Abstract
The p85α subunit of phosphatidylinositol 3-kinase (PI-3k) forms a complex with a protein network associated with oncogenic fusion tyrosine kinases (FTKs) such as BCR/ABL, TEL/ABL, TEL/JAK2, TEL/PDGFβR, and NPM/ALK, resulting in constitutive activation of the p110 catalytic subunit of PI-3k. Introduction of point mutations in the N-terminal and C-terminal SH2 domain and SH3 domain of p85α, which disrupt their ability to bind phosphotyrosine and proline-rich motifs, respectively, abrogated their interaction with the BCR/ABL protein network. The p85α mutant protein (p85mut) bearing these mutations was unable to interact with BCR/ABL and other FTKs, while its binding to the p110α catalytic subunit of PI-3k was intact. In addition, binding of Shc, c-Cbl, and Gab2, but not Crk-L, to p85mut was abrogated. p85mut diminished BCR/ABL-dependent activation of PI-3k and Akt kinase, the downstream effector of PI-3k. This effect was associated with the inhibition of BCR/ABL-dependent growth of the hematopoietic cell line and murine bone marrow cells. Interestingly, the addition of interleukin-3 (IL-3) rescued BCR/ABL-transformed cells from the inhibitory effect of p85mut. SCID mice injected with BCR/ABL-positive hematopoietic cells expressing p85mut survived longer than the animals inoculated with BCR/ABL-transformed counterparts. In conclusion, we have identified the domains of p85α responsible for the interaction with the FTK protein network and transduction of leukemogenic signaling.
Chromosomal translocations are responsible for the appearance of oncogenes encoding fusion tyrosine kinases (FTKs) such as BCR/ABL, TEL/ABL, TEL/JAK2, TEL/PDGFβR, BCR/FGFR1, and NPM/ALK (4, 28). These FTKs (BCR/ABL-related FTKs) share structural and functional similarities. They induce acute and chronic leukemias and lymphomas via activation of multiple signaling pathways which render cells independent of their environment and modulate their response to DNA damage. FTKs allow cells to proliferate in the absence of growth factors, protect cells from apoptosis in the absence of external survival factors, and may promote invasion and metastasis (10, 40, 59). In addition, FTKs can modulate response to DNA damage, rendering cells resistant to genotoxic therapies and causing genomic instability (47).
FTKs form protein networks by interacting directly or indirectly (via adaptor proteins) with numerous signaling molecules, including Ras, phosphatidylinositol (PI) 3-kinase (PI-3k), and STAT5, resulting in their activation, which is essential not only for leukemogenesis but also for resistance to apoptosis induced by DNA damage (10, 40, 52, 59). We and others reported that PI-3k activity is important for the growth of BCR/ABL-positive chronic myelogenous leukemia (CML) cells and NPM/ALK-transformed anaplastic large-cell lymphoma cells; however, it is dispensable for the proliferation of normal hematopoietic precursors (2, 32, 50, 53). In addition, PI-3k inhibitors appear to enhance the effect of imatinib mesylate, a selective inhibitor of BCR/ABL kinase (26, 32). Altogether, these findings made PI-3k an attractive target for novel antileukemia modalities (9, 17).
PI-3k activity is important in FTK-mediated protection from apoptosis in the absence of growth factors, probably due to the activation of Akt resulting in activation of Raf-1 and phosphorylation of Bad (2, 31, 36). However, PI-3k appears to be irrelevant in FTK-mediated protection from apoptosis induced by genotoxic agents (1, 16). In addition, PI-3k plays a role in the stimulation of proliferation of FTK-positive cells, possibly due to a decrease in expression of a cell cycle inhibitor, p27Kip1 (15, 54).
Although multiple forms of PI-3k exist in higher eukaryotes, the class Ia enzymes are primarily responsible for production of D-3 phosphoinositides in response to ligand-dependent receptor stimulation and tyrosine kinase activation (7, 55). Phosphoinositides act as important second messengers in many signal transduction pathways. Class Ia enzymes are heterodimers of adaptor (p85α, p85β, p85γ, and two p85α splice variants, p55α and p50α) and catalytic (p110α, p110β, and p110δ) subunits. The regulatory subunit maintains the catalytic subunit in a low-activity state in quiescent cells and mediates its activation by direct interaction with activated receptor or nonreceptor tyrosine kinase. Thus, loss of adaptor function may be expected to affect catalytic function as well. In accordance with this speculation, BCR/ABL-mediated transformation was inhibited by the downregulation of p85 expression by antisense oligonucleotides or by genetically engineered ablation of p85α and its splice variants, p55α and p50α (24, 50). Therefore, disruption of interaction of p85α with BCR/ABL kinase may exert a detrimental effect on leukemia cells.
The p85α adaptor protein employs the inter-SH2 (iSH2) region to bind the p110 catalytic subunit (27) and employs SH3 and SH2 domains, phosphotyrosine residues, and proline-rich motifs to interact with activated tyrosine kinases (20, 34) including BCR/ABL and NPM/ALK (2, 23) or adaptor proteins such as Shc, c-Cbl, Crk-L, and Gab2 (19, 43, 44), which enhance the association of PI-3k with activated tyrosine kinases (22). Since interaction of p85α with an active tyrosine kinase protein network is essential for stimulation of p110 catalytic activity, disruption of the interaction of the p85α-p110 heterodimer with activated tyrosine kinase should abrogate the biological effects dependent on PI-3k. Here, we have identified the p85α mutant, which forms a heterodimer with p110α but does not interact with the protein networks of BCR/ABL and other FTKs, and described the biological consequences of PI-3k (p85α-p110) dislocation from the BCR/ABL kinase.
MATERIALS AND METHODS
Cell lines.
Murine growth factor-dependent 32Dcl3 and BaF3 cell lines, as well as p210BCR/ABL-, p190BCR/ABL-, TEL/ABL-, TEL/PDGFβR-, TEL/JAK2-, and NPM/ALK-transformed counterparts, were described previously (52). Cells were cultured in Iscove's medium supplemented with 10% fetal bovine serum (FBS), 15% WEHI-conditioned medium as a source of interleukin-3 (IL-3), and penicillin/streptomycin. Human embryonic kidney 293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal calf serum and supplemented with glutamine and penicillin/streptomycin.
Plasmid constructions. (i) Construction of the GST-p85 fusion proteins.
Full-length p85 and p85 fragments were amplified by PCR using bovine p85α cDNA (41) as a template. BamHI and EcoRI restriction sites were introduced at the N and C termini, respectively, and the PCR products were cloned in frame into the pGEX-2T expression vector to generate glutathione S-transferase (GST) fusion proteins. The full-length p85α and the N- and C-terminal SH2 domains (nSH2 and cSH2, respectively) and SH3 domain were obtained as previously described (13). The primers used to amplify the BCR and iSH2 fragments were as follows: 5′-CGAGGATCCGCTCCACCGCTTCTTATCAA-3′ (BCR sense), 5′-CAAGAATTCGGGAGGCAGTGCTGGTGCAGG-3′ (BCR antisense), 5′-CGAGGATCCCAACAGGATCAAGTTGTCAAA-3′ (iSH2 sense), and 5′-CAAGAATTCGTCTTCAGTGTTTTCATTGCC-3′ (iSH2 antisense).
(ii) Site-directed mutagenesis.
A QuikChange XL site-directed mutagenesis kit (Stratagene) was used, according to the manufacturer's instruction, to introduce mutations in the full-length p85α and the SH3 and SH2 domains. The sense primers containing the desired mutations were as follows: SH3 E52D (5′-AAGCCTGAAGATATTGGCTGGTTA-3′), SH3 W55F (5′-GCCAAGCCTGAAGAAATTGGCTTCTTAAATGGCTATAACG-3′), SH3 T72I, (5′-AGGGGAGACTTTCCGGGAATTTACGTAGAATATATTGG-3′), cSH2 R358L (5′-GCTGACGGGACCTTTTTGGTACTAGACGATC-3′), and nSH2 R649L (5′-ACGGCACTTTTCTTGTCCTGGAAAGCAGTAAACAGG-3′). The mutations were confirmed by sequencing.
(iii) Construction of pMIG-p85mutFlag.
The pGEX-2T plasmid containing full-length p85α cDNA or the mutant encoding W55F + T72I + R358L + R649L amino acid substitutions and the pMIGR1-IRES-EGFP retroviral construct were digested with BamHI and BglII, respectively, filled in with Klenow fragment, digested with EcoRI, and ligated, generating p85-internal ribosome entry site (IRES)-green fluorescent protein (GFP) (pMIG-p85wt) and p85mut-IRES-GFP (pMIG-p85mut) retroviral constructs. To insert a Flag tag at the C-terminal end of the p85α constructs, the stop codon was removed by site-directed mutagenesis (QuickChange; Stratagene), and Flag cDNA, followed by the stop codon, was inserted at the EcoRI site. The constructs were verified by sequencing.
Transfection of 293T cells with p210BCR/ABL and p85-Flag constructs.
293T cells were transfected with pMIG-p210BCR/ABL, pMIG-p85wt-Flag, pMIG-p85mut-Flag, or pMIG empty retroviral constructs as described previously using the calcium phosphate protocol (49). Forty-eight hours after transfection, p85-Flag proteins were immunoprecipitated from total cell lysates using anti-Flag M2 affinity gel (Sigma), and the presence of p210BCR/ABL and p85-Flag proteins was assessed by Western analysis.
Pull-down and Western blot analyses.
Cells were solubilized for 20 min on ice in lysis buffer (10 mM HEPES, pH 7.5, 150 mM NaCl, 1% NP-40, 10% glycerol, 5 mM EDTA, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 1 mM Na3VO4, and 10 μg/ml each leupeptin and aprotinin) and centrifuged (16,000 × g for 10 min at 4°C), and the supernatant was collected for experiments. The p190BCR/ABL protein and the kinase-deficient (KD) mutant were in vitro transcribed and translated (IVTT) using pcDNA3-p190BCR/ABL and pcDNA3-p190BCR/ABL-KD plasmids (kindly provided by Justus Duyster [3]) and a TNT-coupled transcription/translation kit (Promega). GST fusion proteins were produced in Escherichia coli DH5α-competent bacteria transformed by the pGEX-2T constructs. GST fusion proteins immobilized on glutathione-Sepharose beads (Pharmacia) were incubated for 2 h or 12 h at 4°C with cell lysates or IVTT reactions, washed, resuspended in 2× Laemmli sample buffer, and boiled for 5 min. The reaction products were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting using the antibodies recognizing c-ABL (Oncogene), p85 and p110α (BD Transduction Laboratories), Akt (Cell Signaling), phospho-Ser473-Akt (Cell Signaling), CrkL (Cell Signaling), Flag (Sigma), GST (Santa Cruz), β-actin (Oncogene), Shc (Santa Cruz), Gab2 (Santa Cruz), c-Cbl (Santa Cruz), PDGFβR (Santa Cruz), ALK (BD Transduction Laboratories), and Jak2 (Santa Cruz).
PI-3k assay.
PI-3k activity was examined in the antiphosphotyrosine immunoprecipitates using [γ-32P]ATP and PI as a substrate as described previously (50). 32P-labeled PI was resolved by thin-layer chromatography and visualized by autoradiography.
Retroviral infections.
Infections with bone marrow cells (BMCs) were performed as described previously (39), with modifications. Briefly, BMCs from C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME) pretreated with 5-fluorouracil or from Gab2−/− mice and Gab2+/+ littermates (18) were stimulated with stem cell factor and IL-3 for 48 h and then cocultivated with the supernatants collected from amphotropic Phoenix packaging cells (American Type Culture Collection, Manassas, VA) transfected with pSRα-p210BCR/ABL and those transfected with p85mut-IRES-GFP or IRES-GFP retroviral particles (if applicable). p210BCR/ABL-32Dcl3 cells (three clones, transformed with pSRα-p210BCR/ABL [51], that are sensitive to the PI-3k inhibitors wortmannin and LY294002) were infected with p85mut-IRES-GFP or IRES-GFP retroviral particles as described above. GFP-positive (GFP+) cells were obtained by sorting after 72 h of continuous infection, expanded for 7 days in the presence of growth factors and G418 (if applicable), and then spun down on Lympholyte-M (Cedarlane Laboratories, Hornby, Ontario, Canada) to eliminate dead cells. GFP+ and G418-resistant GFP+ cells were used for experiments.
Proliferation assays.
Cells (104/0.1 ml) were incubated in the presence or absence of IL-3 as indicated, and their proliferation capability was examined by counting cells using trypan blue as described previously (38). Clonogenic assay was performed in MethoCult H4230 semisolid medium (Stem Cell Technologies, Vancouver, British Columbia, Canada) in the absence of IL-3 as described previously (39, 48). Colonies were counted after 7 days.
Leukemogenesis in SCID mice.
SCID outbred mice (Taconic Farms, Germantown, NJ) were injected with 105 or 106 p210BCR/ABL+E or p210BCR/ABL+p85mut 32Dcl3 cells. Terminally ill mice were sacrificed and examined macroscopically for the development of hematological malignancy as described previously (39). Survival time was calculated by the Kaplan-Meier test.
RESULTS
SH3 and SH2 domains, but not iSH2 and BCR domains, of p85α interact with BCR/ABL kinase.
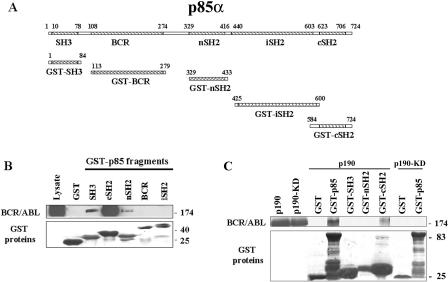
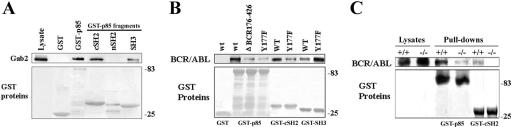
BCR/ABL kinase interacts with the p85α adaptor protein leading to activation of the PI-3k→Akt pathway, which is important for BCR/ABL leukemogenesis. To determine how p85α interacts with BCR/ABL kinase, various p85α domains (Fig. 1A) were fused to GST, immobilized on glutathione-Sepharose beads, and used for pull-down assays with cell lysates prepared from p210BCR/ABL-expressing 32Dcl3 cells. Abundant association was detected between the p85α cSH2 and p210BCR/ABL kinase, whereas the SH3 domain displayed modest binding, and only weak interaction was detected with the nSH2 domain (Fig. 1B). GST alone did not interact with BCR/ABL.
FIG. 1.
Interaction of the p85α fragments with BCR/ABL. GST fusion proteins containing the indicated p85α fragments (A) were used for the pull-down assay along with the total cell lysates isolated from p210BCR/ABL-positive 32Dcl3 cells (B) or with p210BCR/ABL produced by IVTT (C). The reactions were resolved by SDS-PAGE and analyzed by Western blotting using anti-ABL (upper box) and anti-GST (lower box) primary antibodies to detect BCR/ABL and GST proteins, respectively.
To further investigate whether the interactions are direct or indirect, the p190BCR/ABL kinase and p190BCR/ABL-KD mutant were produced in vitro and used for pull-down assays with full-length p85α fused with GST. p190BCR/ABL kinase, but not the kinase-deficient p190BCR/ABL-KD mutant, formed a complex with GST-p85α, suggesting that direct interaction depends on the kinase activity of BCR/ABL (Fig. 1C). To further dissect the mechanisms responsible for this effect, GST fusion proteins containing p85α SH3, nSH2, and cSH2 domains were employed. Interestingly, only cSH2 appears to interact directly with the p190BCR/ABL kinase (Fig. 1C).
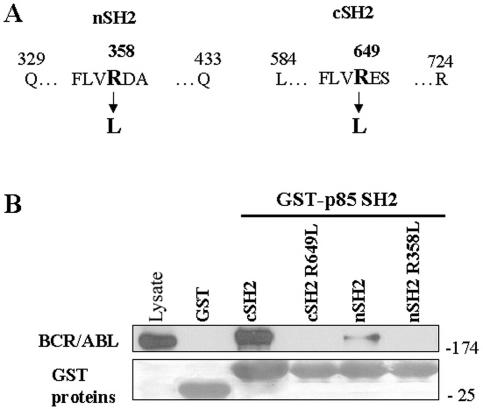
Mutations in the p85α SH3 and SH2 domains disrupt their interactions with p210BCR/ABL.
The FLVRES motif of the SH2 domain is the best-conserved region among all SH2 domains (56), and mutation of the arginine residue prevents the SH2 domain from binding phosphotyrosine-containing sequences (33). Each of the analogous arginine residues in the p85α SH2 domains was mutated to leucine (R358L in the nSH2 domain and R649L in the cSH2 domain [Fig. 2A]). Pull-down experiments in which immobilized GST fusion proteins expressing wild-type or mutated p85α SH2 domains were incubated with cell lysates containing p210BCR/ABL kinase were carried out. Wild-type GST-p85 SH2 domains were able to form complexes with p210BCR/ABL kinase, whereas the mutated domains nSH2 (R358L) and cSH2 (R649L) were not (Fig. 2B). These results demonstrate that these conserved arginine residues within the p85α SH2 domains are required for phosphotyrosine-dependent binding to the BCR/ABL kinase protein network.
FIG. 2.
Mutations in the p85α SH2 domains disrupt the interaction with p210BCR/ABL. GST fusion proteins containing the indicated p85α SH2 domains and their mutants (A) were used for the pull-down assay along with the total cell lysates isolated from p210BCR/ABL-positive 32Dcl3 cells (B). The reactions were resolved as described in the Fig. 1 legend.
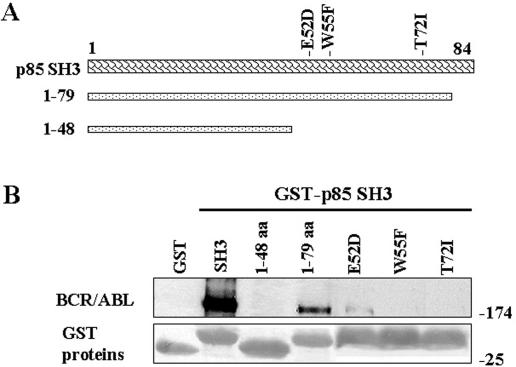
To examine the molecular basis for SH3 association with BCR/ABL, we first generated GST fusion proteins that contained SH3 fragments (shown in Fig. 3A) and applied them in the pull-down assay with cell lysates from p210BCR/ABL 32Dcl3 cells. The fragment containing the N-terminal 79 amino acids (aa) was able to bind BCR/ABL kinase, whereas that containing only 48 amino acids was not (Fig. 3B), suggesting that the interaction depends on the fragment between amino acids 49 and 79. SH3 domains interact with proline-rich sequences (PxxP) and mediate specific protein-protein interactions (57). Three-dimensional structures of the PI-3k SH3 domain have been determined by nuclear magnetic resonance spectroscopy and X-ray crystallography (30), and the amino acid residues E52, W55, and T72 responsible for binding the PxxP motif have been proposed (5, 29, 58), which are in the 49- to 79-aa region (Fig. 3A). Accordingly, E52D, W55F, and T72I mutants of the p85α SH3 domain were generated and applied in pull-downs. W55F and T72I aa substitutions completely abrogated the ability of the p85α SH3 domain to interact with p210BCR/ABL kinase, whereas E52D exerted a partial inhibitory effect (Fig. 3B). Taken together, these results indicate that W55 and T72 are essential for the interaction of the PI-3k SH3 domain with the BCR/ABL kinase protein network. In addition, the p85α W55F mutant expressed in BCR/ABL-positive 32Dcl3 cells exerted a modest (16% ± 5%) but reproducible inhibitory effect on BCR/ABL-mediated growth factor-independent proliferation after 48 h of growth factor starvation.
FIG. 3.
Mutations in the p85α SH3 domain disrupt the interaction with p210BCR/ABL. GST fusion proteins containing the p85α SH3 domain and the indicated fragments or mutants (A) were used for the pull-down assay along with the total cell lysates isolated from p210BCR/ABL-positive 32Dcl3 cells (B). The reactions were resolved as described in the Fig. 1 legend.
Mutations in the SH3 and SH2 domains of the full-length p85α protein abrogate its interaction with BCR/ABL kinase and some adaptor proteins.
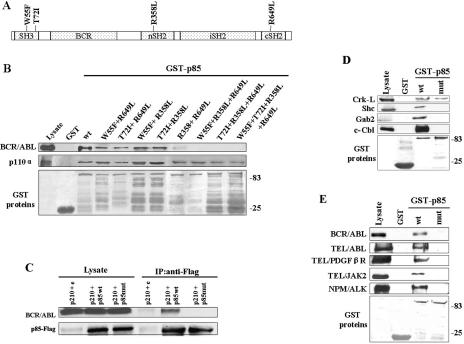
To test whether or not the mutations of the SH2 and SH3 domains in full-length p85α can interfere with its interaction with the BCR/ABL protein network, we generated different p85α mutants (as indicated in Fig. 4A) linked to GST. These GST-p85α mutants and wild-type proteins were used in pull-down assays with lysates from p210BCR/ABL 32Dcl3 cells. Simultaneous introduction of the previously characterized amino acid substitutions (W55F, T72I, R358L, and R649L) in the SH3 and both SH2 domains of full-length p85α (p85mut) disrupted its ability to associate with BCR/ABL kinase in the pull-down assay (Fig. 4B).
FIG. 4.
Mutations in the p85α full-length protein affect the interaction with the p210BCR/ABL protein network. GST fusion proteins containing the p85α wild type (wt) or indicated mutants (A) were used for the pull-down assay along with the total cell lysates isolated from p210BCR/ABL-positive 32Dcl3 cells. (B) The reactions were resolved by SDS-PAGE and analyzed by Western blotting using anti-ABL (top), anti-p110α (middle), and anti-GST (bottom) primary antibodies to detect BCR/ABL, p110α, and GST proteins, respectively. (C) p210BCR/ABL and Flag-p85wt, Flag-p85mut, or empty plasmid were expressed in 293T cells and detected in total cell lysates (Lysate). The presence of BCR/ABL and Flag-p85 proteins in anti-Flag immunoprecipitates was also determined (IP:anti-Flag). (D) GST fusion proteins containing the p85α wild type (wt) or the W55F + T72I + R358L + R649L mutant (mut) were used for the pull-down assay along with the total cell lysates isolated from p210BCR/ABL-positive 32Dcl3 cells. The presence of Crk-L, c-Cbl, Gab2, and Shc in the reactions was determined by Western analysis. (E) GST fusion proteins containing p85wt or p85mut were used for the pull-down assay along with the total cell lysates isolated from BaF3 cells transformed by the indicated FTKs. The reactions were resolved by SDS-PAGE and analyzed by Western blotting using the kinase-specific antibody (upper boxes) and anti-GST antibody (bottom box) to detect the indicated FTKs and GST proteins.
The p85α adaptor subunit is known to form a constitutive heterodimer with the p110α catalytic subunit (13, 21). To investigate whether or not amino acid substitutions in the SH3 and SH2 domains of p85α would affect its binding to p110α, GST-p85α wild-type and mutant proteins were used for the pull-down assay with the lysates from p210BCR/ABL 32Dcl3 cells. The results in Fig. 4B show that the GST-p85α wild type and all mutants are able to interact with p110α. Therefore, it appears that p85α interacts independently with the BCR/ABL protein network via SH3 and SH2 domains and with p110α via the iSH2 region. In addition, since p85mut binds to the p110 catalytic subunit, but not to BCR/ABL kinase, it should exert a dominant-negative mutant (DNM) effect.
To investigate whether amino acid substitutions W55F + T72I + R358L + R649L can disrupt the interaction of p85α with the BCR/ABL protein network in vivo, p85wt-Flag and p85mut-Flag were coexpressed with p210BCR/ABL in 293T cells, and the interaction was assessed in anti-Flag immunoprecipitates. p210BCR/ABL was readily detected in p85wt-Flag but not in p85mut-Flag immunoprecipitate (Fig. 4C). Thus, disruption of the SH3 and SH2 domain binding abilities in p85α severely affected its complex formation ability with p210BCR/ABL.
We also studied whether or not p85mut is able to interact with adaptor proteins such as Crk-L, Gab2, c-Cbl, and Shc, which can mediate the interaction between BCR/ABL kinase and PI-3k (19, 43, 44). Pull-down assays indicated that GST-p85mut loses the ability to associate with Gab2, Shc, and c-Cbl but not with Crk-L (Fig. 4D).
Gab2 was identified as a key determinant in BCR/ABL-mediated activation of PI-3k (43); therefore, its interaction with p85α domains and BCR/ABL kinase was investigated in more detail. Pull-down assays show that cSH2 and SH3 but not the nSH2 domain of p85α interact with Gab2 (Fig. 5A). In addition, Gab2 is a major adaptor protein intermediating between p85α and the BCR/ABL Y177 phosphotyrosine residue (43). The Y177F amino acid substitution reduced the amount of BCR/ABL protein pulled down by GST-cSH2 but not by GST-SH3; the latter interaction even appears enhanced by the Y177F mutation (Fig. 5B). Thus, Gab2 seems to link the cSH2 domain of p85α and phospho-Y177 of BCR/ABL. This hypothesis is further supported by the observation that the interaction between p210BCR/ABL kinase and full-length GST-p85α or GST-cSH2 of p85α is strongly reduced or undetectable, respectively, in the absence of Gab2 (Fig. 5C). Altogether, while Gab2 interacts with both the cSH2 and SH3 domains of p85α, it provides a critical functional link between p85α cSH2 and phospho-Y177 of BCR/ABL.
FIG. 5.
cSH2 and SH3 domains of p85α interact with Gab2. GST fusion proteins containing p85α or the indicated p85α fragments were used for the pull-down assay along with the total cell lysates isolated from (A) p210BCR/ABL-positive 32Dcl3 cells, (B) 32Dcl3 cells expressing the p190BCR/ABL wild type (wt) or indicated mutants, and (C) GFP+ p210BCR/ABL-positive Gab2+/+ and Gab2−/− cells expressing similar levels of p210BCR/ABL kinase (Lysates). The reactions were examined by Western analysis to detect Gab2 (A, upper box), BCR/ABL (B, C, upper boxes), and GST (lower boxes) proteins.
Since FTKs such as TEL/ABL, TEL/PDGFβR, TEL/JAK2, and NPM/ALK also coimmunoprecipitate with p85 and transduce oncogenic signals through PI-3k (8, 37, 40, 53), we examined if they were also able to interact with p85mut. For this reason, GST-p85 and GST-p85mut fusion proteins were used as bait in pull-down assays with cell lysates obtained from FTK-expressing BaF3 cell lines. Similarly to BCR/ABL, other FTKs also do not interact with p85mut (Fig. 4E). These results demonstrate that SH2 and/or SH3 domains of the p85α adaptor protein are important for the interaction with BCR/ABL-related FTKs.
p85mut inhibits PI-3k signaling and BCR/ABL-dependent leukemogenesis.
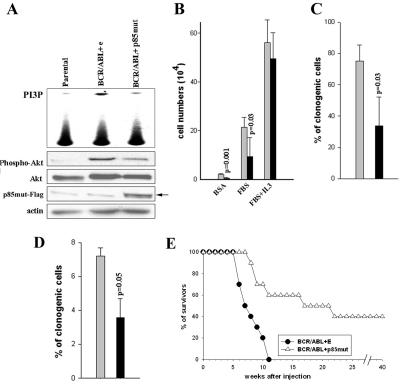
The serine/threonine kinase Akt/protein kinase B is a major downstream mediator of PI-3k activity. PI-3k-dependent activation of Akt is accompanied by its phosphorylation on serine 473 (46). To investigate whether or not p85mut affects PI-3k downstream signaling, serine 473 phosphorylation of Akt was examined by Western analysis in p210BCR/ABL cells cotransfected with p85mut cDNA or empty plasmid. As expected, p85mut expression diminished PI-3k catalytic activity and Akt serine 473 phosphorylation in BCR/ABL cells (Fig. 6A).
FIG. 6.
The p85α mutant inhibits BCR/ABL-mediated leukemogenesis. (A) The p85α mutant inhibits PI-3k activation and phosphorylation of Akt in vivo. 32Dcl3 parental and BCR/ABL-positive counterparts were infected with p85mutFlag-IRES-GFP (p85mut) or empty IRES-GFP retroviral construct (e). PI-3k enzymatic activity was measured in the kinase assay in vitro using phosphatidylinositol as a substrate. Akt activation was assessed by Western analysis detecting the phosphorylated form of Akt; total Akt protein and actin were detected as loading controls. Anti-Flag antibody was used to confirm the expression of p85mut-Flag protein. (B, C) BCR/ABL-expressing 32Dcl3 cells were infected with p85αmutFlag-IRES-GFP (black bars) or empty IRES-GFP (gray bars) retroviral construct. The proliferation potential of GFP+ cells was analyzed by trypan exclusion in the presence of bovine serum albumin, FBS, or FBS plus IL-3 (B) or clonogenic assay (C). (D) Mouse mononuclear bone marrow cells were coinfected with pSRα-BCR/ABL and p85mutFlag-IRES-GFP retroviral construct or IRES-GFP empty construct. The clonogenic ability of GFP+ cells was evaluated in the absence of growth factors. Results in B, C, and D represent means ± standard deviations from three to four independent experiments (P value calculated by Student's t test). % of clonogenic cells represents the percentage of cells plated that formed colonies. (E) Survival of SCID mice injected with cells transfected with BCR/ABL and empty plasmid (BCR/ABL+E), or BCR/ABL and p85mut (BCR/ABL+p85mut). Survival of the animals was monitored weekly.
To investigate whether or not p85mut could affect BCR/ABL-dependent growth, p210BCR/ABL-transformed 32Dcl3 cells were transfected with p85mut-IRES-GFP or GFP retroviral constructs. As shown in Fig. 6B, proliferation of GFP+ cells transfected with p85mut cDNA was inhibited in comparison to that of cells transfected with empty plasmid (GFP only) when cultured in the absence of IL-3; p85mut did not affect proliferation in the presence of IL-3. In concordance, p85mut diminished the clonogenic activity of p210BCR/ABL-expressing 32Dcl3 cells in the absence of IL-3 (Fig. 6C). p85mut exerted a similar effect in p190BCR/ABL-transformed 32Dcl3 cells (data not shown). Finally, murine BMCs were employed to confirm that the inhibitory effect of p85mut could also be observed in primary cells. BMCs cotransfected with p210BCR/ABL and p85mut cDNAs formed fewer colonies in methylcellulose in the absence of IL-3 than cells cotransfected with BCR/ABL and empty vector (Fig. 6D). Altogether, these results suggest that p85mut interferes with BCR/ABL-dependent growth in vitro.
To determine whether or not p85mut also inhibits BCR/ABL-mediated leukemogenesis in vivo, SCID mice were injected with p210BCR/ABL-expressing 32Dcl3 GFP+ cells resulting from transfection with p85mut-IRES-GFP or IRES-GFP retroviral constructs (Fig. 6A to C). All 10 mice inoculated with 105 p210BCR/ABL-32Dcl3 GFP+ cells infected with IRES-GFP (p210BCR/ABL+E) succumbed to leukemia (median survival time, 8.5 weeks) (Fig. 6E). In contrast, 4/10 of those injected with 105 p210BCR/ABL-32Dcl3 GFP+ cells infected with p85mut-IRES-GFP (p210BCR/ABL+p85mut) were still alive after 40 weeks (median survival time, 24.2 weeks [P = 0.0017]). The median survival times of the animals inoculated with 106 p210BCR/ABL+E or p210BCR/ABL+p85mut cells were 5.0 and 8.2 weeks, respectively (P = 0.02). This observation indicates that p85mut interferes with BCR/ABL-mediated leukemogenesis in mice and that the antitumor effect depends on the number of injected leukemia cells. Macroscopic examination of the internal organs (spleen, liver, and kidney) in terminally ill animals indicated the presence of leukemia in essentially all inspected organs.
DISCUSSION
BCR/ABL and other FTKs, such as TEL/ABL, TEL/JAK2, TEL/PDGFβR, and NPM/ALK (BCR/ABL-related FTKs), associate with the p85α subunit of PI-3k, leading to activation of the p110 catalytic subunit (6). Inhibitors of PI-3k, such as wortmannin and LY294002, were proven effective against hematopoietic malignancies expressing BCR/ABL and related FTKs (2, 14, 26, 32, 37, 48, 50, 53). Interestingly, these inhibitors may exert a selective antileukemia/anti-lymphoma effect while sparing normal counterparts (32, 50, 53). Moreover, they enhanced the anti-CML effect of imatinib mesylate (26, 32). Unfortunately, these inhibitors (especially wortmannin) exerted a substantial nonspecific toxicity (12), precluding them from being used as antileukemia drugs in patients. However, compounds more specifically targeting PI-3k or the interactions between PI-3k and FTKs may become novel antileukemia/anti-lymphoma therapeutics. Therefore, studies of the interactions between the p85 subunit of PI-3k and FTKs are needed to pinpoint the mechanisms.
Here, we show that p85α employs multiple domains to interact with BCR/ABL-related FTKs. Studies with BCR/ABL demonstrated that p85α can interact directly and indirectly with BCR/ABL. Direct interaction depends on the kinase activity of BCR/ABL and is mediated by the cSH2 domain, but not nSH2 and SH3 domains, of p85α. This is in agreement with another observation that GST-cSH2, but not the GST-nSH2 domain, pulled down purified c-ABL kinase (35). However, BCR/ABL kinase from leukemia cell lysates was able to interact with the SH3 and both SH2 domains of p85α, implicating indirect interactions via adaptor proteins. BCR/ABL and other FTKs interact directly and indirectly with numerous adaptor proteins to regulate apoptosis and proliferation (4, 42). Adaptors such as Crk-L, Shc, Gab2, and c-Cbl can connect BCR/ABL (and probably also other FTKs) to PI-3k (19, 43, 44), and p85mut loses the capability of interaction with all of them except Crk-L. This result is supported by previous reports showing that adaptor proteins Crk-L, Shc, Gab2, and c-Cbl interact with SH2 and SH3 domains of p85 (11, 19, 22, 43-45). In addition, since the SH3 domain of Crk-L can bind p85 (45), it still forms a complex with p85mut. However, it appears that Crk-L cannot link p85mut with BCR/ABL in the absence of other adaptors, in accordance with a report implicating the Crk-L-c-Cbl complex as a linker between BCR/ABL kinase and p85 (44). Alternatively, the BCR/ABL-Crk-L-p85α association may be weak and thus not detectable in the pull-down assay. In conclusion, BCR/ABL kinase forms a protein complex network involving PI-3k and adaptor proteins. However, due to the high level of complexity of the BCR/ABL-PI-3k proteome (25), members of this complex essential for PI-3k activation are yet to be described. Our studies suggest that Gab2, Shc, and c-Cbl but not Crk-L are required to transfer the activation signal from BCR/ABL kinase to PI-3k and that Gab2 is a key element in the interaction between cSH2 of p85α and phospho-Y177 of BCR/ABL.
Association of the cSH2 domain with the BCR/ABL protein complex seems more abundant than that of nSH2. This effect may reflect differences in the abilities of SH2 domains to interact directly with BCR/ABL (cSH2), Shc (cSH2), and c-Cbl (nSH2 and cSH2) (19, 44). Thus, SH2 domains of p85α may employ different mechanisms to interact with the BCR/ABL kinase protein network.
Mutagenesis of the isolated domains revealed that association of p85α with BCR/ABL was dependent on the SH2- and SH3-mediated interactions with phosphotyrosine and proline-rich motifs, respectively. Simultaneous mutations in the SH3, nSH2, and cSH2 domains of full-length p85α (p85mut) were required to disrupt the p85α-BCR/ABL kinase complex below a detectable level. In addition, BCR/ABL-related FTKs also did not form a detectable complex with p85mut, suggesting that these FTKs may share similar mechanisms of interaction with the p85α subunit and activation of p110 subunit catalytic activity. However, different FTKs may have distinct transformation requirements as indicated by the ability of TEL/JAK2 to induce growth factor-independent myeloid colonies in the absence of Gab2 (43), despite the requirement for PI-3k activation (37). Therefore, mechanisms of FTK-PI-3k interaction and activation may not be universal.
In spite of losing its ability to interact with BCR/ABL and other FTKs, p85mut retains the ability to form a complex with the p110 catalytic subunit, probably because its iSH2 domain remains intact (27). Therefore, it seems that p85mut may be able to work as a DNM of PI-3k. Indeed, we found that expression of p85mut in BCR/ABL-transformed cells diminishes the activation of PI-3k and Akt serine/threonine kinase, one of the PI-3k downstream effectors playing an important role in BCR/ABL leukemogenesis (48). In addition, p85mut inhibits growth factor-independent proliferation of BCR/ABL cells in vitro; however, addition of IL-3 restores the growth of leukemia cells. This observation further verifies a selective and critical role of PI-3k in BCR/ABL-mediated, but not growth factor-dependent, signaling (32, 50). Moreover, expression of p85mut inhibited BCR/ABL-dependent leukemogenesis in SCID mice. This result is in agreement with previous findings that abrogation of BCR/ABL-mediated growth factor independence by mutations in BCR/ABL SH3 and SH2 domains or expression of the DNMs of STAT5 and Akt is associated with impaired leukemogenesis in mice (38, 39, 48). The partial inhibitory effect exerted by p85mut could be due to redundancy in the PI-3k protein family and/or relatively low expression of the p85α mutant in some cells.
Altogether, this work confirms the BCR/ABL-p85 association as a potential target for small molecules designed to disassemble/prevent this interaction and stop malignant growth.
Acknowledgments
This work was supported by RO1 CA83700 to T.S. and PO1 DK50654 to B.G.N. T.S. is a Scholar and M.G.M. is supported by a fellowship from the Hood Foundation/Medical Foundation.
REFERENCES
- 1.Amarante-Mendes, G. P., T. Jascur, W. K. Nishioka, T. Mustelin, and D. R. Green. 1997. Bcr-Abl-mediated resistance to apoptosis is independent of PI 3-kinase activity. Cell Death Differ. 4:548-554. [DOI] [PubMed] [Google Scholar]
- 2.Bai, R. Y., T. Ouyang, C. Miething, S. W. Morris, C. Peschel, and J. Duyster. 2000. Nucleophosmin-anaplastic lymphoma kinase associated with anaplastic large-cell lymphoma activates the phosphatidylinositol 3-kinase/Akt antiapoptotic signaling pathway. Blood 96:4319-4327. [PubMed] [Google Scholar]
- 3.Bassermann, F., T. Jahn, C. Miething, P. Seipel, R. Y. Bai, S. Coutinho, V. L. Tybulewicz, C. Peschel, and J. Duyster. 2002. Association of Bcr-Abl with the proto-oncogene Vav is implicated in activation of the Rac-1 pathway. J. Biol. Chem. 277:12437-12445. [Online.] [DOI] [PubMed] [Google Scholar]
- 4.Blume-Jensen, P., and T. Hunter. 2001. Oncogenic kinase signalling. Nature 411:355-365. [DOI] [PubMed] [Google Scholar]
- 5.Booker, G. W., I. Gout, A. K. Downing, P. C. Driscoll, J. Boyd, M. D. Waterfield, and I. D. Campbell. 1993. Solution structure and ligand-binding site of the SH3 domain of the p85 alpha subunit of phosphatidylinositol 3-kinase. Cell 73:813-822. [DOI] [PubMed] [Google Scholar]
- 6.Calabretta, B., and T. Skorski. 1996. BCR/ABL regulation of PI-3 kinase activity. Leuk. Lymphoma 23:473-476. [DOI] [PubMed] [Google Scholar]
- 7.Cantley, L. C. 2002. The phosphoinositide 3-kinase pathway. Science 296:1655-1657. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, M., M. H. Tomasson, G. F. Barker, T. R. Golub, and D. G. Gilliland. 1996. The TEL/platelet-derived growth factor beta receptor (PDGF beta R) fusion in chronic myelomonocytic leukemia is a transforming protein that self-associates and activates PDGF beta R kinase-dependent signaling pathways. Proc. Natl. Acad. Sci. USA 93:14845-14850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, F., J. T. Lee, P. M. Navolanic, L. S. Steelman, J. G. Shelton, W. L. Blalock, R. A. Franklin, and J. A. McCubrey. 2003. Involvement of PI3K/Akt pathway in cell cycle progression, apoptosis, and neoplastic transformation: a target for cancer chemotherapy. Leukemia 17:590-603. [DOI] [PubMed] [Google Scholar]
- 10.Cross, N. C., and A. Reiter. 2002. Tyrosine kinase fusion genes in chronic myeloproliferative diseases. Leukemia 16:1207-1212. [DOI] [PubMed] [Google Scholar]
- 11.Crouin, C., M. Arnaud, F. Gesbert, J. Camonis, and J. Bertoglio. 2001. A yeast two-hybrid study of human p97/Gab2 interactions with its SH2 domain-containing binding partners. FEBS Lett. 495:148-153. [DOI] [PubMed] [Google Scholar]
- 12.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dhand, R., K. Hara, I. Hiles, B. Bax, I. Gout, G. Panayotou, M. J. Fry, K. Yonezawa, M. Kasuga, and M. D. Waterfield. 1994. PI 3-kinase: structural and functional analysis of intersubunit interactions. EMBO J. 13:511-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dierov, J., Q. Xu, R. Dierova, and M. Carroll. 2002. TEL/platelet-derived growth factor receptor beta activates phosphatidylinositol 3 (PI3) kinase and requires PI3 kinase to regulate the cell cycle. Blood 99:1758-1765. [DOI] [PubMed] [Google Scholar]
- 15.Gesbert, F., W. R. Sellers, S. Signoretti, M. Loda, and J. D. Griffin. 2000. BCR/ABL regulates expression of the cyclin-dependent kinase inhibitor p27Kip1 through the phosphatidylinositol 3-kinase/AKT pathway. J. Biol. Chem. 275:39223-39230. [DOI] [PubMed] [Google Scholar]
- 16.Greenland, C., C. Touriol, G. Chevillard, S. W. Morris, R. Bai, J. Duyster, G. Delsol, and M. Allouche. 2001. Expression of the oncogenic NPM-ALK chimeric protein in human lymphoid T-cells inhibits drug-induced, but not Fas-induced apoptosis. Oncogene 20:7386-7397. [DOI] [PubMed] [Google Scholar]
- 17.Griffin, J. D. 2001. Phosphatidyl inositol signaling by BCR/ABL: opportunities for drug development. Cancer Chemother. Pharmacol. 48:S11-S16. [DOI] [PubMed] [Google Scholar]
- 18.Gu, H., K. Saito, L. D. Klaman, J. Shen, T. Fleming, Y. Wang, J. C. Pratt, G. Lin, B. Lim, J. P. Kinet, and B. G. Neel. 2001. Essential role for Gab2 in the allergic response. Nature 412:186-190. [DOI] [PubMed] [Google Scholar]
- 19.Harrison-Findik, D., M. Susa, and L. Varticovski. 1995. Association of phosphatidylinositol 3-kinase with SHC in chronic myelogeneous leukemia cells. Oncogene 10:1385-1391. [PubMed] [Google Scholar]
- 20.Hellyer, N. J., K. Cheng, and J. G. Koland. 1998. ErbB3 (HER3) interaction with the p85 regulatory subunit of phosphoinositide 3-kinase. Biochem. J. 333:757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hiles, I. D., M. Otsu, S. Volinia, M. J. Fry, I. Gout, R. Dhand, G. Panayotou, F. Ruiz-Larrea, A. Thompson, N. F. Totty, et al. 1992. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell 70:419-429. [DOI] [PubMed] [Google Scholar]
- 22.Jain, S. K., W. Y. Langdon, and L. Varticovski. 1997. Tyrosine phosphorylation of p120cbl in BCR/abl transformed hematopoietic cells mediates enhanced association with phosphatidylinositol 3-kinase. Oncogene 14:2217-2228. [DOI] [PubMed] [Google Scholar]
- 23.Jain, S. K., M. Susa, M. L. Keeler, N. Carlesso, B. Druker, and L. Varticovski. 1996. PI 3-kinase activation in BCR/abl-transformed hematopoietic cells does not require interaction of p85 SH2 domains with p210 BCR/abl. Blood 88:1542-1550. [PubMed] [Google Scholar]
- 24.Kharas, M. G., J. A. Deane, S. Wong, K. R. O'Bosky, N. Rosenberg, O. N. Witte, and D. A. Fruman. 2004. Phosphoinositide 3-kinase signaling is essential for ABL oncogene-mediated transformation of B-lineage cells. Blood 103:4268-4275. [Online.] [DOI] [PubMed] [Google Scholar]
- 25.Kharas, M. G., and D. A. Fruman. 2005. ABL oncogenes and phosphoinositide 3-kinase: mechanism of activation and downstream effectors. Cancer Res. 65:2047-2053. [DOI] [PubMed] [Google Scholar]
- 26.Klejman, A., L. Rushen, A. Morrione, A. Slupianek, and T. Skorski. 2002. Phosphatidylinositol-3 kinase inhibitors enhance the anti-leukemia effect of STI571. Oncogene 21:5868-5876. [DOI] [PubMed] [Google Scholar]
- 27.Klippel, A., J. A. Escobedo, Q. Hu, and L. T. Williams. 1993. A region of the 85-kilodalton (kDa) subunit of phosphatidylinositol 3-kinase binds the 110-kDa catalytic subunit in vivo. Mol. Cell. Biol. 13:5560-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolibaba, K. S., and B. J. Druker. 1997. Protein tyrosine kinases and cancer. Biochim. Biophys. Acta 1333:F217-F248. [DOI] [PubMed] [Google Scholar]
- 29.Koyama, S., H. Yu, D. C. Dalgarno, T. B. Shin, L. D. Zydowsky, and S. L. Schreiber. 1993. Structure of the PI3K SH3 domain and analysis of the SH3 family. Cell 72:945-952. [DOI] [PubMed] [Google Scholar]
- 30.Liang, J., J. K. Chen, S. T. Schreiber, and J. Clardy. 1996. Crystal structure of P13K SH3 domain at 20 angstroms resolution. J. Mol. Biol. 257:632-643. [DOI] [PubMed] [Google Scholar]
- 31.Majewski, M., M. Nieborowska-Skorska, P. Salomoni, A. Slupianek, K. Reiss, R. Trotta, B. Calabretta, and T. Skorski. 1999. Activation of mitochondrial Raf-1 is involved in the antiapoptotic effects of Akt. Cancer Res. 59:2815-2819. [PubMed] [Google Scholar]
- 32.Marley, S. B., J. L. Lewis, H. Schneider, C. E. Rudd, and M. Y. Gordon. 2004. Phosphatidylinositol-3 kinase inhibitors reproduce the selective antiproliferative effects of imatinib on chronic myeloid leukaemia progenitor cells. Br. J. Haematol. 125:500-511. [DOI] [PubMed] [Google Scholar]
- 33.Mayer, B. J., P. K. Jackson, R. A. Van Etten, and D. Baltimore. 1992. Point mutations in the abl SH2 domain coordinately impair phosphotyrosine binding in vitro and transforming activity in vivo. Mol. Cell. Biol. 12:609-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGlade, C. J., C. Ellis, M. Reedijk, D. Anderson, G. Mbamalu, A. D. Reith, G. Panayotou, P. End, A. Bernstein, A. Kazlauskas, et al. 1992. SH2 domains of the p85 alpha subunit of phosphatidylinositol 3-kinase regulate binding to growth factor receptors. Mol. Cell. Biol. 12:991-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller, A. J., A. M. Pendergast, M. H. Havlik, L. Puil, T. Pawson, and O. N. Witte. 1992. A limited set of SH2 domains binds BCR through a high-affinity phosphotyrosine-independent interaction. Mol. Cell. Biol. 12:5087-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neshat, M. S., A. B. Raitano, H. G. Wang, J. C. Reed, and C. L. Sawyers. 2000. The survival function of the Bcr-Abl oncogene is mediated by Bad-dependent and -independent pathways: roles for phosphatidylinositol 3-kinase and Raf. Mol. Cell. Biol. 20:1179-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen, M. H., J. M. Ho, B. K. Beattie, and D. L. Barber. 2001. TEL-JAK2 mediates constitutive activation of the phosphatidylinositol 3′-kinase/protein kinase B signaling pathway. J. Biol. Chem. 276:32704-32713. [Online.] [DOI] [PubMed] [Google Scholar]
- 38.Nieborowska-Skorska, M., G. Hoser, P. Kossev, M. A. Wasik, and T. Skorski. 2002. Complementary functions of the antiapoptotic protein A1 and serine/threonine kinase pim-1 in the BCR/ABL-mediated leukemogenesis. Blood 99:4531-4539. [DOI] [PubMed] [Google Scholar]
- 39.Nieborowska-Skorska, M., M. A. Wasik, A. Slupianek, P. Salomoni, T. Kitamura, B. Calabretta, and T. Skorski. 1999. Signal transducer and activator of transcription (STAT)5 activation by BCR/ABL is dependent on intact Src homology (SH)3 and SH2 domains of BCR/ABL and is required for leukemogenesis. J. Exp. Med. 189:1229-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okuda, K., T. R. Golub, D. G. Gilliland, and J. D. Griffin. 1996. p210BCR/ABL, p190BCR/ABL, and TEL/ABL activate similar signal transduction pathways in hematopoietic cell lines. Oncogene 13:1147-1152. [PubMed] [Google Scholar]
- 41.Otsu, M., I. Hiles, I. Gout, M. J. Fry, F. Ruiz-Larrea, G. Panayotou, A. Thompson, R. Dhand, J. Hsuan, N. Totty, et al. 1991. Characterization of two 85 kd proteins that associate with receptor tyrosine kinases, middle-T/pp60c-src complexes, and PI3-kinase. Cell 65:91-104. [DOI] [PubMed] [Google Scholar]
- 42.Sattler, M., and J. D. Griffin. 2003. Molecular mechanisms of transformation by the BCR-ABL oncogene. Semin. Hematol. 40:4-10. [DOI] [PubMed] [Google Scholar]
- 43.Sattler, M., M. G. Mohi, Y. B. Pride, L. R. Quinnan, N. A. Malouf, K. Podar, F. Gesbert, H. Iwasaki, S. Li, R. A. Van Etten, H. Gu, J. D. Griffin, and B. G. Neel. 2002. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 1:479-492. [DOI] [PubMed] [Google Scholar]
- 44.Sattler, M., R. Salgia, K. Okuda, N. Uemura, M. A. Durstin, E. Pisick, G. Xu, J. L. Li, K. V. Prasad, and J. D. Griffin. 1996. The proto-oncogene product p120CBL and the adaptor proteins CRKL and c-CRK link c-ABL, p190BCR/ABL and p210BCR/ABL to the phosphatidylinositol-3′ kinase pathway. Oncogene 12:839-846. [PubMed] [Google Scholar]
- 45.Sattler, M., R. Salgia, G. Shrikhande, S. Verma, E. Pisick, K. V. Prasad, and J. D. Griffin. 1997. Steel factor induces tyrosine phosphorylation of CRKL and binding of CRKL to a complex containing c-kit, phosphatidylinositol 3-kinase, and p120(CBL). J. Biol. Chem. 272:10248-10253. [DOI] [PubMed] [Google Scholar]
- 46.Scheid, M. P., P. A. Marignani, and J. R. Woodgett. 2002. Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol. Cell. Biol. 22:6247-6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skorski, T. 2002. Oncogenic tyrosine kinases and the DNA-damage response. Nat. Rev. Cancer 2:351-360. [DOI] [PubMed] [Google Scholar]
- 48.Skorski, T., A. Bellacosa, M. Nieborowska-Skorska, M. Majewski, R. Martinez, J. K. Choi, R. Trotta, P. Wlodarski, D. Perrotti, T. O. Chan, M. A. Wasik, P. N. Tsichlis, and B. Calabretta. 1997. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. EMBO J. 16:6151-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Skorski, T., P. Kanakaraj, D. H. Ku, M. Nieborowska-Skorska, E. Canaani, G. Zon, B. Perussia, and B. Calabretta. 1994. Negative regulation of p120GAP GTPase promoting activity by p210bcr/abl: implication for RAS-dependent Philadelphia chromosome positive cell growth. J. Exp. Med. 179:1855-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skorski, T., P. Kanakaraj, M. Nieborowska-Skorska, M. Z. Ratajczak, S. C. Wen, G. Zon, A. M. Gewirtz, B. Perussia, and B. Calabretta. 1995. Phosphatidylinositol-3 kinase activity is regulated by BCR/ABL and is required for the growth of Philadelphia chromosome-positive cells. Blood 86:726-736. [PubMed] [Google Scholar]
- 51.Skorski, T., M. Nieborowska-Skorska, P. Wlodarski, M. Wasik, R. Trotta, P. Kanakaraj, P. Salomoni, M. Antonyak, R. Martinez, M. Majewski, A. Wong, B. Perussia, and B. Calabretta. 1998. The SH3 domain contributes to BCR/ABL-dependent leukemogenesis in vivo: role in adhesion, invasion, and homing. Blood 91:406-418. [PubMed] [Google Scholar]
- 52.Slupianek, A., G. Hoser, I. Majsterek, A. Bronisz, M. Malecki, J. Blasiak, R. Fishel, and T. Skorski. 2002. Fusion tyrosine kinases induce drug resistance by stimulation of homology-dependent recombination repair, prolongation of G2/M phase, and protection from apoptosis. Mol. Cell. Biol. 22:4189-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slupianek, A., M. Nieborowska-Skorska, G. Hoser, A. Morrione, M. Majewski, L. Xue, S. W. Morris, M. A. Wasik, and T. Skorski. 2001. Role of phosphatidylinositol 3-kinase-Akt pathway in nucleophosmin/anaplastic lymphoma kinase-mediated lymphomagenesis. Cancer Res. 61:2194-2199. [PubMed] [Google Scholar]
- 54.Slupianek, A., and T. Skorski. 2004. NPM/ALK downregulates p27(Kip1) in a PI-3K-dependent manner. Exp. Hematol. 32:1265-1271. [DOI] [PubMed] [Google Scholar]
- 55.Vivanco, I., and C. L. Sawyers. 2002. The phosphatidylinositol 3-kinase AKT pathway in human cancer. Nat. Rev. Cancer 2:489-501. [DOI] [PubMed] [Google Scholar]
- 56.Waksman, G., S. E. Shoelson, N. Pant, D. Cowburn, and J. Kuriyan. 1993. Binding of a high affinity phosphotyrosyl peptide to the Src SH2 domain: crystal structures of the complexed and peptide-free forms. Cell 72:779-790. [DOI] [PubMed] [Google Scholar]
- 57.Weng, Z., R. J. Rickles, S. Feng, S. Richard, A. S. Shaw, S. L. Schreiber, and J. S. Brugge. 1995. Structure-function analysis of SH3 domains: SH3 binding specificity altered by single amino acid substitutions. Mol. Cell. Biol. 15:5627-5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yu, H., J. K. Chen, S. Feng, D. C. Dalgarno, A. W. Brauer, and S. L. Schreiber. 1994. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell 76:933-945. [DOI] [PubMed] [Google Scholar]
- 59.Zou, X., and K. Calame. 1999. Signaling pathways activated by oncogenic forms of Abl tyrosine kinase. J. Biol. Chem. 274:18141-18144. [DOI] [PubMed] [Google Scholar]