Abstract
Respiratory syncytial virus (RSV) is a major cause of respiratory illness in infants, immunocompromised patients, and the elderly. New antiviral agents would be important tools in the treatment of acute RSV disease. RSV encodes its own RNA-dependent RNA polymerase that is responsible for the synthesis of both genomic RNA and subgenomic mRNAs. The viral polymerase also cotranscriptionally caps and polyadenylates the RSV mRNAs at their 5′ and 3′ ends, respectively. We have previously reported the discovery of the first nonnucleoside transcriptase inhibitor of RSV polymerase through high-throughput screening. Here we report the design of inhibitors that have improved potency both in vitro and in antiviral assays and that also exhibit activity in a mouse model of RSV infection. We have isolated virus with reduced susceptibility to this class of inhibitors. The mutations conferring resistance mapped to a novel motif within the RSV L gene, which encodes the catalytic subunit of RSV polymerase. This motif is distinct from the catalytic region of the L protein and bears some similarity to the nucleotide binding domain within nucleoside diphosphate kinases. These findings lead to the hypothesis that this class of inhibitors may block synthesis of RSV mRNAs by inhibiting guanylylation of viral transcripts. We show that short transcripts produced in the presence of inhibitor in vitro do not contain a 5′ cap but, instead, are triphosphorylated, confirming this hypothesis. These inhibitors constitute useful tools for elucidating the molecular mechanism of RSV capping and represent valid leads for the development of novel anti-RSV therapeutics.
Human respiratory syncytial virus (RSV) is the leading cause of severe lower respiratory tract infection in infants and a major cause of morbidity and mortality in elderly and immunocompromised adults (8, 10). Only two products are approved for treatment or prevention of RSV infection: the nucleoside analog ribavirin, available for treatment of hospitalized patients, and the monoclonal antibody palivizumab (Synagis), for prophylaxis (14, 30). However, these agents have significant limitations, and new therapeutics with improved efficacy and safety are needed (22, 29).
Viruses in the Mononegavirales order, such as respiratory syncytial virus (RSV), have a nonsegmented negative sense RNA genome which serves as template for transcription of subgenomic mRNAs and the positive sense replication intermediate. The enzyme responsible for both of these processes, the RSV RNA-dependent RNA polymerase, is comprised of at least five viral components including the genomic RNA and the L, N, P, and M2-1 proteins (8). Together these components form a ribonucleoprotein (RNP) complex that is capable of de novo initiation and synthesis of positive and negative sense genomic RNA (“replicase” activity) as well as subgenomic mRNAs (“transcriptase” activity). The RNP complex isolated from RSV-infected cells has been shown to possess an RNA-dependent RNA polymerase (RdRp), mRNA polyadenylation, and mRNA capping activities in vitro (4, 5, 27). Multiple protein-protein and protein-RNA interactions mediate the assembly and activity of this complex (12, 13, 16, 20, 21, 26, 33, 38, 41). Thus, the RSV RNP is a complex that is rich in potential antiviral targets.
We along with others have previously reported the preparation of RNP that is capable of synthesis of RSV mRNAs in vitro (4, 17, 18, 27). We have used such a crude enzymatic activity to develop an assay that specifically measures the synthesis of RSV transcripts in vitro through capture of the mRNA poly(A) tails (27). A novel inhibitor of RSV polymerase was discovered through screening of this poly(A) capture assay (Fig. 1, compound A). Inhibition by this compound was noncompetitive with nucleotides in vitro and prevented the replication of RSV in cell culture assays (27). Here we show that compounds with greater potency against RSV transcriptase are also more potent inhibitors of viral replication in cell culture and have activity in a mouse model for RSV infection (Fig. 1, compounds B to E, and Table 1). Isolation and characterization of virus with reduced susceptibility to this class of inhibitor have led to the identification of a new motif in the RSV L protein. We show that inhibition of mRNA guanylylation by these small molecules can prevent the synthesis of full-length RSV transcripts. We believe that this is a unique mechanism of action for the inhibition of a viral transcriptase and that this mechanism is most likely responsible for their antiviral activity in cells and animals.
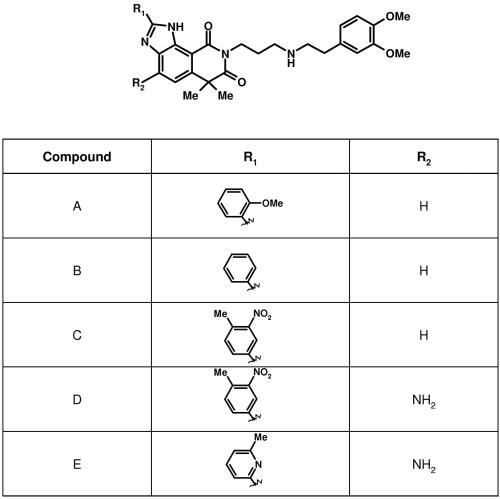
FIG. 1.
Molecular structure of RSV polymerase inhibitors. Compounds were synthesized using a method similar to that initially described for compound A (1). Details will be published elsewhere (B. Simoneau, unpublished data).
TABLE 1.
Biological profile of RSV polymerase inhibitors as compared to ribavirina
| Compound | Polymerase IC50 (μM) | Antiviral EC50 (μM) | Cytotoxicity CC50 (μM) | Selectivity (CC50/EC50) | Mouse model log titer reduction (N)b |
|---|---|---|---|---|---|
| B | 10.4 | 2.1 | 16 | 8 | ND |
| A | 4.5 | 1.3 | 7.7 | 6 | ND |
| C | 0.88 | 0.25 | 7.6 | 30 | 0.12 (6) |
| E | 0.66 | 0.10 | 10.7 | 107 | 0.38* (12) |
| D | 0.089 | 0.021 | 8.4 | 400 | 0.60* (12) |
| Ribavirin | ND | 6.9 | 200 | 29 | 0.04 (12) |
RSV polymerase assays were performed using the poly-A capture assay and antiviral activity was measured by ELISA. Cytotoxicity was determined using an MTT assay. Values shown are the average of at least three determinations.
The effect of inhibitors on RSV lung titers in mice was determined using the number of animals shown in parentheses. Vehicle-treated animals (n = 24) exhibited lung titers of 3.52 ± 0.04 log. The doses administered were as follows: compound C, 0.4 mg/kg/day; compound D, 0.4 mg/kg/day and compound E, 4.1 mg/kg/day. Ribavirin (Sigma) was administered at a dose of 10 mg/kg/day in vehicle. *, significantly different from vehicle; ND, not determined.
MATERIALS AND METHODS
Animals, cells, and viruses.
Cell culture reagents and media were obtained from Gibco BRL. BALB/c mice were purchased from Charles Rivers Laboratories. HEp-2 cells and the A2 and Long strains of RSV were from the American Type Culture Collection. HEp-2 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin sulfate, and 100 μg/ml kanamycin sulfate at 37°C in an atmosphere of 5% CO2. Virus stocks were grown in HEp-2 cells, and virus titers were determined by a standard plaque assay.
RSV polymerase assays.
Isolation of RSV RNP from virus-infected HEp-2 cells, transcription reactions, and RSV poly(A) capture assay were described previously (27). Compounds were tested in serial threefold dilutions. Nonlinear regression analysis using SAS software (SAS Institute, Cary, NC) was employed to determine the inhibitor concentration needed to give 50% reduction of enzyme activity (IC50).
Analysis of RNA products by gel electrophoresis.
Transcription reactions were performed as previously described (27) but contained 15 μCi of [α-33P]CTP (3,000 Ci/mmol) or ∼50 nM CTP. RNA products were incubated with 5 μg oligo(dT)15 and then with 7 U of RNase H to remove poly(A) tails. The reactions were stopped with 80 μg of tRNA and 20 mM EDTA, and the transcripts were extracted with Trizol LS (Invitrogen) as per the manufacturer's protocol. Transcripts were treated with formamide and run on a 6% polyacrylamide-7 M urea gel and detected using a Storm PhosphorImager (Molecular Dynamics).
Analysis of cap structure by HPLC.
RSV mRNA was made in in vitro transcription reactions as previously described (27), except reaction mixtures contained 5 μM S-adenosylmethionine (SAM), 400 μM ATP, 400 μM UTP, 10 μM CTP, 50 μM GTP, and 100 μCi of [α-32P]GTP (3,000 Ci/mmole; NEN) with or without 100 μM compound E in 4% dimethyl sulfoxide. Incubation proceeded for 6 h at 30°C. RNA was purified on ProbeQuant G-50 columns (Amersham). 32P-labeled NS2 RNA was prepared with the MaxiScript Kit (Ambion) using a 102-bp DNA template corresponding to the 5′ end of the RSV NS2 open reading frame in the presence of 100 μM GTP and 70 μCi of [α-32P]GTP (3,000 Ci/mmole) and then purified on G-50 columns and precipitated with 0.8 M LiCl, 100 μg/ml glycogen, and 50% isopropanol at −20°C; samples were washed twice with ice-cold 70% ethanol, dried, and resuspended in 4 mM potassium phosphate, pH 5.5. Samples were digested with 0.1 mg/ml nuclease P1 or 5 U of RNase T2. T2-digested samples were subsequently digested with 5 μg of calf intestinal phosphatase (CIP; Roche) in 20 mM Tris-Cl, pH 8.5, 100 mM NaCl. Samples were analyzed on a Partisil 5 SAX column using a gradient from 4 to 1,000 mM KPO4, pH 5.5, over 60 min at 1 ml/min (37). Products containing 32P were detected on a Berthold radioactivity monitor.
Antiviral assay.
Antiviral activity was assessed in an enzyme-linked immunosorbent assay (ELISA) as described previously (25, 27) using an anti-RSV F monoclonal antibody (Serotec MCA 490, clone B016). HEp-2 cells in DMEM containing 2% FBS were infected with RSV Long at a multiplicity of infection (MOI) of 0.1 and incubated for 48 h in the presence or absence of serial dilutions of inhibitors. Compound solutions in dimethyl sulfoxide (final concentration, 1%) were filtered through 0.22-μm-pore-size μStar LB filters (Costar), and concentrations were verified by high-pressure liquid chromatography (HPLC). The concentration of compound required to inhibit virus replication by 50% (EC50) was calculated as specified for the IC50 determinations. Cytotoxicity was determined using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (25) and results were expressed as 50% cytotoxic concentrations (CC50).
Isolation and sequencing of resistant viruses.
Two resistant viral isolates, Cr-1 and Cr-19, were selected by serial passage of RSV Long at an MOI of 1 in the presence of 2 to 4 μM compound C on confluent HEp-2 cells. Likewise, the resistant isolate Br-1 was selected at an MOI of 0.1 in the presence of 20 μM compound B. In each case, control viruses were grown in parallel to the same passage without compound. All resistant isolates were plaque purified in the presence of inhibitor. RNA was isolated from viral particles obtained from cells infected with the various resistant viruses using Trizol LS. cDNA was prepared by reverse-transcription PCR for DNA sequence determination, using Superscript II (Invitrogen) according to the manufacturer's protocol.
Cloning and production of RSV L with an I1381S substitution.
Standard molecular biology techniques (39) were employed for the construction of all plasmids [pMini-RSV-Luc3, pCR-N12, pCR-P29, pcDNA-M2-1, and wild-type pcDNA-L(Long)] for expression of RSV proteins and minigenomic RNA. To produce mutated pcDNA-L(I1381S), the L genes were amplified by PCR, cloned using the Echo cloning system (Invitrogen), and recombined with the mammalian expression vector pcDNA3.1E.
Minigenome reporter assay.
HEp-2 cells were plated at 250,000 cells per 12-well dish in DMEM-10% FBS and incubated overnight. Cells were infected with vaccinia vTF7.3 at an MOI of 5 PFU per cell in Opti-MEM containing 2% FBS, followed immediately by cotransfection with 200 ng of pLuc3, 200 ng of pN12, 100 ng of pP29, 50 ng of pM2, and 50 ng of either pcDNA-L(Long) wild-type or I1381S plasmids using Lipofectamine Plus (Invitrogen) according to the manufacturer's directions. At 16 h posttransfection the medium was removed, and the cells were incubated for 2 h with 2 μg per ml actinomycin D. The cells were washed with phosphate-buffered saline (PBS) and incubated in Opti-MEM containing 2% FBS with or without compound. All incubations were conducted at 37°C in 5% CO2 humidified air. Cells were lysed 40 h posttransfection, and luciferase activity was assayed using the Promega luciferase assay system according to manufacturer's protocol.
Murine RSV model.
Female BALB/c mice (5 weeks of age; 14 to 16 g) were infected intranasally with RSV A2 (5,000 syncytia forming units). RSV polymerase inhibitors or ribavirin was dissolved in vehicle [saline 0.9% (wt/vol) and 2.1% (wt/vol) Tween-80, pH 7] at the highest soluble concentrations. Treatments with RSV polymerase inhibitors or ribavirin were administered intranasally 3 and 6 h following inoculation with virus and then three times per day for 3 days. Lung viral titers at day 4 postinfection were determined by plaque assay (34). Statistical differences were determined by analysis of variance, followed by a Student-Newman-Keuls test for multiple comparisons, with P < 0.05 considered statistically significant.
Bioinformatics analyses.
Polymerase sequences from eight Mononegavirales viruses (Ebola, measles, mumps, Newcastle disease virus, rabies, RSV, Sendai, and vesicular stomatitis virus) were used with the algorithm MEME (Multiple EM for Motif elicitation; http://meme.sdsc.edu/meme/website/intro.html) (2) to find conserved motifs. Use of the full-length sequences of these eight polymerases led to the identification of the motifs already known to play a role in the polymerase (A′ to D) (32, 36). Further subdivision of the sequences into regions N-terminal and C-terminal to these known motifs led to the identification of other motifs (see Fig. 3 for motifs 1N to 9N, 1C to 10C, and methyltransferase; detailed alignments showing the positions of all motifs are available upon request). These motifs were named based on their level of conservation, with 1 being the most conserved and 10 being the least conserved. MEME creates a profile (based on the frequency of each amino acid at each position) of the various motifs it finds, and these profiles can then be used to search other databases for their occurrence in other proteins. This search was undertaken with each of the profiles from the various motifs using the MAST (motif alignment search tool; http://meme.sdsc.edu/meme/website/intro.html) algorithm. Only motif 5C led to interesting hits other than Mononegavirales polymerases, namely, multiple plant nucleoside diphosphate kinases (NDKs).
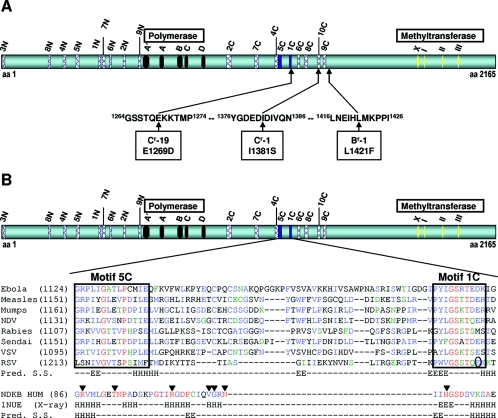
FIG. 3.
Mapping of RSV polymerase inhibitor resistance mutations to a central region of RSV L having similarity to NDKs. (A) Single amino acid changes in three resistant RSV isolates mapped to a central region of RSV L at amino acids 1269, 1381, and 1421. The highlighted motifs have known functions for polymerase activity (black boxes) (32) and methyltransferase activity (yellow diamonds) (11). (B) The L protein sequences from the indicated viruses were used as input for the program MEME. The position of the starting amino acid is in parentheses. Alignment of the 5C and 1C motifs (blue boxes) is shown with the similar region in human NDK (P22392). This NDK sequence (NDKB HUM) is colored according to the conservation of residues found in an alignment of the following NDKs for which an X-ray structure was obtained: P52174, P22887, P15266, and P22392. Residues in red are invariant, residues in blue are highly conserved, and those in green are weakly conserved. Arrows indicate residues that contact nucleotides in NDKs: arginine and threonine contact the β-phosphate and histidine binds the γ-phosphate (31). The circled residue in the RSV sequence is mutated in Cr-19. NDV, Newcastle disease virus; VSV, vesicular stomatitis virus; Pred. S.S., predicted secondary structure; 1NUE (X-ray), experimental secondary structure according to the 1NUE PDB record; H, helix; E, extended.
Secondary structure analysis was performed by submitting either the RSV or the human NDK sequence to the Jpred server (9) (http://www.compbio.dundee.ac.uk/∼www-jpred/).
RESULTS
Lead optimization and antiviral activity.
We have previously discovered a nonnucleoside inhibitor of the RSV RdRp activity via high-throughput screening of our corporate compound collection using a novel RSV polymerase poly(A) capture assay (Fig. 1, compound A structure) (27). Lead optimization studies have resulted in more potent analogs of compound A (Table 1 and Fig. 1). In particular, compound D was 51 times more potent than compound A in the RSV polymerase assay. In addition, this series of compounds exhibited significant antiviral activity with the same rank order of potency in a cell-based ELISA virus replication assay against both Long (Table 1) and A2 (data not shown) RSV strains. Significantly, compound D was over 300 times more active than ribavirin, with an EC50 of 21 nM and a selectivity index of 400. RSV polymerase inhibitors were also active in viral plaque reduction assays, with EC50s typically twofold higher than in ELISA (data not shown). The inhibitory effect of RSV polymerase inhibitors was lost when added 9 to 10 h postinfection (data not shown), consistent with the effect of compounds on postadsorption/fusion events and the onset of viral transcription (8). The potential for antiviral activity in vivo was assessed by testing selected RSV polymerase inhibitors in female BALB/c mice infected with RSV strain A2. A significant reduction in pulmonary titers of RSV was observed with compounds D and E after intranasal administration of these two compounds (Table 1). In contrast, topically applied ribavirin was ineffective at reducing viral lung titers. This lack of efficacy correlates with previous observations that a reduction of lung viral titers in mice by ribavirin is dependent on the amount of viral inoculum employed (7). That is, the higher the inoculum is, the greater the reduction of lung viral titers at a given dose of drug. In contrast, the efficacy of compounds D and E shown here indicates that inhibition of RSV polymerase can lead to reduction of RSV replication in vivo.
Mechanism of action of RSV polymerase inhibitors.
To confirm that the compounds inhibited the RSV polymerase within cells, selected compounds were tested in an RSV minigenome assay (23, 24, 45) which measures viral RNA-dependent RNA polymerase activity independently of viral replication. The RSV polymerase activity is reconstituted via cotransfection of T7-vaccinia virus-infected HEp-2 cells with T7-based plasmids that express RSV N, P, M2-1, and L genes, along with a minigenome plasmid resulting in luciferase activity in the absence of any other RSV proteins. Consistent with inhibition of RSV transcription, compounds C and D inhibited luciferase activity in this assay, with EC50 values of 450 and 33 nM, respectively.
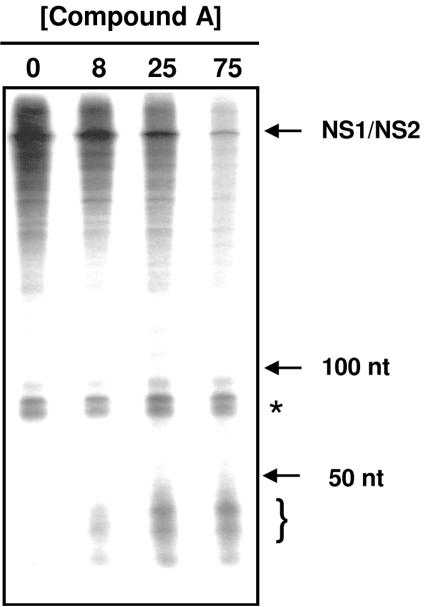
In our initial characterization of an inhibitor of this class (27), we have already shown that inhibition by compound A was noncompetitive versus nucleotides. Additionally, we showed that the synthesis of all major RSV mRNA species produced by viral RNP in vitro was equally inhibited by this compound (27). Interestingly, when RNA products produced by in vitro transcription reactions were analyzed on 6% polyacrylamide-7 M urea gels, shorter heterogeneous RNAs were observed only in the presence of compound, especially when low concentrations of [α-33P]CTP (Fig. 2) were used. Moreover, in a standard DE81 filter binding assay, compound A achieved less than 50% inhibition at the highest concentration tested (27), a result consistent with the production of short RNA species which would be retained on DE81 filters and mask the inhibition. Although there are alternative explanations for the appearance of short RNAs in the presence of inhibitor, such as increased degradation due to lack of poly(A) tails or increased synthesis of the leader RNA, it is plausible that compound A caused iterative abortive mRNA transcription leading to increased amounts of short RNA species.
FIG. 2.
Effect of compound A on RSV transcripts. Transcription reactions were performed with 50 nM [α-33P]CTP in the presence of 0 to 75 μM of compound A. Denatured transcripts were run on a 6% polyacrylamide-7 M urea gel. The bracket indicates the position of small RNA transcripts produced only in the presence of compound A. The asterisk indicates nonspecific products by contaminating nucleotidyl transferase. The relative migration of 50- and 100-nucleotide (nt) markers and that of RSV transcripts are indicated.
Isolation of inhibitor-resistant RSV and identification of the RNP subunit targeted by the inhibitor.
To identify the molecular target of the inhibitors, three independent drug-resistant RSV mutants were selected in the presence of the inhibitors B and C. Resistant viral isolates were obtained after 5 passages with compound C and after 11 passages with compound B. Plaque-purified resistant isolates Cr-1, Cr-19, and Br-1 profiled against selected inhibitors using the ELISA RSV replication assay were approximately eight-, four- and threefold resistant to compound C, respectively. All three isolates were cross-resistant to other RSV polymerase inhibitors within the same chemical class, but resistant viruses were still sensitive to ribavirin, indicating a distinct antiviral mechanism of action (data not shown). Furthermore, all mutant viruses replicated with near wild-type growth kinetics, and titers close to wild-type virus were obtained (data not shown). The RSV N, P, M2-1, and L genes from the resistant isolates were amplified by reverse-transcription-PCR and sequenced in order to determine which subunit of the RNP complex was targeted by the inhibitors. Each of the three resistant strains harbored a different single nucleotide substitution in the L gene, resulting in one of the following amino acid changes: I1381S, E1269D, or L1421F (Fig. 3A). No mutations in the other RNP subunit genes from the inhibitor-resistant isolates were detected when compared to virus passaged in the absence of inhibitor (data not shown). Next, the L genes from both wild-type RSV Long and resistant isolate Cr-1 were tested in the minigenome assay to verify that the mutation was responsible for reduced susceptibility. As shown in Table 2, the inhibitory activity of compounds C and D was reduced by eight- and ninefold, respectively, when they were tested in the presence of the mutated L gene. This loss of sensitivity was similar to that seen in the RSV replication assay or in in vitro transcription assays using RNP isolated from HEp-2 cells infected with Cr-1. These results confirm that the amino acid substitution I1381S in the L protein reduced RSV sensitivity to these compounds and suggest that this class of RSV polymerase inhibitors may bind to a specific region of the L protein, defined in part by a 152-amino-acid segment (Fig. 3A) that encodes the three resistance mutations.
TABLE 2.
Resistance profile of RSV polymerase inhibitors
| Compound | Polymerase IC50 (μM)a | Antiviral EC50 (μM)b | Minigenome EC50 (μM)c |
|---|---|---|---|
| C | 6.2 (7) | 2.0 (8) | 3.6 (8) |
| D | 1.1 (12) | 0.40 (19) | 0.30 (9) |
RSV polymerase poly-A capture assays were performed with RSV RNP isolated from cells infected with RSV Long Cr-1.
Antiviral activity against RSV Cr-1 was measured by ELISA.
Activity in the minigenome assay was measured using the mutated pcDNA-LI1381S plasmid. Values in parenthesis indicate the n-fold increase compared to wild-type (mutant/WT).
Identification of a new motif in the RSV L.
In an attempt to assign a function to the putative inhibitor-binding region, we used MEME (2) to identify regions conserved within L proteins from closely related viruses of the Mononegavirales order. Several motifs were identified, including those conserved among polymerases (32, 36) and methyltransferases (11) (M. Cartier, unpublished data). Using the MAST (3) algorithm to search the public protein database with each motif's MEME profile revealed that two motifs (Fig. 3B, motifs 5C and 1C) close to the region containing the three resistance mutations showed some similarity to a region in NDKs (31), important for nucleotide binding, suggesting that this region of the L proteins might also participate in nucleotide binding. Since RSV polymerase inhibitors increased the appearance of short RNAs (Fig. 2), we hypothesized that the inhibitors might prevent cotranscriptional capping of viral mRNAs required for full elongation of transcripts (5, 8). Although the precise mechanism of mRNA capping in viruses of the Mononegavirales order is not known (40), perturbations in cap formation can affect downstream transcriptional events for some of these viruses (19, 42). Cap methylation is not essential for RSV transcription since S-adenosylhomocysteine does not affect transcription although it does inhibit SAM-dependent methylation of RSV transcripts (5) (M. Liuzzi, unpublished results). It was therefore more likely that RSV polymerase inhibitors affected cotranscriptional guanylylation of mRNAs.
Effect of RSV polymerase inhibitors on RSV mRNA cap formation.
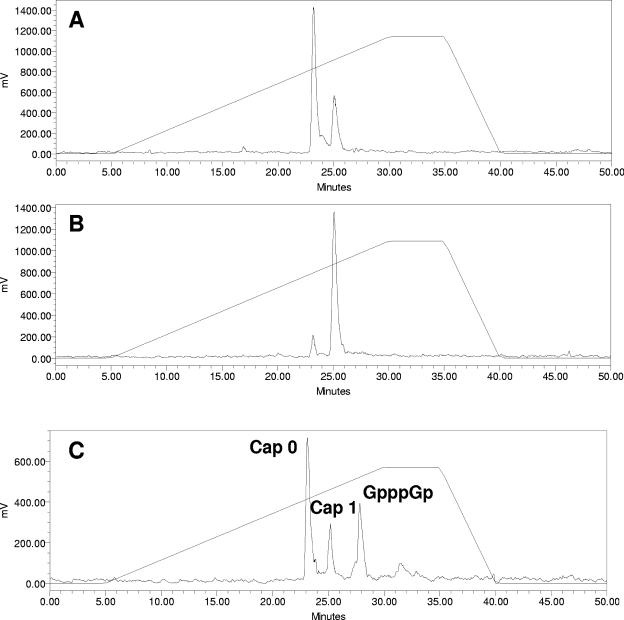
Prior to evaluating the effect of inhibitors on RSV mRNA cap formation, we assessed whether our RSV polymerase preparation was capable of catalyzing the formation of mRNA caps. For this, RSV transcripts were prepared in vitro in the presence of [3H]SAM and purified on Oligotex resin. Labeled transcripts were then treated with RNase T2, and the digested material was analyzed by strong anion exchange HPLC. As a reference, the expected products from the various enzymatic treatments used in these experiments on different RNAs are indicated in Table 3. Digestion of RSV transcripts with RNase T2 produced two peaks at 23 and 25 min (Fig. 4). The elution patterns of these two peaks were consistent with cap 0 (7mGpppGp) and cap 1 (7mGpppGmpGp) structures, as determined by elution profiles of standards produced by RNase T2 digestion of NS2 RNA produced with T7 RNA polymerase and capped with vaccinia virus capping enzyme in the presence of [α-32P]GTP and cold SAM (Fig. 4C). Interestingly, and contrary to a previous report by Barik (5), the RSV RNP produced both cap 0 and cap 1 structures, and the relative abundance of the two caps could be modulated by the concentration of SAM used in the experiment: in the presence of 0.2 μM SAM the transcripts were predominantly cap 0, but in the presence of 10 μM SAM they were mostly cap 1 (Fig. 4A and B). Combined, these results indicate that our RSV polymerase preparation is fully capable of guanylylating and methylating RSV mRNAs to form both cap 0 and cap 1 structures.
TABLE 3.
Expected products from enzymatic treatment of capped and uncapped RNAs
| Enzyme | RNA
|
||
|---|---|---|---|
| Cap0 | Cap1 | Uncapped RNA | |
| None | 7mGpppGpGpGp... | 7mGpppGmpGpGp... | pppGpGpGp... |
| Ribonuclease T2 | 7mGpppGp + 3′-GMP | 7mGpppGmpGp + 3′-GMP | pppGp + 3′-GMP |
| Ribonuclease T2 plus CIP | 7mGpppG + guanosine + Pi | 7mGpppGmpG + guanosine + Pi | Guanosine + Pi |
| Nuclease P1 | 7mGpppG + 5′-GMP | 7mGpppGm + 5′-GMP | pppG + 5′-GMP |
FIG. 4.
RSV polymerase preparations produce both cap 0 and cap 1. HPLC analyses of RNase T2-digested RNA on a Partisil 5 SAX column using a gradient of potassium phosphate (pH 5.5) from 4 to 800 mM. (A) Capped RSV RNA prepared by in vitro transcription in the presence of 0.2 μM [3H]SAM and purified on Oligotex resin followed by treatment with RNase T2 and analysis by HPLC. (B) Capped RSV RNA prepared by in vitro transcription in the presence of 10 μM [3H]SAM was treated as in panel A. (C) Capped NS2 RNA was produced by in vitro transcription using T7 RNA polymerase followed by incubation in the presence of vaccinia virus capping enzyme and [α-32P]GTP. This RNA was then purified on an RNeasy column, treated with RNase T2, and analyzed as in panel A. The peaks labeled cap 0 and cap 1 refer to 7mGpppGp and 7mGpppGmpGp, respectively.
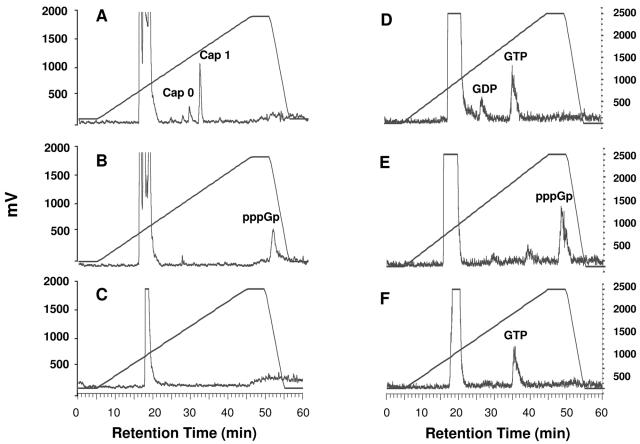
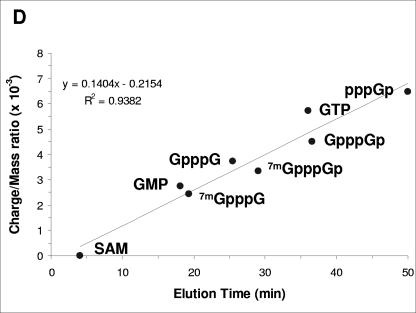
To investigate whether polymerase inhibitors affected RSV mRNA cap formation, we labeled the transcripts with [α-32P]GTP in the absence and presence of compound E. Since all RSV transcripts start with a guanine nucleotide (8), we reasoned that using labeled GTP instead of [3H]SAM would allow the determination of the status of the 5′ end of the transcripts whether they were modified by a cap or not. Labeled transcripts were isolated and treated with either RNase T2, alone or in combination with CIP or with nuclease P1 (NP1), and the digested material was then analyzed by strong anion exchange HPLC (Fig. 5) (Table 3 shows the activities of these enzymes in relation to the status of the 5′ end of transcripts). In the absence of inhibitor, digestion with RNase T2 produced a large peak at ∼20 min, corresponding to nucleoside monophosphates, and, as expected, two smaller peaks at 30 and 33 min corresponding to cap 0 (7mGpppGp) and cap 1 (7mGpppGmpGp) structures (Fig. 5A) in comparison to standards run under identical conditions (Fig. 6). In contrast to the cap 0 and cap 1 species observed in the absence of RSV polymerase inhibitor, in the presence of compound E, a different peak appeared at ∼50 min (Fig. 5B). Additional treatment with CIP eliminated this unknown peak, indicating the presence of unprotected phosphate groups (Fig. 5C). Lastly, NP1 digestion of RSV transcripts made in the presence of inhibitor yielded primarily GTP and some GDP (Fig. 5D). This result suggested that the short transcripts were phosphorylated and that the structure of the peak at ∼50 min might be guanosine tetraphosphate (pppGp). As a control, an uncapped RNA synthesized with T7 RNA polymerase that was digested with either RNase T2 or NP1 to produce pppGp and GTP, respectively, verified this hypothesis (Fig. 4E and F). Additionally, a plot of the charge-to-mass ratio of several standards compared to HPLC retention time under identical conditions supports the idea that the peak at 50 min corresponds to pppGp (Fig. 6D). Altogether, these results indicate that RSV polymerase inhibitors prevent mRNA guanylylation.
FIG. 5.
RSV polymerase inhibitors prevent RSV mRNA guanylylation. RSV transcripts were labeled with [α-32P]GTP in the presence or absence of inhibitor (compound E), digested with nuclease, and analyzed by HPLC as described in the legend of Fig. 4, except that the potassium phosphate gradient used in these experiments (and in those shown in Fig. 6) was 4 to 1,000 mM, instead of 4 to 800 mM. (A) RNase T2 digestion of RSV mRNA in absence of inhibitor. (B) RNase T2 digestion of RSV mRNA in presence of inhibitor. (C) RNase T2 and CIP treatment of RSV mRNA in presence of inhibitor. (D) NP1 digestion of RSV mRNA in presence of inhibitor. (E) T7 polymerase synthesized-NS2 RNA after RNase T2 digestion. (F) T7 polymerase-synthesized NS2 RNA after NP1 digestion. Cap 0 represents 7mGpppGp and cap 1 represents 7mGpppGmpGp. Note that although other inhibitors were more potent, compound E was used in this experiment due to better compound solubility. The y axis, in units of mV, is a measure of radioactivity by the Berthold monitor, which converts radioisotope decay to pulses of electricity (1,000 mV is approximately 8,500 cpm). The retention time of the peak in panel B varied from 48.3 to 50.9 min over seven experiments, whereas the retention time of the peak in panel E varied from 48 to 49 min.
FIG. 6.
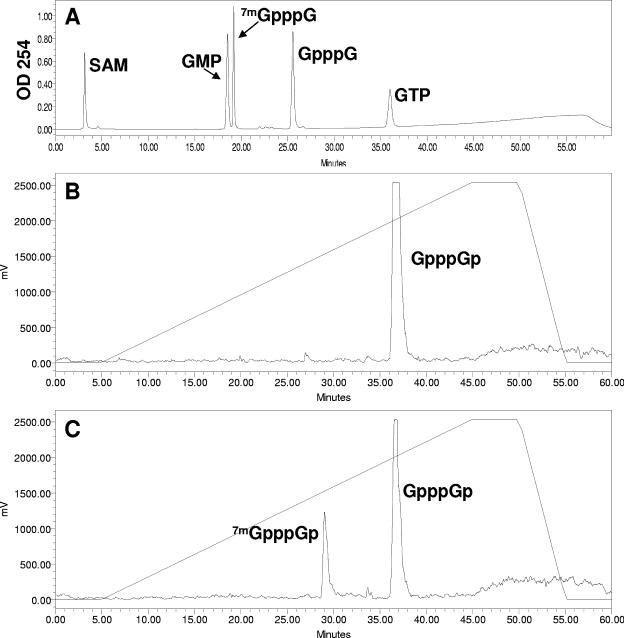
Analyses of labeled and unlabeled cap and nucleotide standards. (A) Unlabeled commercially available standards (as indicated) were chromatographed on a Partisil 5 SAX column as described in the legend of Fig. 5, except that the peaks were detected by UV absorbance at 254 nm. Each product was analyzed separately in order to determine the identity of the peaks when all standards were chromatographed together as presented in the panel. (B) Capped NS2 RNA was produced by in vitro transcription using T7 RNA polymerase followed by incubation in the presence of human capping enzyme (obtained from A. Shatkin, University of Medicine and Dentistry of New Jersey) and [α-32P]GTP. The RNA was purified on an RNeasy column treated with RNase T2 and analyzed as described in the legend of Fig. 5. (C) Capped NS2 RNA prepared with human capping enzyme, human methyltransferase (obtained from A. Shatkin), and [α-32P]GTP was purified on an RNeasy column and analyzed as in panel B. Note that methylation of the guanylated RNA in panel B is incomplete in panel C. Human methyltransferase only methylates the capping guanine resulting in cap 0 (35). (D) Plot of retention time from HPLC column (x axis) versus calculated charge/mass ratio (y axis) for standards from panels A, B, and C. Also included in the plot is the retention time of the product shown in Fig. 5D. Linear regression of the plot produces the formula shown with R2 = 0.9382, indicating a reasonable fit of the data points.
DISCUSSION
The RSV polymerase is comprised of at least five viral components which together form an RNP complex, with RdRp activity required for the synthesis of both viral genomic RNA (replicase activity) and subgenomic mRNAs (transcriptase activity). As opposed to the viral replication products, RSV mRNAs are cotranscriptionally capped (i.e., guanylated and methylated) at their 5′ ends and polyadenylated at their 3′ end by the RNP complex (4, 5, 8, 27, 44). These modifications are necessary for translation of the viral mRNAs by the host protein synthesis machinery. The RNP complex functions exclusively in the cytoplasm of the RSV-infected cells (8, 40). Since the host proteins responsible for the capping of mRNAs are located in the nucleus of the cell, capping of viral mRNAs by the RNP complex is essential for the synthesis of RSV proteins. These multiple activities of the RSV transcriptase make it an attractive target in screening for potential antiviral agents. A recent report of a distinct chemical class of RSV replication inhibitors with resistance mutations mapping to Y1631 of the L gene confirms this point (43).
The RSV poly(A) capture assay, described previously (27), had the potential to detect inhibition at any step of the RSV transcription cycle that is required for the synthesis of full-length polyadenylated mRNAs. Using this assay, we screened our compound collection, leading to the discovery of a unique inhibitor of RSV transcription with interesting properties: this nonnucleoside inhibitor was noncompetitive with nucleotides but was able to completely inhibit the synthesis of all major mRNA species in vitro. However, this compound only partially inhibited RNA synthesis when measured in a nonspecific capture format such as a DE81 filter binding assay. Consistent with this observation, short RNAs (<50 nucleotides) were detected when transcripts produced in the presence of inhibitor were analyzed on polyacrylamide gels. These observations suggest that inhibition of an early postinitiation step in the transcription cycle could result in the synthesis of high levels of short abortive transcripts. A similar interplay between 5′ capping and transcription elongation has been proposed for vesicular stomatitis virus (19, 42).
The synthesis of compounds with greater antiviral potency or greater selectivity index (CC50/EC50) (Table 1) permitted the isolation of inhibitor-resistant virus. Mapping of the mutations conferring resistance to inhibitors identified a region in the central portion of RSV L with similarity to NDKs. These are ubiquitous proteins that play a role in maintaining the balance of intracellular nucleotide pools by exchanging gamma-phosphate groups from NTP to NDP. The sequence similarity to NDKs and the partial overlap of the inhibitor resistance mutations to this region of L lead to the hypothesis that this NDK motif is involved in nucleotide binding, which may be important for mRNA guanylylation. Unfortunately, we were unable to show a direct interaction of guanine nucleotides with the L protein or this portion of the L protein. Although the Mononegavirales polymerases lack the histidine in motif 1C that is critical to catalytic function of authentic NDK proteins, the rest of the 1C motif, in particular, is very well conserved as Hy-Hy-G-S-Po-T/S (where Hy is hydrophobic and Po is polar). Therefore, the NDK similarity region of L protein could participate in the guanylylation reaction without directly catalyzing the guanine nucleotide exchange reaction. For example, this region could coordinate the triphosphorylated 5′ end of the nascent transcript and/or the guanine nucleotide for their interaction in the guanylylation reaction.
Based on the presence of inhibitor resistance mutations within and near the NDK-similarity region of L, we predict that inhibitors of RSV transcriptase may bind to the L protein in this region. Through this interaction, these inhibitors would be able to prevent the guanylylation of RSV mRNAs. Thus, the NDK similarity region may be directly or indirectly involved in mRNA guanylylation.
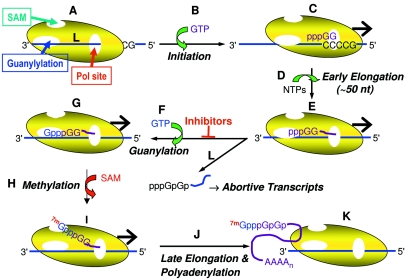
The data presented here and previously (27) support a model for RSV transcription where mRNA guanylylation is an essential step in establishing an efficient transcription elongation complex. The RSV transcriptase could be pictured as a complex with separate active sites for polymerization, guanylylation, and methylation (or SAM binding) (Fig. 7A). Transcription initiation occurs at a gene start sequence (Fig. 7B and C), and transcripts are extended in early elongation complexes (Fig. 7D) until they are about 45 to 50 nucleotides long, the size of the aborted RNAs observed in the presence of inhibitor A on the polyacrylamide gel presented in Fig. 2. Extension of the RNA to this length may position the 5′ end of the transcript within the guanylylation site (or NDK-similarity region) (Fig. 7E). Simultaneous binding of the 5′ end of the nascent transcript and guanine nucleotide would cause cotranscriptional formation the GpppGp cap at the 5′ end of the mRNA (Fig. 7F and G). The precise mechanism of guanine nucleotide transfer is currently unknown. Following guanylylation, the 5′ end of the mRNA would be transferred to the SAM binding site, allowing for cotranscriptional methylation of the guanylylated RNA (Fig. 7H and I). However, methylation of the mRNA is not essential for elongation, since S-adenosylhomocysteine does not inhibit transcription but only inhibits cotranscriptional methylation of the RSV mRNAs (5) (M. Liuzzi and R. S. McCollum, unpublished results). The capped and methylated mRNA is then elongated until the transcriptase encounters a gene end sequence, where polyadenylation of the 3′ end of the transcript occurs (15, 44). In contrast, the presence of inhibitor blocks guanylylation (Fig. 7L), preventing further elongation and resulting in release of short transcripts. Thus, the action of inhibitor causes the iterative synthesis of short uncapped transcripts that are detected in the DE81 filter binding assay and on polyacrylamide gels (Fig. 2) but not in the poly(A) capture assay since they are not polyadenylated.
FIG. 7.
Model for cotranscriptional guanylylation and action of RSV transcriptase inhibitors. See text for details.
Several predictions can be made based on this model. The length of the RNA as it reaches the guanylylation site and its sequence are crucial determinants for whether it is subject to guanylylation and distinguish it from the leader RNA. Thus, the leader RNA is not capped and is extended as a replication product without guanylylation of its 5′ end (8). Dissection of the leader region points to elements required for either replication or transcription (28). In addition, the “switch” from transcriptase to replicase activity could be the action of a natural inhibitor of guanylylation, with the result that RNAs containing the leader sequence are elongated past the first (and probably all) gene start sequence(s). The identity of the molecular switch remains to be determined but could be the M2-2 protein since the genetic ablation of M2-2 expression greatly enhances the synthesis of mRNA over antigenome (6).
In conclusion, the RSV polymerase inhibitors described here represent a novel class of nonnucleoside antiviral agents with a unique mechanism of action. Our results demonstrate that the guanylylation activity of the RSV RNP complex is an attractive target in the search for new anti-RSV agents. Most importantly, the optimized leads exhibited antiviral activity in vivo. These novel inhibitors may therefore provide a good starting point for the development of anti-RSV therapeutics for the treatment and prevention of RSV infections in humans.
Acknowledgments
We thank Jacques Archambault and George Kukolj for critical comments on the manuscript.
REFERENCES
- 1.Austel, A., E. Kutter, U. Heider, W. Eberlein, W. Korginger, C. Lillie, W. Diederen, and W. Haarmann. November 1979. Imidazoisoquinoline-diones and salts thereof. U.S. Patent 4,176,184.
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers, p. 28-36. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 3.Bailey, T. L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48-54. [DOI] [PubMed] [Google Scholar]
- 4.Barik, S. 1992. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s). J. Virol. 66:6813-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik, S. 1993. The structure of the 5′ terminal cap of the respiratory syncytial virus mRNA. J. Gen. Virol. 74:485-490. [DOI] [PubMed] [Google Scholar]
- 6.Bermingham, A., and P. L. Collins. 1999. The M2-2 protein of human respiratory syncytial virus is a regulatory factor involved in the balance between RNA replication and transcription. Proc. Natl. Acad. Sci. USA 96:11259-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolger, G., N. Lapeyre, N. Dansereau, L. Lagace, G. Berry, K. Klosowski, T. Mewhort, and M. Liuzzi. 2005. Primary infection of mice with high titer inoculum respiratory syncytial virus: characterization and response to antiviral therapy. Can. J. Physiol. Pharmacol. 83:198-213. [DOI] [PubMed] [Google Scholar]
- 8.Collins, P. L., R. M. Chanock, and B. R. Murphy. 2001. Respiratory syncytial virus, p. 1443-1485. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 9.Cuff, J. A., M. E. Clamp, A. S. Siddiqui, M. Finlay, and G. J. Barton. 1998. JPred: a consensus secondary structure prediction server. Bioinformatics 14:892-893. [DOI] [PubMed] [Google Scholar]
- 10.Falsey, A. R., and E. E. Walsh. 2000. Respiratory syncytial virus infection in adults. Clin. Microbiol. Rev. 13:371-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferron, F., S. Longhi, B. Henrissat, and B. Canard. 2002. Viral RNA-polymerases—a predicted 2′-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 27:222-224. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, J., B. Garcia-Barreno, A. Vivo, and J. A. Melero. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243-247. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Barreno, B., T. Delgado, and J. A. Melero. 1996. Identification of protein regions involved in the interaction of human respiratory syncytial virus phosphoprotein and nucleoprotein: significance for nucleocapsid assembly and formation of cytoplasmic inclusions. J. Virol. 70:801-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, C. B., J. T. McBride, E. E. Walsh, D. M. Bell, C. L. Gala, S. Hildreth, L. G. Ten Eyck, and W. J. Hall. 1983. Aerosolized ribavirin treatment of infants with respiratory syncytial viral infection. A randomized double-blind study. N. Engl. J. Med. 308:1443-1447. [DOI] [PubMed] [Google Scholar]
- 15.Hardy, R. W., S. B. Harmon, and G. W. Wertz. 1999. Diverse gene junctions of respiratory syncytial virus modulate the efficiency of transcription termination and respond differently to M2-mediated antitermination. J. Virol. 73:170-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hengst, U., and P. Kiefer. 2000. Domains of human respiratory syncytial virus P protein essential for homodimerization and for binding to N and NS1 protein. Virus Genes 20:221-225. [DOI] [PubMed] [Google Scholar]
- 17.Herman, R. C. 1989. Synthesis of respiratory syncytial virus RNA in cell-free extracts. J. Gen. Virol. 70:755-761. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y. T., R. R. Romito, B. P. De, and A. K. Banerjee. 1993. Characterization of the in vitro system for the synthesis of mRNA from human respiratory syncytial virus. Virology 193:862-867. [DOI] [PubMed] [Google Scholar]
- 19.Hunt, D. M., and K. L. Hutchinson. 1993. Amino acid changes in the L polymerase protein of vesicular stomatitis virus which confer aberrant polyadenylation and temperature-sensitive phenotypes. Virology 193:786-793. [DOI] [PubMed] [Google Scholar]
- 20.Khattar, S. K., A. S. Yunus, P. L. Collins, and S. K. Samal. 2001. Deletion and substitution analysis defines regions and residues within the phosphoprotein of bovine respiratory syncytial virus that affect transcription, RNA replication, and interaction with the nucleoprotein. Virology 285:253-269. [DOI] [PubMed] [Google Scholar]
- 21.Khattar, S. K., A. S. Yunus, and S. K. Samal. 2001. Mapping the domains on the phosphoprotein of bovine respiratory syncytial virus required for N-P and P-L interactions using a minigenome system. J. Gen. Virol. 82:775-779. [DOI] [PubMed] [Google Scholar]
- 22.Krilov, L. R. 2002. Palivizumab in the prevention of respiratory syncytial virus disease. Expert. Opin. Biol. Ther. 2:763-769. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, L., R. Fearns, and P. L. Collins. 1996. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J. Virol. 70:6143-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuo, L., H. Grosfeld, J. Cristina, M. G. Hill, and P. L. Collins. 1996. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J. Virol. 70:6892-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawetz, C., and M. Liuzzi. 1998. The antiviral activity of the ribonucleotide reductase inhibitor BILD 1351 SE in combination with acyclovir against HSV type-1 in cell culture. Antivir. Res. 39:35-46. [DOI] [PubMed] [Google Scholar]
- 26.Mason, S. W., E. Aberg, C. Lawetz, R. DeLong, P. Whitehead, and M. Liuzzi. 2003. Interaction between human respiratory syncytial virus (RSV) M2-1 and P proteins is required for reconstitution of M2-1-dependent RSV minigenome activity. J. Virol. 77:10670-10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason, S. W., C. Lawetz, Y. Gaudette, F. Do, E. Scouten, L. Lagace, B. Simoneau, and M. Liuzzi. 2004. Polyadenylation-dependent screening assay for respiratory syncytial virus RNA transcriptase activity and identification of an inhibitor. Nucleic Acids Res. 32:4758-4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGivern, D. R., P. L. Collins, and R. Fearns. 2005. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J. Virol. 79:2449-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meanwell, N. A., and M. Krystal. 2000. Respiratory syncytial virus: recent progress towards the discovery of effective prophylactic and therapeutic agents. Drug Discov. Today 5:241-252. [DOI] [PubMed] [Google Scholar]
- 30.Meissner, H. C., R. C. Welliver, S. A. Chartrand, B. J. Law, L. E. Weisman, H. L. Dorkin, and W. J. Rodriguez. 1999. Immunoprophylaxis with palivizumab, a humanized respiratory syncytial virus monoclonal antibody, for prevention of respiratory syncytial virus infection in high-risk infants: a consensus opinion. Pediatr. Infect. Dis. J. 18:223-231. [DOI] [PubMed] [Google Scholar]
- 31.Morera, S., M. L. Lacombe, Y. Xu, G. LeBras, and J. Janin. 1995. X-ray structure of human nucleoside diphosphate kinase B complexed with GDP at 2 Å resolution. Structure 3:1307-1314. [DOI] [PubMed] [Google Scholar]
- 32.Muller, R., O. Poch, M. Delarue, D. H. Bishop, and M. Bouloy. 1994. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J. Gen. Virol. 75:1345-1352. [DOI] [PubMed] [Google Scholar]
- 33.Murray, J., C. Loney, L. B. Murphy, S. Graham, and R. P. Yeo. 2001. Characterization of monoclonal antibodies raised against recombinant respiratory syncytial virus nucleocapsid (N) protein: identification of a region in the carboxy terminus of N involved in the interaction with P protein. Virology 289:252-261. [DOI] [PubMed] [Google Scholar]
- 34.Pastey, M. K., T. L. Gower, P. W. Spearman, J. E. Crowe, Jr., and B. S. Graham. 2000. A RhoA-derived peptide inhibits syncytium formation induced by respiratory syncytial virus and parainfluenza virus type 3. Nat. Med. 6:35-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pillutla, R. C., Z. Yue, E. Maldonado, and A. J. Shatkin. 1998. Recombinant human mRNA cap methyltransferase binds capping enzyme/RNA polymerase IIo complexes. J. Biol. Chem. 273:21443-21446. [DOI] [PubMed] [Google Scholar]
- 36.Poch, O., B. M. Blumberg, L. Bougueleret, and N. Tordo. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 71:1153-1162. [DOI] [PubMed] [Google Scholar]
- 37.Ramadevi, N., N. J. Burroughs, P. P. Mertens, I. M. Jones, and P. Roy. 1998. Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc. Natl. Acad. Sci. USA 95:13537-13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Samal, S. K., M. K. Pastey, T. H. McPhillips, and S. B. Mohanty. 1993. Bovine respiratory syncytial virus nucleocapsid protein expressed in insect cells specifically interacts with the phosphoprotein and the M2 protein. Virology 193:470-473. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Shuman, S. 1997. A proposed mechanism of mRNA synthesis and capping by vesicular stomatitis virus. Virology 227:1-6. [DOI] [PubMed] [Google Scholar]
- 41.Slack, M. S., and A. J. Easton. 1998. Characterization of the interaction of the human respiratory syncytial virus phosphoprotein and nucleocapsid protein using the two-hybrid system. Virus Res. 55:167-176. [DOI] [PubMed] [Google Scholar]
- 42.Stillman, E. A., and M. A. Whitt. 1999. Transcript initiation and 5′-end modifications are separable events during vesicular stomatitis virus transcription. J. Virol. 73:7199-7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sudo, K., Y. Miyazaki, N. Kojima, M. Kobayashi, H. Suzuki, M. Shintani, and Y. Shimizu. 2005. YM-53403, a unique anti-respiratory syncytial virus agent with a novel mechanism of action. Antivir. Res. 65:125-131. [DOI] [PubMed] [Google Scholar]
- 44.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61-119. [DOI] [PubMed] [Google Scholar]
- 45.Yu, Q., R. W. Hardy, and G. W. Wertz. 1995. Functional cDNA clones of the human respiratory syncytial (RS) virus N, P, and L proteins support replication of RS virus genomic RNA analogs and define minimal trans-acting requirements for RNA replication. J. Virol. 69:2412-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]