Abstract
Understanding the pattern of linkage disequilibrium (LD) in the human genome is important both for successful implementation of disease-gene mapping approaches and for inferences about human demographic histories. Previous studies have examined LD between loci within single genes or confined genomic regions, which may not be representative of the genome; between loci separated by large distances, where little LD is seen; or in population groups that differ from one study to the next. We measured LD in a large set of locus pairs distributed throughout the genome, with loci within each pair separated by short distances (average 124 bp). Given current models of the history of the human population, nearly all pairs of loci at such short distances would be expected to show complete LD as a consequence of lack of recombination in the short interval. Contrary to this expectation, a significant fraction of pairs showed incomplete LD. A standard model of recombination applied to these data leads to an estimate of effective human population size of 110,000. This estimate is an order of magnitude higher than most estimates based on nucleotide diversity. The most likely explanation of this discrepancy is that gene conversion increases the apparent rate of recombination between nearby loci.
Introduction
With the completion of a reference human genome sequence in sight (International Human Genome Sequencing Consortium 2001), attention is shifting to sequence variation. Most of this variation is in the form of single-nucleotide polymorphisms (SNPs). Large numbers of SNPs throughout the human genome have now been discovered and mapped (The International SNP Map Working Group 2001). SNP-discovery efforts have been carried out with the belief that these polymorphisms either will turn out to be the causative mutations in human complex disease or can be used as markers to map such mutations by linkage disequilibrium (LD) (Collins et al. 1997). The success of LD-mapping approaches based on a genomewide map of SNPs will depend in large part on the extent of LD between loci. LD can be influenced by a number of factors. These include forces that act on individual genomic regions (such as natural selection and the rates of mutation and recombination), as well as forces that have affected the entire genome (effective population size, population expansions and bottlenecks, population subdivision, and gene flow). Thus, patterns of LD are expected to vary from one region of the genome to another, as well as among populations.
Previous studies have demonstrated a nonuniform distribution of LD on a chromosomal scale, using estimates of LD based on microsatellite CEPH-Genethon data (Huttley et al. 1999), as well as population differences in the extent of LD (Eaves et al. 2000; Goddard et al. 2000; Kidd et al. 2000; Taillon-Miller et al. 2000). Most current estimates of LD are derived from disparate populations or single genomic regions (Clark et al. 1998; Fullerton et al. 2000; Kidd et al. 2000; Nickerson et al. 2000; Tishkoff et al. 2000) and are still too few to provide general insights into the patterns and distribution of LD, although data from larger populations and regions are beginning to emerge (e.g., Dunning et al. 2000; Abecasis et al. 2001). Variation in LD among studies is illustrated by a summary of five large single-gene studies (Pzreworski et al. 2000), in which three show complete LD over at least 2.5 kb, while two show rapid decline of LD. Thus, even as dense SNP maps are becoming available, key questions remain concerning the distribution of LD between SNPs throughout the genome.
We set out to examine LD in a genomewide collection of locus pairs. For practical reasons, we chose pairs separated by short distances. SNPs are typically detected by comparing the sequences of a short unique stretch of DNA among individuals (such stretches of DNA are commonly known as “sequence-tagged sites” [STSs]). To examine LD between SNPs, we drew on three large-scale SNP-discovery efforts to assemble a collection of 103 STSs that each contained multiple SNPs (a total of 325 SNPs). The STSs are distributed throughout the genome (including chromosomes 1–19, 22, and X). We sequenced these STSs in a globally representative sample of 47 individuals (94 chromosomes) and measured LD between SNPs on the same STS. We report that significantly more SNP pairs show incomplete LD than would be expected on the basis of accepted values of effective human population size and recombination rate, and we discuss possible explanations for this observation.
Subjects, Material, and Methods
Samples and Loci
The study sample comprised 10 individuals from Europe, 19 from Asia and the Pacific Islands, 4 from the Americas, and 14 from central and northeast Africa. Many of these samples overlap with those described by Cargill et al. (1999). One common chimpanzee (Pan troglodytes) was also sequenced.
All STS regions were chosen with prior knowledge that they contained at least two SNPs. These STS regions, and the SNPs within them, were ascertained in three separate studies. Forty-one STSs (group 1) were chosen from the STSs surveyed by Wang et al. (1998). One-third of the STSs in that survey were derived from random genomic sequence, and two-thirds were from the 3′ ends of expressed sequence tags, primarily the untranslated regions of genes. These STSs have a size range of 58–430 bp (average 232 bp). Twenty-eight STSs (group 2) were derived from the genes sequenced by Cargill et al. (1999) and are located primarily within coding sequence. These regions are defined by a primer pair designed to amplify primarily coding sequence from genomic DNA, and they have a size range of 175–725 bp (average 436 bp). The remaining 34 regions (group 3) were derived from a reduced representation shotgun (RRS) library used by The SNP Consortium (Altshuler et al. 2000). These are random genomic regions with a size range of 160–540 bp (average 354 bp).
Genotyping
All markers were genotyped by ABI sequencing of each STS. Sequences were base-called and assembled, and polymorphic sites were identified by the PolyPhred program with default settings (Nickerson et al. 1997). Results were visually inspected and verified by two observers. Sequencing was performed in the forward direction only; however, extensive confirmatory resequencing and data-validation procedures were carried out. All SNPs for which the minor allele was seen only once or twice in the sample, as well as all newly discovered SNPs, were resequenced three times to confirm genotypes. Additionally, on average, 22% of all individuals were fully resequenced at least two times and had their genotypes revalidated for each STS region. We discarded from further analyses those SNPs for which the minor allele was seen only once in our sample (35 SNPs), as well as two insertion/deletions.
Analysis of Data
Given the current views on the history of the human population, pairs of SNPs spaced at very short intervals are expected to exist in complete LD. That is, one would expect to observe only three of the four possible haplotypes. The fourth haplotype could arise through breakdown of LD by recombination, recurrent mutation, or gene conversion. We sought to determine the frequency with which all four haplotypes could be observed for nearby loci.
LD is often measured by one of several coefficients that reflect departures of two-locus haplotype frequencies from those expected if the loci were independent (Devlin and Risch 1995; Jorde 2000). These measures are not ideal for the present study, because (a) they are not sensitive to small departures from complete LD that might occur at very short distances and (b) they are sensitive to allele frequency. To quantify the breakdown of LD, we used a measure, RM, which counts the minimum number of recombination events in the history of the sample that are necessary to explain the observed data (Hudson and Kaplan 1985). This measure has previously been used to estimate the rate of recombination in the apolipoprotein AI-CIII-AIV gene cluster (Antonarakis et al. 1988). A recombination event is inferred when all four haplotypes are observed for a pair of loci. For a single STS, RM is the number of nonoverlapping SNP pairs that show all four haplotypes. When tabulating such events, we conservatively considered only locus pairs for which all four haplotypes were unambiguously observed in the data (that is, not hidden in double heterozygotes). For those locus pairs considered most likely to have a hidden fourth haplotype present (∼10% of STSs), haplotypes were resolved empirically by use of allele-specific PCR. There were no instances in which the fourth haplotype was uncovered through allele-specific PCR.
The inference of a recombination event from the observation of four haplotypes is valid only under the infinite-sites model (Kimura 1969), because recurrent mutation can also result in the presence of all four haplotypes. Although most SNPs are thought to arise from unique mutational events, some sites (including CpG dinucleotides and some repetitive sequences) are believed to be highly mutable and subject to recurrent mutation (Templeton et al. 2000). To obtain a conservative estimate of RM, we removed all CpGs and sites that fell within mononucleotide repeats of >5 bp (85 SNPs or ∼30%) from the data set. The inclusion of these sites would have raised the observed RM from 7 to 19.
Coalescent Simulations
We used the coalescent approach (Hudson 1983) to simulate the history of our sample for different values of 4Nr. We then calculated the mean and the distribution of RM.
For each value of 4Nr, we carried out two sets of simulations. In the first set (type I simulations), random genealogies of our sample were generated for each STS, using the coalescent model with recombination (Hudson 1983). A simulated genealogy was accepted if, at the positions where SNPs are observed on the corresponding STS, the trees contained branches such that mutations on those branches would match the number of observations of each allele in the sample. For the accepted genealogies, mutations were placed on these branches, and the resulting genotype observations were added to the simulated data set. If several choices of a branch were available, a single branch was randomly selected, with probability proportional to its length. For each value of 4Nr, 1,000 genealogies were generated for each STS. This set of simulations exactly matched the SNP number, allele frequencies, and SNP spacing of each STS. The software used to carry out these simulations is available on our Web site.
A concern with this set of simulations is that for each STS, the number of SNPs and their allele frequencies and positions were assigned a priori and placed on randomly generated genealogies. In fact, given the size of our sample and the levels of human-nucleotide diversity, most genomic regions of the size considered here would not be expected to contain multiple SNPs. By choosing regions that do contain multiple SNPs, we are likely to enrich for regions of the genome that have older genealogies with more opportunity for mutation. Such deep genealogies would also allow more opportunity for recombination, potentially biasing the real data set to higher RM values than are predicted by our simulation.
To address this potential bias, we carried out a second set of simulations (type II). These simulations were performed using the program mksamples, which is distributed by R. Hudson through his Web site (Hudson 1983). For each value of 4Nr, we generated random genealogies and placed mutations on these genealogies according to a Poisson process with the rate set by the observed nucleotide diversity in humans (4Nμ=8×10-4). As in the real data set, we then discarded all STSs that did not contain at least two SNPs with both alleles observed at least twice in the simulated sample. Most genealogies were rejected by these criteria. Only those genealogies with two, three, or four SNPs and with all SNP alleles observed at least twice were retained for further analysis. We then selected a subset of these genealogies with multiple SNPs that matched the average SNP number, SNP spacing, and SNP allele-frequency distribution of our data set. Specifically, for STSs with two SNPs, we generated 107 genealogies for segments of 285 bp each, and we then selected those with exactly two SNPs separated by ⩾45 bp (average spacing 125 bp [same as in the real data set]). We then binned these simulated STSs according to the geometric average of the two minor-allele frequencies. The geometric average was chosen because it is the square root of the frequency of the rarest haplotype under linkage equilibrium. A similar procedure was followed for STSs with three and four SNPs—except that (a) for STSs with three SNPs, segment length was 365 bp; (b) the largest distance between two SNPs was set to a minimum of 50 bp; and (c) the smallest distance was set to a minimum of 10 bp. For STSs with four SNPs, segment length was 420 bp, the largest distance between two SNPs was set to a maximum of 300 bp, and the smallest distance was set to a minimum of 10 bp. The average distance between SNPs was 130 bp and 121 bp, compared with 132 and 108 bp for the real data, on STSs with three and four SNPs, respectively. The arithmetic mean of the geometric average of minor-allele frequencies for each SNP pair was used to define the frequency bins. We calculated the distribution of RM by randomly selecting simulated STSs from the binned set, such that the number of STSs with two, three, or more SNPs, as well as the number in each frequency bin, matched the real data set. The allele-frequency distributions of the real and simulated data sets are given in table 1. The nucleotide diversity for both sets was 2.5×10-3. As expected, this is much higher than for the genome as a whole, because we selected for regions of high polymorphism. We then combined the RM values for the 68 simulated STSs. This sampling was repeated 1,000 times to obtain the distribution of RM. This set of simulations, together with the ones including gene conversion (see below) required generating a total of 750 million genealogies and produced results very similar to those obtained from type I simulations.
Table 1.
SNP Minor Allele-Frequency Distributions for the Real and Simulated Data Sets
|
Percentage of Alleles in |
||
| FrequencyRange | RealData | SimulatedData |
| 0–.1 | 30 | 31 |
| .1–.2 | 14 | 16 |
| .2–.3 | 21 | 16 |
| .3–.4 | 14 | 16 |
| .4–.5 | 21 | 21 |
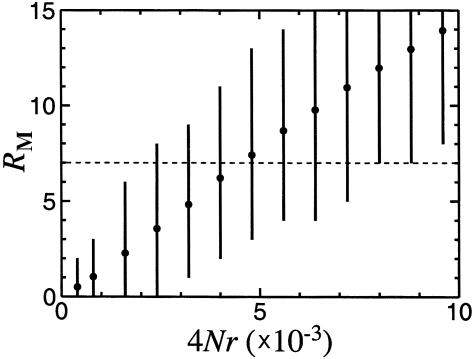
To assess the accuracy of matching between the real and simulated data sets—and also to understand the magnitude of the bias introduced by use of random genealogies versus those conditioned on polymorphism—we carried out type I simulations for simulated data sets generated by type II simulations for values of 4Nr between 4×10-4 and 9.6×10-3. We found that type I simulations produced estimates of RM that were ∼10% lower than the actual values in the simulated (type II) data, indicating that only a small bias in observed recombination is caused by greater genealogy depth of regions with higher polymorphism. For example, for 4Nr=4.8×10-3, the type I simulation underestimated the actual value by 11% (expected RM values for type I and type II simulations were 6.7 and 7.5 for the 68 STSs). Because of the similarity of results from the two types of simulations, we conservatively use the results of type II simulations in figure 1 and the text.
Figure 1.
Expected number of obligate recombination events (RM) as a function of population recombination parameter 4Nr. Dashed line indicates the observed value of RM=7. Each blackened circle is an average over 1,000 simulated replicates of the data. Vertical bars show the values that fall at 2.5%–97.5% of the simulated distribution.
To simulate the effect of variation in recombination rate, we assumed that the rate is increased by a factor of 1/α for a fraction α of the genome, while the rest of the genome does not recombine. Note that the overall rate of recombination per genome is kept fixed at its known value by this assumption. We assigned STSs to groups with recombination rate r/α or 0 with probabilities α and 1−α, respectively. We then computed the expected number of recombination events. The simulations were carried out for a range of 1/α=1.5 to 20.
Simulations with gene conversion were carried out with mksamples, which implements the model of Wiuf and Hein (2000). We used parameter track_len = 500 for the average conversion tract length, measured in base pairs, and carried out type II simulations, as described above, with the ratio f of conversion to recombination events ranging from 0 to 12.
Results
Our final data set consists of 68 STS regions with more than one SNP, of which 38 contain two SNPs, 19 contain three SNPs, 10 contain four SNPs, and 1 contains five SNPs (a total of 178 SNPs). Of the 165 overlapping pairs of SNPs on the same STS, 12 unambiguously show all four haplotypes. When the overlaps between pairs on the same STS are taken into account, at least seven obligate recombination events on six STSs are needed to explain the observed data (i.e., RM = 7). Every event is supported by at least two unambiguous observations of each of the four haplotypes. Observations of the rarest haplotype are distributed across samples of different geographic origin. The actual number of recombination events in the history of the sample is likely much higher than seven, because RM is known to provide an underestimate (Hudson and Kaplan 1985).
We next sought to estimate population parameters consistent with the observation of RM=7. Under the standard neutral model (Przeworski et al. 2000), the expected number of recombination events observed in a sample is determined by the parameter 4Nr, where N is the effective population size and r is the recombination rate per nucleotide per generation. The average recombination rate in humans, 1 cM per Mb, corresponds to r=10-8 and, together with the frequently cited value of N=10,000 (e.g., Harpending et al. 1998, Harris and Hey 1999), yields 4Nr=4×10-4. We used the coalescent approach (Hudson 1983) to simulate the history of our sample for different values of 4Nr between 4×10-4 and 9.6×10-3 and calculated the mean and the distribution of RM. With 4Nr=4×10-4, the expected value of RM is <1; 98% of the replicates give RM ⩽ 2; and the highest value observed in 1,000 simulated replicates of the data is RM=5 (a value that is observed once). A mean value of RM=7 is obtained at 4Nr≈4.4×10-3 (fig. 1). We can confine 4Nr to the range of 2.4×10-3 to 8.8×10-3 with 95% confidence (fig. 1).
Discussion
Our estimate of 4Nr=4.4×10-3, together with a recombination rate r=10-8, leads to a recombination-based estimate of effective human population size N = 110,000. This estimate is 11-fold higher than the frequently cited value of N = 10,000 (e.g., Harpending et al. 1998; Harris and Hey 1999). If r=10-8, then N = 10,000 is clearly excluded by our data. Estimates of effective population size based on sequence diversity rely on the nucleotide mutation rate, which, in humans, is estimated to be on the order of 2×10-8 per base pair per generation (Drake et al. 1998; Nachman et al. 1998). For a typical nucleotide diversity of ∼8×10-4 (Wang et al. 1998; Cargill et al. 1999; Halushka et al. 1999; Przeworski et al. 2000; The International SNP Map Working Group 2001), the estimate N=110,000 would imply a mutation rate of ∼2×10-9, an order of magnitude too low. On the basis of estimates derived from diversity of HLA, mitochondria, and the Y chromosome, Ayala (1995) argued that the effective long-term human population size is closer to 100,000 than 10,000, which would be consistent with our recombination-based estimate. However, most other recent estimates of N are consistent with a lower value of 10,000. Why are we seeing significantly more apparent recombination events than would be predicted from the generally accepted values of N and r?
While a larger ancestral effective population size is one explanation for our observation of a high RM, it is not the only one. There are several possible reasons for the discrepancy between observed and predicted values of RM, some of which we think are unlikely. Our analysis would overestimate the frequency of historical recombination if some of what we consider obligate recombination events were, despite the exclusion of sites known to be highly mutable, the result of recurrent mutation. In principle, two tests could be used to discriminate between recombination and recurrent mutation. First, the probability of recombination increases with distance between two SNPs, while the probability of recurrent mutation does not. The seven events occur on STSs that are somewhat longer than average (369 bp vs. 330 bp), but this difference is not statistically significant. Second, when three nearby sites are considered, observation of all four haplotypes at sites 1 and 2 and at sites 2 and 3 can be explained by a single recurrent mutation at the middle site but would require the unlikely occurrence of two recombination events. None of the four STSs that contain three or more SNPs and show all four haplotypes for at least one pair of sites fall into this category. Two can be explained with either a single recurrent mutation or a single recombination, one can be explained with a single recombination but would require recurrent mutation at two or more sites, and one requires two events of either type. Once again, these data are not sufficient to provide statistically significant evidence against recurrent mutation, but they are consistent with a recombination-only explanation.
An additional concern is that in choosing regions containing two or more SNPs, we may have chosen regions that have a higher mutation rate (although the studies reporting these SNPs did not find an unexpectedly high fraction of STSs with multiple SNPs [Wang et al. 1998; Cargill et al. 1999; Altshuler at al. 2000]). We examined the human-chimpanzee divergence rates for our regions. Eighty-four percent of all STSs were successfully amplified and sequenced in the chimpanzee, and overall human-chimpanzee sequence divergence was 1%. While the mean human-chimpanzee divergence was lower for group 2 STSs, which lie primarily in coding regions, this difference was not significant, suggesting no overall differences in mutation rates between the groups. Most importantly, the divergence rate is similar to that reported for the genome as a whole (Nachman et al. 1998, Hacia et al. 1999).
Another potential explanation for our data is that recombination across the genome is highly nonuniform and that our STSs with obligate recombination events fall into high-recombination regions. An extreme version of this model would postulate that the genome consists of nonrecombining blocks separated by regions of high recombination. Because the overall rate of recombination per genome is well established, our results would then suggest that the high-recombination regions constitute ∼10% of the genome. We carried out simulations to estimate the effect of such variation in recombination rate on our estimate of 4Nr. The results showed that for a given 4Nr, the expected value of RM is actually lower when recombination rate is variable, and thus an even higher value of 4Nr would be required to explain the observed data.
As described above, the SNP markers in the present study were derived from three different studies, each of which differed in the total number and the relative population diversity of the samples in which the SNPs were originally discovered. The bias that results from the variability in the ascertainment of SNPs can influence allele frequency and demographic inference. Among the three source groups we surveyed, bias is greatest in groups 1 and 3, where a much smaller sample was used for initial SNP discovery, and is least in group 2, because of considerable overlap between the initial discovery sample and the sample used in the present study. We might expect the groups with a smaller discovery sample to contain a greater proportion of common, older SNPs, which are more likely to have experienced recombination events. We therefore examined whether RM values differed among groups. Of the seven obligate recombination events, one was on an STS from group 1, two on STSs from group 2, and four on STSs from group 3. These numbers were not significantly different from expectation, given the total amount of sequence from each source (6,613 bp, 5,657 bp, and 10,184 bp, respectively), suggesting that we do not see a systematic bias in our measurement of RM. The specific effects of ascertainment are complex and are considered for these data in greater detail in another manuscript (J. Wakeley, R. Nielsen, K. Ardlie, S. N. Liu-Cordero, and E. S. Lander, unpublished data).
We believe that the most likely explanation for our observed excess of historical recombination is gene conversion. There is growing recognition that gene conversion can be a factor in shaping fine-structure patterns of LD. A high rate of conversion events in HLA-DPB1 was reported by Zangenberg et al. (1995). Analyses by Andolfatto and Nordborg (1998), Wiuf (2000), and Wiuf and Hein (2000) demonstrate that, at intragenic distances, gene conversion, rather than crossing over, is likely to be the dominant force that breaks up sites, and that gene conversion might account for the demonstrated lack of intralocus associations found in Drosophila melanogaster. Gene conversion increases the rate of exchange for closely linked sites but has negligible effects for more distant sites—as intersite distance increases, gene-conversion events that affect one of the sites become rare compared with crossovers between the sites. Indeed, gene conversion should contribute significantly to the apparent rate of recombination only when the distance between two sites is not appreciably greater than the length of a gene-conversion tract (Andofatto and Nordborg 1998).
A rough idea of the effects of gene conversion can be obtained from the following highly simplified model (see also Andofatto and Nordborg 1998; Wiuf 2000; Wiuf and Hein 2000). Assume that H Holliday junctions are formed for each reciprocal crossing-over event in a genome, and that each junction is accompanied by a tract of conversion of fixed length L. It is then easy to show that the apparent rate of recombination between sites separated by distances d<L is 2Hdr. For sites separated by distances d>L, the apparent rate of recombination is dr+Lr(2H-1), which is of order dr for d>>L (as would be expected without gene conversion). Note that the apparent rate for d<L is independent of L. The length L of gene-conversion tracts is generally thought to be in the range of 350–1,000 bp, and thus at the distances between loci considered in this article, the apparent rate of recombination should be enhanced by a factor of 2H. The value of H is not known in humans but tends to fall in the range of 1–5 for organisms (Saccharomyces cerevisiae, Neurospora crassa, and D. melanogaster) in which it has been measured (Foss et al. 1993). A value of H=5 should lower our estimate of 4Nr 10-fold, bringing it in line with expectation from the generally accepted values of N and r. We tested this prediction by performing simulations that incorporate the gene-conversion model of Wiuf and Hein (2000) with 4Nr=4×10-4. The results indicate that a ratio of conversion events to recombination events in the range of 3–10 is consistent with our data, with a ratio of 6 providing the best fit.
A recent review of the literature found RM=55 in a total of 71,824 bp of sequence from 15 independent regions—a rate of historical recombination events even higher than our observation of RM=7 in 22,454 bp (Przeworski et al. 2000). The difference could reflect the fact that the analysis of Przeworski et al. included longer regions and did not exclude highly mutable sites. Przeworski and Wall (2001) reanalyzed publicly available polymorphism data for nine loci and found evidence for more recombination than would be expected on the basis of estimates of recombination rates derived from an integration of genetic and physical maps. A model that incorporated gene conversion showed a better fit to the data than did a model of crossing-over only but was not sufficient to completely explain the data. These authors considered values H=1 and H=2; a higher value of H could potentially explain their observations.
A final possibility is that a high value of RM might also reflect the effect of population subdivision and/or admixture. A realistic demographic model for humans is likely to be complex, and population structure has been documented for humans, particularly in African populations (Tishkoff et al. 2000; J. Wakeley, R. Nielsen, K. Ardlie, S. N. Liu-Cordero, and E. S. Lander, unpublished data). A high value of RM is unlikely under population structure alone, as haplotypes in different subpopulations should have a low chance of recombining with one another (Przeworski and Wall 2001). However, it could result from recent admixture of formerly subdivided populations, combined with some amount of recombination or gene conversion. Such an example is provided by the dpp locus in Drosophila (Richter et al. 1997), where a lack of LD at short distances, together with a clade structure of haplotypes, indicated that the population surveyed was a mixture of several divergent haplotypes that had recently recombined, probably through gene conversion, although insufficiently to bring the population to linkage equilibrium.
The present study provides a genomewide look at LD over short distances. We point out that factors such as gene conversion make it difficult to extrapolate properties of LD from short to long distances and, probably, from one region to another. A more-complete characterization of LD in the human genome will thus require both a better understanding of molecular processes—such as mutation, recombination, and conversion—and the use of direct genomewide measurements covering a large range of distances.
Acknowledgments
We thank M. Goldis for technical assistance and H. Coller, L. Hartwell, R.C. Lewontin, D. Nickerson, M. Olson, D. Page, M. Przeworski, D. Reich, G. Smith, J. Wakeley, G. Wong, and two anonymous referees for helpful discussions and comments on the manuscript. This work was supported in part by grants from the National Institutes of Health to E.S.L and L.K. L.K. is a James S. McDonnell Centennial Fellow.
Electronic-Database Information
The URLs for data in this article are as follows:
- Hudson Lab Home Page, http://home.uchicago.edu/∼rhudson1 (for Richard Hudson’s simulation software)
- Kruglyak Lab Home Page, http://www.fhcrc.org/labs/kruglyak (for polymorphism data and simulation software)
References
- Abecasis GR, Noguchi E, Heinsmann A, Traherne JA, Bhattacharyya S, Leaves NI, Anderson GG, Zhang Y, Lench NJ, Carey A, Cardon LR, Moffatt MF, Cookson WOC (2001) Extent and distribution of linkage disequilibrium in three genomic regions. Am J Hum Genet 68:191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Pollara VJ, Cowles CR, Van Etten WJ, Baldwin J, Linton L, Lander ES (2000) A human SNP map generated by reduced representation shotgun sequencing. Nature 407:513–516 [DOI] [PubMed] [Google Scholar]
- Andofatto P, Nordborg M (1998) The effect of gene conversion on intralocus associations. Genetics 148:1397–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonarakis SE, Oettgen P, Chakravarti A, Halloran SL, Hudson RR, Feisee L, Karathanasis SK (1988) DNA polymorphism haplotypes of the human apolipoprotein APOA1-APOC3-APOA4 gene cluster. Hum Genet 80:265–273 [DOI] [PubMed] [Google Scholar]
- Ayala FJ (1995) The myth of Eve: molecular biology and human origins. Science 270:1930–1936 [DOI] [PubMed] [Google Scholar]
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES (1999) Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 22:231–238 [DOI] [PubMed] [Google Scholar]
- Clark AG, Weiss KM, Nickerson DA, Taylor SL, Buchanan A, Stengaird J, Salomaa V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (1998) Haplotype structure and population genetic inferences from nucleotide-sequence variation in human lipoprotein lipase. Am J Hum Genet 63:595–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Guyer MS, Chakravarti A (1997) Variations on a theme: cataloging human DNA sequence variation. Science 278:1580–1581 [DOI] [PubMed] [Google Scholar]
- Devlin B, Risch N (1995) A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29:311–322 [DOI] [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF (1998) Rates of spontaneous mutation. Genetics 148:1667–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning AM, Durocher F, Healey CS, Teare MD, McBride SE, Carlomagno F, Xu C-F, Dawson E, Rhodes S, Ueda S, Lai E, Luben RN, Van Rensburg EJ, Mannermaa A, Kataja V, Rennart G, Dunham I, Purvis I, Easton D, Ponder BAJ (2000) The extent of linkage disequilibrium in four populations with distinct demographic histories. Am J Hum Genet 67:1544–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaves IA, Merriman TR, Barber RA, Nutland S, Tuomilehto-Wolf E, Tuomilehto J, Cucca F, Todd JA (2000) The genetically isolated populations of Finland and Sardinia may not be a panacea for linkage disequilibrium mapping of common diseases. Nat Genet 25:320–323 [DOI] [PubMed] [Google Scholar]
- Foss E, Lande R, Stahl FW, Steiberg CM (1993) Chiasma interference as a function of genetic distance. Genetics 133:681–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fullerton SM, Clark AG, Weiss KM, Nickerson DA, Taylor SL, Stengard JH, Salomaa, V, Vartiainen E, Perola M, Boerwinkle E, Sing CF (2000) Apolipoprotein E variation at the sequence haplotype level: implications for the origin and maintenance of a major human polymorphism. Am J Hum Genet 67:881–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard KA, Hopkins PJ, Hall JM, Witte JS (2000) Linkage disequilibrium and allele-frequency distributions for 114 single-nucleotide polymorphisms in five populations. Am J Hum Genet 66:216–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacia JG, Fan JB, Rydeo O, Jin L, Edgemon K, Ghandour G, Mayer RA, Sun B, Hsie L, Robbins CM, Brody LC, Wang D, Lander ES, Lipshutz R, Fodor SP, Collins FS (1999) Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat Genet 22:164–167 [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bently K, Hsie L, Shen N, Weder A, Cooper R, Lipshutz R, Chakravarti A (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22:239–247 [DOI] [PubMed] [Google Scholar]
- Harpending HC, Batzer MA, Gurven M, Jorde LB, Rogers AR, Sherry ST (1998) Genetic traces of ancient demography. Proc Natl Acad Sci USA 95:1961–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EE, Hey J (1999) X chromosome evidence for ancient human histories. Proc Natl Acad Sci USA 96:3320–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR (1983) Properties of a neutral allele model with intragenic recombination. Theor Popul Biol 23:183–201 [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL (1985) Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111:147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttley GA, Smith MW, Carrington M, O'Brien SJ (1999) A scan for linkage disequilibrium across the human genome. Genetics 152:1711–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature 409:860–921 [DOI] [PubMed] [Google Scholar]
- International SNP Map Working Group, The (2001) A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409:928–933 [DOI] [PubMed] [Google Scholar]
- Jorde LB (2000) Linkage disequilibrium and the search for complex disease genes. Genome Res 10:1435–1444 [DOI] [PubMed] [Google Scholar]
- Kidd JR, Pakstis AJ, Zhao H, Lu R-B, Okonofua E, Odunsi A, Grigorenko E, Bonne-Tamir B, Friedlaender J, Schulz LO, Parnas J, Kidd KK (2000) Haplotypes and linkage disequilibrium at the phenylalanine hydroxylase locus, PAH, in a global representation of populations. Am J Hum Genet 66:1882–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M (1969) The rate of molecular evolution considered from the standpoint of population genetics. Proc Natl Acad Sci USA 63:1181–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Bauer VL, Crowell SL, Aquadro CF (1998) DNA variability and recombination rates at X-linked loci in humans. Genetics 150:1133–1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Taylor SL, Fullerton SM, Weiss KM, Clark AG, Stengard JH, Salomaa V, Boerwinkle E, Sing CF (2000) Sequence diversity and large-scale typing of SNPs in the human apolipoprotein E gene. Genome Res 10:1532–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DA, Tobe VO, Taylor SL (1997) PolyPhred: automating the detection and genotyping of single nucleotide substitutions using fluorescence-based resequencing. Nucleic Acids Res 25:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przeworski M, Hudson RR, Di Rienzo A (2000) Adjusting the focus on human variation. Trends Genet 16:296–302 [DOI] [PubMed] [Google Scholar]
- Przeworski M, Wall JD (2001) Why is there so little linkage disequilibrium in humans? Genet Res 77:143–151 [DOI] [PubMed] [Google Scholar]
- Richter B, Long M, Lewontin RC, Nitasaka E (1997) Nucleotide variation and conservation at the dpp locus, a gene controlling early development in Drosophila. Genetics 145:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon-Miller P, Bauer Sardina I, Saccone NL, Putzel J, Laitinen T, Cao A, Kere J, Pilia G, Rice JP, Kwok P-Y (2000) Juxtaposed regions of extensive and minimal linkage disequilibrium in human Xq25 and Xq28. Nat Genet 25:324–328 [DOI] [PubMed] [Google Scholar]
- Templeton AR, Clark AG, Weiss KM, Nickerson DA, Boerwinkle E, Sing CF (2000) Recombinational and mutational hotspots within the human lipoprotein lipase gene. Am J Hum Genet 66:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Pakstis AJ, Stoneking M, Kidd JR, Destro-Bisol G, Sanjantila A, Lu R-B, Deinard AS, Sirugo G, Jenkins T, Kidd KK, Clark AG (2000) Short tandem-repeat polymorphism/Alu haplotype variation at the PLAT locus: implications for modern human origins. Am J Hum Genet 67:901–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DG, Fan JB, Siao CJ, Berno A, Young P, Sapolsky R, Ghandour G, et al (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280:1077–1082 [DOI] [PubMed] [Google Scholar]
- Wiuf C (2000) A coalescence approach to gene conversion. Theor Popul Biol 57:357–367 [DOI] [PubMed] [Google Scholar]
- Wiuf C, Hein J (2000) The coalescent with gene conversion. Genetics 155:451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zangenberg G, Huang M-M, Arnheim N, Erlich H (1995) New HLA-DPB1 alleles generated by interallelic gene conversion detected by analysis of sperm. Nat Genet 10:407–414 [DOI] [PubMed] [Google Scholar]