. Ying, and M. A. Rudnicki. 1998. Strain-dependent embryonic lethality in mice lacking the retinoblastoma-related p130 gene. Development 125:4669-4679. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. H., B. O. Williams, G. Mulligan, S. Mukai, R. T. Bronson, N. Dyson, E. Harlow, and T. Jacks. 1996. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 10:1621-1632. [DOI] [PubMed] [Google Scholar]
- 26.Leone, G., J. DeGregori, Z. Yan, L. Jakoi, S. Ishida, R. S. Williams, and J. R. Nevins. 1998. E2F3 activity is regulated during the cell cycle and is required for the induction of S phase. Genes Dev. 12:2120-2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2:RESEARCH0032. [DOI] [PMC free article] [PubMed]
- 28.Ludlow, J. W., C. L. Glendening, D. M. Livingston, and J. A. DeCarprio. 1993. Specific enzymatic dephosphorylation of the retinoblastoma protein. Mol. Cell. Biol. 13:367-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maiti, B., J. Li, A. de Bruin, F. Gordon, C. Timmers, R. Opavsky, K. Patil, J. Tuttle, W. Cleghorn, and G. Leone. 2005. Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J. Biol. Chem. 280:18211-18220. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Balbas, M. A., A. Dey, S. K. Rabindran, K. Ozato, and C. Wu. 1995. Displacement of sequence-specific transcription factors from mitotic chromatin. Cell 83:29-38. [DOI] [PubMed] [Google Scholar]
- 31.Moberg, K., M. A. Starz, and J. A. Lees. 1996. E2F-4 switches from p130 to p107 and pRB in response to cell cycle reentry. Mol. Cell. Biol. 16:1436-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morkel, M., J. Wenkel, A. J. Bannister, T. Kouzarides, and C. Hagemeier. 1997. An E2F-like repressor of transcription. Nature 390:567-568. [DOI] [PubMed] [Google Scholar]
- 33.Odom, D. T., N. Zizlsperger, D. B. Gordon, G. W. Bell, N. J. Rinaldi, H. L. Murray, T. L. Volkert, J. Schreiber, P. A. Rolfe, D. K. Gifford, E. Fraenkel, G. I. Bell, and R. A. Young. 2004. Control of pancreas and liver gene expression by HNF transcription factors. Science 303:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogawa, H., K. Ishiguro, S. Gaubatz, D. M. Livingston, and Y. Nakatani. 2002. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science 296:1132-1136. [DOI] [PubMed] [Google Scholar]
- 35.Petersen, B. O., C. Wagener, F. Marinoni, E. R. Kramer, M. Melixetian, E. L. Denchi, C. Gieffers, C. Matteucci, J. M. Peters, and K. Helin. 2000. Cell cycle- and cell growth-regulated proteolysis of mammalian CDC6 is dependent on APC-CDH1. Genes Dev. 14:2330-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polager, S., Y. Kalma, E. Berkovich, and D. Ginsberg. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437-446. [DOI] [PubMed] [Google Scholar]
- 37.Rayman, J. B., Y. Takahashi, V. B. Indjeian, J. H. Dannenberg, S. Catchpole, R. J. Watson, H. te Riele, and B. D. Dynlacht. 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 16:933-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rempel, R. E., M. T. Saenz-Robles, R. Storms, S. Morham, S. Ishida, A. Engel, L. Jakoi, M. F. Melhem, J. M. Pipas, C. Smith, and J. R. Nevins. 2000. Loss of E2F4 activity leads to abnormal development of multiple cellular lineages. Mol. Cell 6:293-306. [DOI] [PubMed] [Google Scholar]
- 39.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein, G. H. 1979. T98G: an anchorage-independent human tumor cell line that exhibits stationary phase G1 arrest in vitro. J. Cell Physiol. 99:43-54. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, Y., J. B. Rayman, and B. D. Dynlacht. 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 14:804-816. [PMC free article] [PubMed] [Google Scholar]
- 44.Tamrakar, S., and J. W. Ludlow. 2000. The carboxyl-terminal region of the retinoblastoma protein binds non-competitively to protein phosphatase type 1alpha and inhibits catalytic activity. J. Biol. Chem. 275:27784-27789. [DOI] [PubMed] [Google Scholar]
- 45.Trimarchi, J. M., B. Fairchild, R. Verona, K. Moberg, N. Andon, and J. A. Lees. 1998. E2F-6, a member of the E2F family that can behave as a transcriptional repressor. Proc. Natl. Acad. Sci. USA 95:2850-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valdez, B. C., D. Henning, R. B. So, J. Dixon, and M. J. Dixon. 2004. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA 101:10709-10714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vanderluit, J. L., K. L. Ferguson, V. Nikoletopoulou, M. Parker, V. Ruzhynsky, T. Alexson, S. M. McNamara, D. S. Park, M. Rudnicki, and R. S. Slack. 2004. p107 regulates neural precursor cells in the mammalian brain. J. Cell Biol. 166:853-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wei, Y., L. Yu, J. Bowen, M. A. Gorovsky, and C. D. Allis. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99-109. [DOI] [PubMed] [Google Scholar]
- 49.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitfield, M. L., G. Sherlock, A. J. Saldanha, J. I. Murray, C. A. Ball, K. E. Alexander, J. C. Matese, C. M. Perou, M. M. Hurt, P. O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13:1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao, K. W., X. Li, Q. Zhao, Y. Huang, D. Li, Z. G. Peng, W. Z. Shen, J. Zhao, Q. Zhou, Z. Chen, P. J. Sims, T. Wiedmer, and G. Q. Chen. 2004. Protein kinase Cdelta mediates retinoic acid and phorbol myristate acetate-induced phospholipid scramblase 1 gene expression: its role in leukemic cell differentiation. Blood 104:3731-3738. [DOI] [PubMed] [Google Scholar]