Abstract
Human prion diseases have inherited, sporadic, and acquired etiologies. The appearance of the novel acquired prion disease, variant Creutzfeldt-Jakob disease (vCJD), and the demonstration that it is caused by the same prion strain as that causing bovine spongiform encephalopathy, has led to fears of a major human epidemic. The etiology of classical (sporadic) CJD, which has a worldwide incidence, remains obscure. A common human prion-protein–gene (PRNP) polymorphism (encoding either methionine or valine at codon 129) is a strong susceptibility factor for sporadic and acquired prion disease. However, a quantitative-trait–locus study of prion incubation periods in mice has demonstrated an important factor that is close to Prnp but is independent of its coding sequence or that of the nearby prion-like doppel gene (Prnd). We have analyzed the PRNP locus for such tightly linked susceptibility factors. Fifty-six polymorphic sites have been identified within 25 kb of the PRNP open reading frame, including sites within the PRNP promoter and the PRNP 3′ untranslated region. These have been characterized in 61 Centre d’Étude du Polymorphisme Humain (CEPH) families, demonstrating extensive linkage disequilibrium around PRNP and the existence of 11 major European PRNP haplotypes. Haplotype frequencies estimated in healthy U.K. control individuals were very similar to those deduced in the CEPH families. A common haplotype was overrepresented in patients with sporadic CJD (sCJD). Through use of a log-linear modeling approach to simultaneously model Hardy-Weinberg and linkage disequilibria, a significant independent association was found between sCJD and a polymorphism upstream of PRNP exon 1 (P=.005), in addition to the strong susceptibility conferred by codon 129 (P=2×10-8). However, although our sample size was necessarily small, no association was found between these polymorphisms and vCJD or iatrogenic CJD, in keeping with their having distinct disease mechanisms. In addition, there was no evidence of a PRNP founder effect in the first reported geographical cluster of vCJD.
Introduction
The appearance of variant Creutzfeldt-Jakob disease (vCJD), which is caused by the same prion strain as that causing bovine spongiform encephalopathy (BSE) (Collinge et al. 1996b; Bruce et al. 1997; Hill et al. 1997), has led to fears of a major human epidemic (Collinge 1999). There are three well-documented episodes of human population exposure to infectious prions: administration of prion-contaminated cadaver-derived growth hormone, the epidemic of kuru (MIM 245300) in Papua New Guinea, and, more recently, exposure of the European population to BSE-contaminated material. An extraordinarily wide range of incubation periods has been observed in these episodes (Collinge 1999; Brown et al. 2000). In the case of the kuru epidemic, with which we have the longest experience, there are some individuals who appear to be completely resistant to prion disease, despite numerous exposures, and there is a range in the incubation period, in those affected, of 5 years to >40 years (Collinge 1999). The determinants of this variability in susceptibility and incubation time are not well understood.
A common human prion-protein–gene (PRNP [MIM 176640]) polymorphism, encoding either methionine or valine at codon 129, is a strong susceptibility factor for prion disease. Methionine homozygotes comprise 37% of the U.K. population, whereas valine homozygotes comprise 12% (Owen et al. 1990). Patients with sporadic CJD (sCJD [MIM 123400]) are predominantly methionine or valine homozygotes (Palmer et al. 1991), whereas all patients with vCJD to date have been methionine homozygotes (Collinge et al. 1996a; Zeidler et al. 1997). Valine-homozygous individuals are more susceptible to iatrogenic growth hormone–associated prion disease (Collinge et al. 1991). The age at onset of six-octapeptide–repeat insertion prion disease is earlier for methionine homozygotes, compared with heterozygotes (Baker et al. 1991; Collinge et al. 1992). Homozygotes of either allele have a shortened incubation period for kuru (Cervenakova et al. 1998).
Recent evidence points to genetic susceptibility factors near mouse Prnp, independent of its coding sequence or that of the nearby Prnd gene. A quantitative-trait–locus (QTL) mapping study of inbred mouse strains has identified a region of chromosome 2, encompassing Prnp, as one of three major QTLs that determine prion-disease incubation time after intracerebral inoculation with mouse-adapted scrapie (Lloyd et al. 2001). It has been known for some time that transgenic mouse models with additional copies of Prnp demonstrate an inverse correlation between PrP expression level and incubation time (Prusiner et al. 1990). Susceptibility factors for human prion disease might therefore include sequence variation in the PRNP promoter or variation in another tightly linked regulatory region. There is, as yet, no evidence that sequence variation in the recently identified PRND (MIM 604263) gene, situated 25 kb downstream of PRNP, plays any role in prion-disease pathogenesis (Mead et al. 2000; Peoc'h et al. 2000). However, the extent of linkage disequilibrium (LD) at the locus is not known, potentially complicating the assessment of individual polymorphisms.
Material and Methods
Samples
CEPH family samples were obtained from the Foundation Jean Dausset, Paris. Variant, sporadic, and iatrogenic CJD samples were obtained from patients seen at the U.K. National Prion Clinic, St. Mary’s Hospital, London. All cases of sporadic and vCJD were neuropathologically confirmed. DNA was extracted from either blood or brain tissue, by use of commercially available kits. Chimpanzee (Pan troglodytes) blood was provided by the Institute of Zoology, London. Informed consent was obtained for all patient samples used in the study. The St. Mary’s Hospital Ethics Committee gave ethical approval.
Single-Nucleotide Polymorphism (SNP) Identification
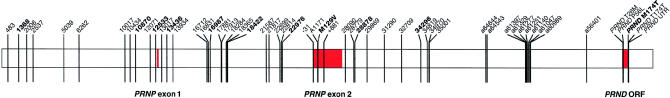
SNPs of the PRNP locus were detected by automated fluorescent sequencing of samples from eight CEPH parents (133101, 133102, 136201, 136202, 134701, 134702, 1329401, and 1329402). A single chimpanzee was also sequenced. Initially, the available human genomic sequence was derived from a 35-kb clone (GenBank accession number U29185) centered on PRNP, over which 32 kb was sequenced in eight CEPH parents. Subsequent to flanking genomic sequence becoming available (GenBank accession number AL133396), an additional 3 kb was resequenced in the intergenic region between PRNP and PRND, including the PRND open reading frame (ORF) (situated 25 kb 3′ to the PRNP ORF). A large number of individuals (16 CEPH parents and >80 patients with prion disease) have been sequenced for PRNP exon 1 and the PRNP promoter (−750 to +240, relative to transcription start site), as well as for PRNP exon 2, including the entire 3′ UTR. Primers were designed to amplify PCR products that were 800–1,050 bp in length; the oligonucleotide sequences and conditions are available on request. PCR, product purification, and sequencing were performed in a 96-well format (96-well plates supplied by Anachem). PCR products were purified using a 96-well QIAGEN PCR-purification kit. The sequencing reaction was performed with Perkin Elmer Big Dye Sequence Terminator Ready Reaction Mix. The resulting sequencing products were washed with 70% ethanol and were then precipitated in 95% ethanol. Precipitated sequencing products were then resuspended in 2 μl of formamide loading buffer and were denatured for 2 min, before electrophoresis on a denaturing 4.2% acrylamide gel (Sequagel XR, National Diagnostics) on an ABI-377 automated sequencer for 9 h. Electropherogram traces were visually compared, to identify SNPs in heterozygous individuals. Fifty-six SNPs were characterized and were designated according to their position on clone U29185 or on clone AL133396 (in which case the position is prefixed with “a”); the relative locations are shown in figure 1. SNPs at sites of particular interest were given specific names; for example, ORF polymorphism PRNP M129V, an SNP that is 31 bp 5′ to PRNP exon 2, was designated “−31,” and a PRNP 3′ UTR SNP was designated “+881.”
Figure 1.
Line drawing of the PRNP-PRND locus, including the location of SNPs relative to PRNP exons 1 and 2
SNP Genotyping
Eleven of fifty-six SNPs (shown in boldface type in fig. 1) were genotyped in 652 individuals from the CEPH families (parents, all available grandparents, and at least four children from each of the 61 families), to determine SNP phase in each of the 244 CEPH founder chromosomes. The 11 SNPs were chosen for genotyping because they were highly polymorphic, were evenly distributed across the region of interest, and altered a convenient restriction digest site. New primers were designed to amplify PCR products of ∼200–400 bp, including the SNP. Restriction digests (enzymes supplied by New England Biolabs or by Roche) were performed in a 20-μl reaction volume by use of 2–5 μl of neat PCR product. Digested PCR products were electrophoresed on agarose gel at 3 V/cm and were stained with 100 ng/ml ethidium bromide in a 3×96-well format. Genotyping reactions at six sites (13436, 16987, PRNP M129V, 28878, 34296, and PRND M174T) were internally controlled for failure to digest with an invariant restriction site found near the SNP. Genotypes for the 11 SNPs were also determined for unrelated patients with prion disease (43 with vCJD, 93 with sCJD, and 32 with iatrogenic CJD).
Subsequently, to estimate the degree of LD in regions flanking the PRNP haplotypes (nucleotide positions [nps] 1368–28878 on clone U29185), 17/56 SNPs were genotyped in the 122 CEPH parents, either by restriction digest or by automated sequencing. Genotyping of at least one child in each family determined the phase relationship between these 17 SNPs and the known haplotypes. Finally, 23/56 SNPs, all located within the region of PRNP haplotypes (nps 1368–28878 on clone U29185), were genotyped in a selection of 20 CEPH individuals, 9 patients with vCJD, and 2 patients with sCJD. This genotyping was performed to determine whether the PRNP haplotypes might be significantly subdivided when all SNPs were considered.
Statistical Genetic Analysis
Pairwise LD was assessed for all combinations of the 26 SNPs genotyped in all CEPH founders. D′, a measure of LD, was calculated using the DNA Sequence Polymorphism program (Rozas and Rozas 1999). Strong LD facilitated the deduction of haplotypes between np 1368 and np 28878. Haplotypes were estimated in the U.K. control samples by empirical allocation based on the CEPH haplotype frequencies compatible with the genotype data for each individual. An analysis using an estimation-maximization (EM) algorithm gave similar results in the two control samples (data not shown). Since the prion-disease groups displayed marked Hardy-Weinberg disequilibrium at most of the markers, EM algorithms may not produce reliable results. Haplotypes of the prion-disease samples were therefore estimated empirically on the basis of haplotype frequencies that were found in the U.K. control group. Comparisons of haplotype counts in control and patient groups were made using standard contingency-table methods (Agresti 1990).
To test for independent associations between SNPs and prion disease, distinct from hitchhiking effects with codon 129, a series of log-linear two-locus models were fitted to joint genotype counts in patients and control individuals (Weir and Wilson 1986; Tiret et al. 1991; Weir 1996). Through use of the parameter nomenclature introduced by Weir (1996), each pair of markers, A and B, was fitted, in a series of nested models, with parameters to model allele frequencies (MA and MB), Hardy-Weinberg disequilibria (MAA and MBB), linkage disequilibria (SAB and QAB), and trigenic disequilibria (MAAB and MABB); a separate set of parameters is specified for the patients and control individuals. The saturated model, therefore, includes eight parameters to model the nine joint genotype counts that can be distinguished (i.e., quadrigenic disequilibria were undefined). Models were fitted using an Excel spreadsheet (Microsoft), which is available from the authors on request; log likelihoods were maximized numerically through use of the “Solver” function, and multiple initial parameter values and alternative “Solver” settings were used to reduce the possibility of accepting local—rather than global—function minima. The goodness-of-fit of nested models was evaluated using likelihood-ratio tests with reference to χ2 distributions with degrees of freedom equal to the difference in the number of parameters being estimated.
Results
Identification of SNPs
Fifty-six SNPs were identified in the eight CEPH parents (figs. 1 and 2; table 1), including one triallelic SNP and one single-base–insertion polymorphism. As expected, transitions (39/54) outnumbered transversions (15/54). Twenty-two SNPs occurred at mutation hot-spot motifs: eighteen were at CpG sites, two were in mononucleotide runs that were more than four bases long, and two were at DNA polymerase α-arrest motifs.
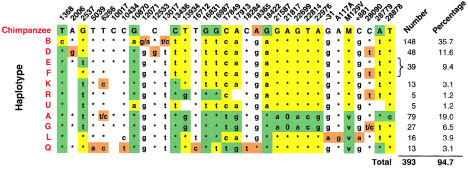
Figure 2.
Haplotype table of the CEPH parents and CEPH grandparents (nontransmitted haplotypes only). Two bases at a single site separated by a slash (/) indicates that there were rare haplotypes in which the second of the two bases shown was found. An asterisk (*) indicates identity with the chimpanzee base. Suspected substitutions are shown in orange. B = most common haplotype with methionine at codon 129, shown in yellow. D = methionine haplotype with a guanine substitution at np 12533, near the PRNP promoter, and thymine at np 28090, near the PRNP 3′ UTR; this haplotype may have been generated by the substitution of guanine at np 12533 on an F (483c) haplotype. E = methionine haplotype similar to B, without substitutions at np 1368 and np 10870 and without the promoter substitution at np 12533 but including the substitution at np 483 that is not found on valine haplotypes; alternatively, E may be a recombinant between B and A, with a break point between np 10870 and np 13436, but here one must account for the presence of cytosine at np 483, only found on methionine haplotypes. U and F = uncommon methionine haplotypes similar to E but with 5′ substitutions; E and F were not distinguished in CEPH grandparents, and therefore the frequencies are combined. E was found in 15 CEPH parents, and F was found in 5 CEPH parents. K and R = two methionine haplotypes whose origin could be best explained by a single 5′ recombination event between A and B; these haplotypes were probably generated by different but nearby recombination break points. In the case of R, the recombination may have occurred on a D or F haplotype with 28090t, with the breakpoint between np 16987 and np 18284; in the case of K, the 5′ recombinant chromosome was probably a rare subset of A, with a substitution of 6266c and a breakpoint between np 13426 and np 13934. A = most common haplotype with valine at codon 129, shown in green. G = valine haplotype similar to A in the 5′ direction from M129V but similar to B in the 3′ direction from M129V. L = valine haplotype, which, like Q below, has a pattern of SNPs similar to the chimpanzee; also found are substitutions unique to this haplotype, including the A117A PRNP ORF silent polymorphism, a polymorphism 31 bp 5′ to the PRNP translation start site (−31), and a common PRNP 3′ UTR polymorphism 881 bp 3′ to the PRNP translation start site (+881), shown in purple. Q = valine haplotype, similar to L and, again, with a number of substitutions found only on this haplotype, shown in blue (nps 5039, 10434, 16713, 18284, 18365, and 31290). Only 8/244 haplotypes, all on valine chromosomes, each found in one parental chromosome only, could not be placed in one of these eight haplotypes. These are not shown.
Table 1.
SNPs Identified
| Namea | SNPb | Method ofGenotypingc | No. of CEPHChromosomes Genotypedd | Frequency ofUncommon Allele(%) |
| 483 | c/T | SfcI | 244+ | 19 |
| 1368 | C/t | PvuII | 652 | 35 |
| 2006 | a/g | Sequencing | 16 | Singleton |
| 2537 | G/t | ItaI | 44 | 50 |
| 5039 | T/a | MaeII | 48 | 4 |
| 6262 | A/g | XmnI | 62 | 8 |
| 10017 | c/g | Sequencing | 14 | Singleton |
| 10434 | c/t | Sequencing | 44 | 5 |
| 10870 | g/A | Tsp509I | 652 | 36 |
| 12071 | a/g | DGGE | 80 | 2 |
| 12533 | c/G | BstNI | 652 | 13 |
| 13017 | t/c | Sequencing | 42 | Singleton |
| 13436 | T/c | PstI | 652 | 42 |
| 13934 | T/g | TfiI | 42 | 31 |
| 16712 | t/c | Sequencing | 40 | 5 |
| 16831 | t/G | BstNI | 244+ | 39 |
| 16987 | G/t | AluI | 652 | 35 |
| 17881 | a/g | Sequencing | 44 | 39 |
| 17913 | c/t | Sequencing | 44 | 39 |
| 18284 | c/t | Sequencing | 38 | 5 |
| 18365 | g/a | Sequencing | 38 | 5 |
| 18422 | a/G | MaeII | 652 | 37 |
| 21587 | a/g | Sequencing | 44 | 34 |
| 21817 | insert A | Sequencing | 44 | 34 |
| 22699 | g/a | Sequencing | 48 | 38 |
| 22814 | t/c | Sequencing | 44 | 41 |
| 22976 | a/G | AccI | 652 | 30 |
| −31 | g/a | Sequencing | 160 | 4 |
| A117A | A/g | PvuII | 160 | 4 |
| M129V | A/g | NspI | 652 | 38 |
| +881 | g/a | Sequencing | 160 | 4 |
| 28090 | c/T | AluI | 244+ | 25 |
| 28779 | t/a | Sequencing | 48 | 33 |
| 28878 | C/t | AluI | 652 | 23 |
| 29869 | a/T | Tsp509I | 244+ | 28 |
| 31290 | a/C | PvuII | 48 | 4 |
| 32709 | a/g | Sequencing | 48 | 4 |
| 34296 | a/G | HincII | 652 | 36 |
| 34819 | t/C | RsaI | 48 | 10 |
| 34970 | t/c | Sequencing | 48 | Singleton |
| 35001 | g/c | Sequencing | 48 | 6 |
| a64644 | g/c | Sequencing | 42 | 14 |
| a64543 | t/C | HpyCH4IV | 244+ | 19 |
| a61397 | c/t | Sequencing | 244+ | 22 |
| a61329 | c/t | Sequencing | 244+ | 22 |
| a61277 | c/t | Sequencing | 244+ | 18 |
| a61251 | a/g | Sequencing | 244+ | 18 |
| a61140 | c/a | Sequencing | 244+ | 22 |
| a61047 | g/t | Sequencing | 244+ | Singleton |
| a60989 | c/g/a | Sequencing | 244+ | 15/8 |
| a56401 | C/g | Eco57I | 244+ | 9 |
| PRND T26M | c/t | Sequencing | 244+ | 2 |
| PRND P56L | c/t | Sequencing | 244+ | 1 |
| PRND M174T | c/T | NlaIII | 652 | 48 |
| PRND T174T | a/g | Sequencing | 244+ | 1 |
| PRND UTR | t/c | Sequencing | 244+ | 49 |
Name corresponds with the location of the SNP on U29185 (no prefix) or on AL133396 (prefixed by “a”).
The nucleotide to the left of the slash was the more frequent allele; the nucleotide denoted by a capital letter is the allele that is cut by the restriction enzyme shown in the “Method of Genotyping” column.
DGGE = Denaturing gradient gel electrophoresis.
244+ = Genotyping of all CEPH parents and at least one child, to experimentally determine SNP phase in the parents.
Characterization of SNP Haplotypes
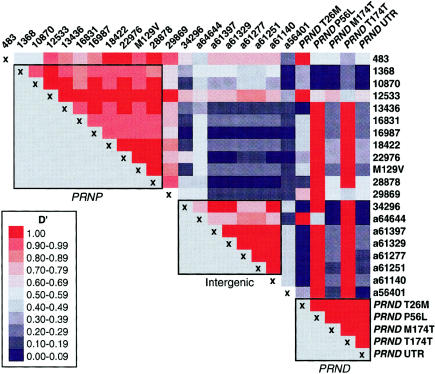
Strong LD was observed between all SNPs identified between np 1368 and np 29869 (designated by their location on clone U29185). 483c was found on a proportion of all methionine haplotypes, and 34296g was found on a proportion of all major haplotypes. These two polymorphic sites, therefore, marked the boundaries of the PRNP haplotypes (fig. 2). Between PRNP and PRND, a second region of strong LD developed, comprising 10 SNPs extending over 12 kb of genomic sequence. This region of LD is delimited in the 3′ direction by a polymorphism near the putative PRND transcription start site (a56401). Farther 3′, within the PRND gene, the common PRND ORF polymorphism, M174T, and the 3′ UTR polymorphism (38 bp 3′ to M174T) are, unsurprisingly, in strong LD with each other but not with the intergenic haplotypes. These neighboring areas of strong LD are illustrated in figure 3, which shows plotting of D′ between pairs of SNPs.
Figure 3.
Table of pairwise D′ (calculated using DNAsp), showing pairwise LD between SNPs
In the region of strong LD around PRNP, between np 1368 and np 29686, 34 SNPs were identified. Nine of these SNPs (shown in boldface in fig. 1) were used for haplotyping in all the CEPH families. There was no phase ambiguity in any family. The PRNP haplotypes were empirically placed into 11 groups (fig. 2). Rare haplotypes—which were identical, except for a substitution at a single site, to a more common haplotype—were also included in the larger group. The likely nucleotide substitution at each site was predicted by comparison with the chimpanzee sequence.
The two most common haplotypes, A (valine) and B (methionine), representing >55% of the total haplotype diversity, are opposite in state at all highly polymorphic loci. This suggests that the northern European PRNP genealogy is characterized by two major branches. Haplotypes Q and L are remarkable in that their patterns of SNPs are strikingly similar to that observed in the chimpanzee, with the exception of a substitution of valine for methionine at PRNP codon 129. This chimpanzee haplotype pattern, together with a number of nucleotide substitutions found only on haplotypes Q and L, suggests that these two uncommon haplotypes also represent deep branches of the PRNP genealogy. 5′ recombination events of A on B were assumed to have generated haplotypes K, R, and, possibly, U and E, since these haplotypes change state from being like an A haplotype to being like a B haplotype, at a single putative ancestral recombination break point. Recombinant haplotypes were observed on methionine haplotypes, but none of the complementary recombinant haplotypes were observed on a valine haplotype.
To test whether the above haplotypes of 9 SNPs between np 1368 and np 29869 were representative of the true haplotype diversity in the CEPH families, an additional 23 SNPs in the interval between np 1368 and np 29869 were genotyped in a collection of 20 CEPH individuals chosen to represent the previously established haplotype diversity. SNPs at np 16731, np 28090, and np 29869 were genotyped and phased in all CEPH parents. The extra genotypes in the additional individuals were consistent with the previously established groups of haplotypes, demonstrating the validity of the restricted SNP set. Only two new haplotypes were generated by the additional genotyping: a variant of haplotype E, with 2537g and 28090t, found in 5/244 haplotypes (designated “haplotype F”), and a variant of haplotype B, with 16831g, found in 1/244 haplotypes.
Association Study
Prion-disease samples (43 vCJD, 95 sCJD, and 32 iatrogenic CJD) were genotyped at 11 sites, including the 9 SNPs used to define PRNP haplotypes in the CEPH families, shown in boldface type in figure 1. Haplotype frequencies determined in the CEPH families, together with estimated haplotype frequencies from the U.K. control population, from the patient group with sCJD, and from the patient group with vCJD, are shown in table 2. There is no significant difference in haplotype frequencies between the CEPH families and U.K. control population (P=.89). Haplotype frequencies in the prion disease groups are very different from both control populations (e.g., CEPH vs. sCJD, P=4×10-4; CEPH vs. vCJD, P<10-6). The haplotype distribution is markedly skewed toward methionine haplotypes, a pattern predicted by the reported susceptibility to prion diseases that is associated with the M129V polymorphism. The strongest effect of M129V in sCJD is the resistance to prion disease shown by heterozygotes (table 3; χ2=21.6, P<.0001). In combination with the strong LD at the PRNP locus, this leads to a striking Hardy-Weinberg disequilibrium in the prion-disease groups, at all sites between np 1368 and np 28878. It is therefore necessary to model Hardy-Weinberg disequilibrium and LD between markers, to assess the independent statistical association of polymorphisms at PRNP. It is important to differentiate between those markers that associate with prion disease because of a hitchhiking effect with codon 129 and alleles that are independent prion-disease–susceptibility factors. We applied a log-linear two-locus modeling approach, to tackle this problem (Weir and Wilson 1986; Tiret et al. 1991; Weir 1996) in a joint genotype analysis of a test SNP and M129V (Tiret et al. 1991), comparing genotype frequencies in patients with sCJD and in CEPH founders. There was no evidence of trigenic disequilibria in the analyses of SNP 1368 with M129V (table 4; comparison of models 1 and 2, P=.84). There was also no evidence (P=.67) of Hardy-Weinberg disequilibrium in the case or control groups at SNP 1368, nor was there evidence of Hardy-Weinberg disequilibrium in the control group at M129V, and identical patterns of LD were consistent in the case and control groups (comparison of models 1 and 3). This parsimonious model (model 3) was compared with nested model 4, in a likelihood-ratio test of the independent association between the cytosine allele at SNP 1368 and sCJD (χ2=7.9, 1 df, P=.005). A comparison of hierarchical models 2 and 5 provides a test for the independent association between M129V and susceptibility to sCJD (χ2=47.1, 6 df, P=2×10-8). Similar results were found in a complementary analysis of the patients with sCJD and 269 healthy white individuals from the U.K. population (table 3); SNP 1368 was found to be independently associated with the risk of developing sCJD (P=.003). 1368c is nearly always found on a 129M chromosome (haplotype B), and, therefore, we propose that this haplotype is associated with sporadic prion disease. Since 1368c is very rarely coupled with 129V (only one 1368c 129V haplotype in patients with sCJD), the association applies to methionine-homozygous sCJD. Analysis of variance indicated that there was no difference in age at onset of disease or in disease duration between the three genotypes at SNP 1368. All other sites, including SNP 34296 and PRND M174T, showed no independent evidence of association once the association at M129V had been modeled (data not shown).
Table 2.
Estimated Haplotype Frequencies in Case and Control Groups
|
Frequency (%) ina |
||||
| Haplotype | CEPH Individuals(n=412) | U.K. ControlPopulation (n=190) | Patients with sCJD (n=186) | Patients with vCJD (n=86) |
| B | 148 (35.7) | 70 (36.8) | 100 (53.8) | 50 (58.1) |
| D | 48 (11.6) | 22 (11.6) | 18 (9.8) | 12 (13.9) |
| E/Fb | 39 (9.4) | 19 (10.0) | 15 (8.1) | 17 (19.7) |
| K | 13 (3.1) | 7 (3.7) | 4 (2.1) | 2 (2.3) |
| R | 5 (1.2) | 0 | 3 (1.6) | 3 (3.5) |
| U | 5 (1.2) | 1 (0.5) | 0 | 0 |
| Other M | 4 (1.0) | 2 (1.1) | 6 (3.2) | 2 (2.3) |
| A | 79 (19.0) | 34 (18.4) | 27 (14.5) | 0 |
| G | 27 (6.5) | 10 (5.3) | 4 (2.1) | 0 |
| L | 16 (3.9) | 9 (4.7) | 1 (0.5) | 0 |
| Q | 13 (3.1) | 6 (3.2) | 6 (3.2) | 0 |
| Other V | 15 (3.6) | 10 (5.3) | 2 (1.1) | 0 |
In the CEPH founders, phase assignments were unambiguously deduced in genotyped families; in the other groups, assignments were deduced by empirical allocation based on frequencies determined in control populations.
E and F could not always be distinguished in the disease groups, and, therefore, a combined total is shown.
Table 3.
Joint Genotype Table at Sites 1368 and M129V
|
Frequency in |
|||||
| Genotype(1368/M129V) | CEPHIndividuals | U.K. Control Population | Patientswith sCJD | Age at Onset (Range)(years) | Median Disease Duration (Range)(mo) |
| tt/vv | 25 | 31 | 15 | 57 (37–77) | 6 (2–15) |
| ct/vv | 4 | 3 | 1 | 58 | 4 |
| cc/vv | 0 | 0 | 0 | … | … |
| tt/mv | 34 | 55 | 3 | 65 (61–65) | 7 (3–11) |
| ct/mv | 59 | 60 | 5 | 71 (47–79) | 14 (2–23) |
| cc/mv | 1 | 0 | 0 | … | … |
| tt/mm | 14 | 23 | 4 | 63 (56–79) | 2 (1–3) |
| ct/mm | 45 | 51 | 29 | 69 (43–86) | 3 (1–22) |
| cc/mm | 23 | 46 | 36 | 66 (41–83) | 3 (1–17) |
| Total | 205 | 269 | 93 | ||
Table 4.
Log-Linear Two-Locus Analysis of SNP 1368 and M129V
|
Parametera |
|||||||||||||||
| Model | MA1 | MA2 | MB1 | MB2 | MAA1 | MAA2 | MBB1 | MBB2 | SAB1 | SAB2 | QAB1 | QAB2 | TrigenicDisequilibrium | Devianceb | No. of Parameters |
| 1 | * | * | * | * | * | * | * | * | * | * | * | * | * | .00 | 16 |
| 2 | * | * | * | * | * | * | * | * | * | * | * | * | 1 | 1.42 | 12 |
| 3 | * | * | * | * | 1 | 1 | 1 | * | * | SAB1 | * | QAB1 | 1 | 6.73 | 7 |
| 4 | * | MA1 | * | * | 1 | 1 | 1 | * | * | SAB1 | * | QAB1 | 1 | 14.63 | 6 |
| 5 | * | * | * | MB1 | 1 | * | 1 | 1 | * | SAB1 | * | QAB1 | 1 | 48.56 | 6 |
Parameters MA and MB model allele frequencies—and MAA and MBB model Hardy-Weinberg disequilibrium—for the 1368 and M129V markers, respectively. SAB and QAB model LD between alleles at the two markers. Parameters for the CEPH and sCJD strata are labeled with subscripts 1 and 2, respectively. Trigenic disequilibrium includes two parameters, MAAB and MABB, for each stratum (cases and controls). An asterisk (*) indicates that the parameter was estimated by maximum likelihood in the analysis. “1” indicates that the parameter was fixed (null effect) in the analysis. Otherwise, the parameters were equated across the two strata.
Deviance is the −2 loge-likelihood difference with saturated model 1.
Founder Haplotypes
A cluster of vCJD near the Leicestershire village of Queniborough in the United Kingdom has been investigated for local environmental risk factors (Cousens et al. 2001). An alternative hypothesis is that a rare PRNP vCJD-susceptibility allele is found at a high frequency in this locality, because of a founder effect, with the allele occurring recently enough to be geographically localized but not so recently that family relationships may be readily identified. Three samples from patients with vCJD from the Leicestershire cluster were available for this study. The individual genotype combinations in these three individuals were incompatible with a shared PRNP susceptibility allele.
Discussion
We have sequenced 35 kb of the PRNP locus, to identify informative SNPs in the northern European population. This was followed by RFLP analysis of the CEPH families, to build a haplotype table of a region of strong LD around PRNP. In the northern European population, PRNP haplotypes with methionine at codon 129 were characterized by a 10-kb region of complete LD around the PRNP ORF. 5′ substitutions and multiple presumed recombination events between the common A and B haplotypes provided the haplotype diversity. The haplotypes with valine at codon 129 were more diverse than those with methionine at codon 129, with the uncommon Q and L haplotypes being very similar to the chimpanzee haplotype. We have found a significant association between a PRNP upstream polymorphism and sCJD, by genotyping 168 patients with prion disease of different etiologies.
The etiology of sCJD remains obscure; it is possible that some cases result from environmental exposure to prions, but this mechanism is unlikely, on epidemiological grounds, to explain the majority of cases (Brown et al. 1987). Alternative possibilities include a stochastic conversion of normal prion protein to the infectious form or a somatic PRNP mutation leading to a cell or cluster of cells that translate a mutant prion protein predisposed to convert to an infectious prion. Haplotype association could act through enhanced expression of PRNP, which fits most easily with a hypothesis of stochastic protein misfolding. High levels of expression of PRNP may make the initial conversion of PrPc to the infectious form more likely. One might expect altered disease phenotypes with overexpression; however, no major differences were seen in age at onset or in duration of disease, when analyzed by polymorphism. This dissociation of disease susceptibility and phenotype suggests that different biological mechanisms account for these variables. Factors such as PrPSc strain type and variation of other genes affecting disease pathways may have a greater role in the generation of variation in clinical phenotypes.
In regard to the potential confounding variable of population stratification, there are numerous genotyping studies of the codon 129 polymorphism in Europe, including studies in France, Germany, Italy, Spain, and the United Kingdom (Owen et al. 1990; Salvatore et al. 1994; Berr et al. 1998; Windl et al. 1999; Zimmermann et al. 1999). There is a very small range of valine allele frequencies in these countries (0.348–0.377). Additionally, there were no significant differences between estimated haplotype frequencies or allele frequencies at 11 SNPs, genotyped in 95 U.K. blood donors and the CEPH families. An association between 1368c and sCJD was demonstrated with either the CEPH families or the healthy U.K. population used as a control group.
Geographical and temporal coincidence, together with molecular and biological strain-typing studies, have clearly linked the etiology of vCJD with the epidemic of BSE in the United Kingdom (Collinge et al. 1996b; Bruce et al. 1997; Hill et al. 1997). The extent of human genetic susceptibility to BSE prions in the U.K. population is not only important for modeling the potential size of the epidemic of vCJD in the future but also for identifying high-risk individuals who might be in a presymptomatic phase of illness. Such individuals could be targeted, should a treatment become available, and they represent a risk of potential transmission of human prions via blood transfusion or surgical instruments (Collinge 1999). vCJD has, so far, been diagnosed only in those who are homozygous for methionine at codon 129 of the PRNP ORF (Collinge et al. 1996a; Zeidler et al. 1997). The present study shows that this genetic association is not related to a unique susceptibility haplotype, since the complete spectrum of northern European methionine-haplotype diversity was observed in the vCJD group. However, all methionine haplotypes were identical over a 10-kb region, which includes the PRNP ORF. It remains possible, therefore, that the predilection for vCJD in methionine homozygotes may be related to variation in intronic regulatory regions, in addition to the codon 129 amino acid change. As the epidemic of vCJD progresses, we expect to see valine-homozygous and, later, valine-heterozygous cases (Collinge 1999). If so, then the greater intronic diversity of the valine haplotypes observed in the CEPH families, including methionine-haplotype–like bases in the PRNP intron, might allow this possibility to be tested.
The vCJD cases to date are presumably those in individuals with both susceptibility to prion disease and a short incubation time after exposure to BSE. It might be expected that these individuals have not only disease-susceptibility alleles but also short-incubation-period alleles, at all human prion-disease QTLs. We were unable to demonstrate an association between PRNP polymorphisms and vCJD; this may relate to a lack of statistical power due to the relatively small numbers of patients with vCJD available for analysis to date. Alternative explanations of the observed variation in incubation time in vCJD include a BSE dosage effect, other unidentified environmental factors, and QTLs on other chromosomes. There has been interest in the evident geographical clustering of vCJD, in a village in Leicestershire, United Kingdom (Cousens et al. 2001), that might reflect shared environmental and/or genetic risk factors. There is no evidence of a PRNP founder effect in this cluster, discounting the hypothesis that the clustering may relate to a high-risk PRNP vCJD-susceptibility allele.
Attention has been focused on the recently identified prion-protein–like gene PRND, although there is no evidence that coding variation has a role in human prion disease (Mead et al. 2000; Peoc'h et al. 2000). Uncommon alleles in the PRND ORF, such as P56L and T174T, tend to occur on single extended haplotypes, presumably reflecting their recent evolutionary origin. An extensive region of LD was identified across the intergenic region between PRND and PRNP. Neither the intergenic haplotype, the PRND M174T polymorphism, nor any of the extended haplotypes associated with uncommon PRND alleles were associated with any prion-disease variables. Investigation of PRND may prove to be more useful for understanding normal prion-protein function, rather than prion disease pathogenesis.
The identification of PRNP upstream polymorphisms that confer susceptibility to human prion disease is in keeping with the findings of a major QTL on mouse chromosome 2 (Lloyd et al. 2001). It should be noted, however, that the present study does not provide evidence that polymorphisms upstream of Prnp are responsible for the mouse chromosome 2 QTL. It is important that an association be tested in different human populations, particularly those with potentially greater intronic haplotype diversity. Further studies are underway to characterize a mechanism of susceptibility, by deletion analysis in transgenic mice.
Acknowledgments
Ray Young assisted with figure design. Colin Brown, of the National Blood Service, provided healthy U.K. population DNA. S.M. is a Wellcome Trust Training Fellow.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for clones U29185 and AL133396 [gi:6562003])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for kuru [MIM 245300], PRNP [MIM 176640], CJD [MIM 123400], and PRND [MIM 604263])
References
- Agresti A (1990) Categorical data analysis. John Wiley & Sons, New York [Google Scholar]
- Baker HE, Poulter M, Crow TJ, Frith CD, Lofthouse R, Ridley RM, Collinge J (1991) Amino acid polymorphism in human prion protein and age at death in inherited prion disease. Lancet 337:1286–1286 [DOI] [PubMed] [Google Scholar]
- Berr C, Richard F, Dufouil C, Amant C, Alperovitch A, Amouyel P (1998) Polymorphism of the prion protein is associated with cognitive impairment in the elderly: the EVA study. Neurology 51:734–737 [DOI] [PubMed] [Google Scholar]
- Brown P, Cathala F, Raubertas RF, Gajdusek DC, Castaigne P (1987) The epidemiology of Creutzfeldt-Jakob disease: conclusion of a 15-year investigation in France and review of the world literature. Neurology 37:895–904 [DOI] [PubMed] [Google Scholar]
- Brown P, Preece M, Brandel JP, Sato T, McShane L, Zerr I, Fletcher A, Will RG, Pocchiari M, Cashman NR, D'Aignaux JH, Cervenakova L, Fradkin J, Schonberger LB, Collins SJ (2000) Iatrogenic Creutzfeldt-Jakob disease at the millennium. Neurology 55:1075–1081 [DOI] [PubMed] [Google Scholar]
- Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock CJ (1997) Transmissions to mice indicate that ‘new variant' CJD is caused by the BSE agent. Nature 389:498–501 [DOI] [PubMed] [Google Scholar]
- Cervenakova L, Goldfarb LG, Garruto R, Lee HS, Gajdusek DC, Brown P (1998) Phenotype-genotype studies in kuru: implications for new variant Creutzfeldt-Jakob disease. Proc Natl Acad Sci USA 95:13239–13241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collinge J (1999) Variant Creutzfeldt-Jakob disease. Lancet 354:317–323 [DOI] [PubMed] [Google Scholar]
- Collinge J, Beck J, Campbell T, Estibeiro K, Will RG (1996a) Prion protein gene analysis in new variant cases of Creutzfeldt-Jakob disease. Lancet 348:56–56 [DOI] [PubMed] [Google Scholar]
- Collinge J, Brown J, Hardy J, Mullan M, Rossor MN, Baker H, Crow TJ, Lofthouse R, Poulter M, Ridley R, Owen F, Bennett C, Dunn G, Harding AE, Quinn N, Doshi B, Roberts, GW, Honavar M, Janota I, Lantos PL (1992) Inherited prion disease with 144 base pair gene insertion. II. Clinical and pathological features. Brain 115:687–710 [DOI] [PubMed] [Google Scholar]
- Collinge J, Palmer MS, and Dryden AJ (1991) Genetic predisposition to iatrogenic Creutzfeldt-Jakob disease. Lancet 337:1441–1442 [DOI] [PubMed] [Google Scholar]
- Collinge J, Sidle KCL, Meads J, Ironside J, Hill AF (1996b) Molecular analysis of prion strain variation and the aetiology of ‘new variant' CJD. Nature 383:685–690 [DOI] [PubMed] [Google Scholar]
- Cousens S, Smith PG, Ward H, Everington D, Knight RSG, Zeidler M, Stewart G, Smith-Bathgate EAB, Macleod MA, Mackenzie J, Will RG (2001) Geographical distribution of variant Creutzfeldt-Jakob disease in Great Britain, 1994–2000. Lancet 357:1002–1007 [DOI] [PubMed] [Google Scholar]
- Hill AF, Desbruslais M, Joiner S, Sidle KCL, Gowland I, Collinge J (1997) The same prion strain causes vCJD and BSE. Nature 389:448–450 [DOI] [PubMed] [Google Scholar]
- Lloyd SE, Onwuazor ON, Beck JA, Mallinson G, Farrall M, Targonski P, Collinge J, Fisher EMC (2001) Identification of multiple quantitative trait loci linked to prion disease incubation period in mice. Proc Natl Acad Sci USA 98:6279–6283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead S, Beck J, Dickinson A, Fisher EMC, Collinge J (2000) Examination of the human prion protein-like gene Doppel for genetic susceptibility to sporadic and variant Creutzfeldt-Jakob disease. Neurosci Lett 290:117–120 [DOI] [PubMed] [Google Scholar]
- Owen F, Poulter M, Collinge J, Crow TJ (1990) Codon 129 changes in the prion protein gene in Caucasians. Am J Hum Genet 46:1215–1216 [PMC free article] [PubMed] [Google Scholar]
- Palmer MS, Dryden AJ, Hughes JT, Collinge J (1991) Homozygous prion protein genotype predisposes to sporadic Creutzfeldt-Jakob disease. Nature 352:340–342 [DOI] [PubMed] [Google Scholar]
- Peoc'h K, Guerin C, Brandel JP, Launay JM, Laplanche JL (2000) First report of polymorphisms in the prion-like protein gene (PRND): implications for human prion diseases. Neurosci Lett 286:144–148 [DOI] [PubMed] [Google Scholar]
- Prusiner SB, Scott M, Foster D, Pan KM, Groth D, Mirenda C, Torchia M, Yang SL, Serban D, Carlson GA, Raeber AJ (1990) Transgenetic studies implicate interactions between homologous PrP isoforms in scrapie prion replication. Cell 63:673–686 [DOI] [PubMed] [Google Scholar]
- Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175 [DOI] [PubMed] [Google Scholar]
- Salvatore M, Genuardi M, Petraroli R, Masullo C, D'Alessandro M, Pocchiari M (1994) Polymorphisms of the prion protein gene in Italian patients with Creutzfeldt-Jakob disease. Hum Genet 94:375–379 [DOI] [PubMed] [Google Scholar]
- Tiret L, Amouyel P, Rakotovao R, Cambien F, Ducimetiere P (1991) Testing for association between disease and linked marker loci: a log-linear-model analysis. Am J Hum Genet 48:926–934 [PMC free article] [PubMed] [Google Scholar]
- Weir BS (1996) Genetic data analysis. Sinauer Associates, Sunderland, MA [Google Scholar]
- Weir BS, Wilson SR (1986) Log-linear models for linked loci. Biometrics 42:665–670 [PubMed] [Google Scholar]
- Windl O, Giese A, Schulz-Schaeffer W, Zerr I, Skworc K, Arendt S, Oberdieck C, Bodemer M, Poser S, Kretzschmar HA (1999) Molecular genetics of human prion diseases in Germany. Hum Genet 105:244–252 [DOI] [PubMed] [Google Scholar]
- Zeidler M, Stewart G, Cousens SN, Estebeiro K, Will RG (1997) Codon 129 genotype and new variant CJD. Lancet 350:668–668 [DOI] [PubMed] [Google Scholar]
- Zimmermann K, Turecek PL, Schwarz HP (1999) Genotyping of the prion protein gene at codon 129. Acta Neuropathol (Berl) 97:355–358 [DOI] [PubMed] [Google Scholar]