Abstract
The life cycle of human papillomaviruses (HPVs) is tightly linked to the differentiation program of the host's stratified epithelia that it infects. E1∧E4 is a viral protein that has been ascribed multiple biochemical properties of potential biological relevance to the viral life cycle. To identify the role(s) of the viral E1∧E4 protein in the HPV life cycle, we characterized the properties of HPV type 16 (HPV16) genomes harboring mutations in the E4 gene in NIKS cells, a spontaneously immortalized keratinocyte cell line that when grown in organotypic raft cultures supports the HPV life cycle. We learned that E1∧E4 contributes to the replication of the viral plasmid genome as a nuclear plasmid in basal cells, in which we also found E1∧E4 protein to be expressed at low levels. In the suprabasal compartment of organotypic raft cultures harboring E1∧E4 mutant HPV16 genomes there were alterations in the frequency of suprabasal cells supporting DNA synthesis, the levels of viral DNA amplification, and the degree to which the virus perturbs differentiation. Interestingly, the comparison of the phenotypes of various mutations in E4 indicated that the E1∧E4 protein-encoding requirements for these various processes differed. These data support the hypothesis that E1∧E4 is a multifunctional protein and that the different properties of E1∧E4 contribute to different processes in both the early and late stages of the virus life cycle.
Human papillomaviruses (HPVs) are small, double-stranded DNA viruses that infect the stratified epithelium lining the skin, anogenital tract, and oral cavity. Viral infection generally causes hyperproliferative lesions such as warts and condyloma. High-risk, mucosotropic (previously termed anogenital) HPVs, most commonly HPV type 16 (HPV16), are also associated with malignant tumors of the anogenital tract and oral cavity and are now accepted as the major causative agent of cervical cancer (64, 69). The life cycle of HPVs is tightly linked to the differentiation program of the host epithelium. HPVs infect basal keratinocytes, presumably at a site of wounding, and they establish their double-stranded, circular DNA genome as an extrachromosomal nuclear plasmid (replicon) at a low copy number. In these proliferating basal cells, early viral genes are selectively expressed, and viral DNA replication occurs along with cellular chromosomal DNA replication to maintain viral DNA copy numbers in both parent and daughter cells. This stage of the viral life cycle within basal cells is called the nonproductive or early stage because no new virus is made. As the infected cells migrate upward and undergo terminal differentiation, the productive or late stage of the viral life cycle begins. In the suprabasal compartment of the epithelium, the viral DNA is amplified, and this is followed by the expression of the late viral genes, including those encoding the structural proteins that form the capsid. Viral DNA is packaged into these capsids to form progeny virions that accumulate in the most superficial cell layers, where they can be released into the environment (27).
The 7,904-bp HPV16 genome contains eight functional translational open reading frames (ORFs) that encode five early gene products (E1, E2, E5, E6, and E7) and three late gene products (E4, L1, and L2). Two of the early gene products, E1 and E2, are involved in DNA replication and transcription of the viral genome (36). Three other early gene products, E5, E6, and E7, are oncoproteins that alter cell growth and/or differentiation. These oncoproteins have been demonstrated to contribute to HPV associated malignancies, individually and/or cooperatively (4, 37, 42, 55). Of the late gene products, L1 and L2 are the structural proteins that self assemble to form an icosahedral capsid (34). The third late gene product, E1∧E4, is encoded by spliced mRNAs that fuse the E1 and E4 ORFs. The expression of the HPV E1∧E4 protein is most abundant in the productive stage of the viral life cycle, specifically coinciding with the onset of viral DNA amplification (16, 40). Consistent with these observations, studies with cottontail rabbit papillomavirus (CRPV) indicate that E4 plays a crucial role in the productive stage of the viral life cycle (46).
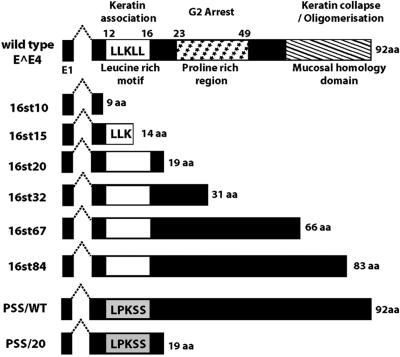
HV16 E1∧E4 protein can be divided into three functional regions (Fig. 1), the N-terminal leucine-rich motif (LLXLL), the central proline-rich charged region, and the C-terminal mucosal homology motif (49, 51). Previous studies have revealed multiple biological properties of HPV E1∧E4 proteins. HPV E1∧E4 proteins can associate with keratin intermediate filament (IF) networks; E1∧E4 of HPV16 and HPV18, but not of HPV1, can cause network collapse. The conserved N-terminal leucine-rich motif (LLXLL) of E1∧E4 is essential for the association with keratin IF networks, and the C terminus of HPV16 and -18 E1∧E4 is required for inducing collapse (15, 49, 50). A second property of HPV E1∧E4 proteins is their ability to induce G2/M cell cycle arrest in keratinocytes. This appears to depend on sequences in the central portion of E1∧E4 that can associate with cyclin B/Cdk1 complexes (9, 10, 35, 43). A third property of HPV16 E1∧E4 is its ability to induce apoptosis. The association of HPV16 E1∧E4 with mitochondria is proposed to be key for its induction of apoptosis (48). HPV16 E1∧E4 protein, but not that of HPV1 or HPV6, can also bind to a DEAD box RNA helicase (E4-DEAD box protein) through its C terminus (13).
FIG. 1.
Schematic representation of E1∧E4 mutant genes. At the top is shown a schematic diagram of HPV16 E1∧E4 indicating certain functional domains and related biological properties. Below are the predicted structures of the E1∧E4 proteins expressed from the different E4 mutant HPV16 genomes characterized in this study.
The majority of studies identifying biological properties of E1∧E4 have been carried out with tissue cultures under conditions in which E1∧E4 alone is highly overexpressed in proliferating, poorly differentiated keratinocytes. This contrasts to its natural expression pattern in the context of the viral life cycle, where it is highly expressed only in the terminally differentiated cellular compartment of a wart. To determine the role of the E1∧E4 protein in the HPV16 life cycle, we generated a collection of E4 mutant HPV16 genomes predicted to express a variety of altered E1∧E4 proteins and analyzed their abilities to carry out the viral life cycle using an organotypic, raft culture system. We discovered that mutations in the E4 ORF affected multiple aspects of HPV16 life cycle and surprisingly were not restricted to the late stage of the viral life cycle. We found that some mutations in the E4 ORF led to alterations in viral DNA replication in poorly differentiated NIKS cells that support the early stage of the viral life cycle. In the context of the late stage of the viral life cycle, mutations in the E4 ORF affected the efficiency with which the HPV16 genome induces DNA synthesis in the suprabasal compartment, affects viral DNA amplification, and alters terminal differentiation. These aspects of the viral life cycle are also modulated to various degrees and in distinct manners by other viral genes including E7 and E5. In that context, we propose a model to account for the individual roles of viral genes in the HPV16 life cycle.
MATERIALS AND METHODS
Construction of E4 mutant HPV16 genomes.
The plasmid pEFHPV-16W12E (20), which contains the full-length extrachromosomal HPV16 genome that was isolated from W12E clonal population 20863 (GenBank accession no. AF125673) or a fragment of the same HPV16 genome containing E4 ORF subcloned into a bacterial plasmid vector, was used as a template DNA for PCR-mediated, site-directed mutagenesis (QuickChange XL Site-Direct Mutagenesis kit; Stratagene, La Jolla, CA) of the E4 gene as instructed by the manufacturer. Plasmid DNAs were recovered from transformed bacteria, and the presence of the desired mutation(s) was confirmed by sequencing. Where necessary, the subgenomic fragment was cloned back into pEFHPV-16W12E. The entire HPV16 genome was completely resequenced to confirm that the derived plasmids contained only the desired mutation(s). The primers containing the following sequences and primers with their complementary sequences were used for introducing each mutation: 16st10, 5′-GTGTTTAGCAGCAACGAAGTAACCTCTCCTGAAATTATTAGGC-3′; 16st15, 5′-CGAAGTATCCTCTCCTGAAATTAGGCAGACATTGGCCAAA-3′; 16st20, 5′-CTGAAATAATAAGGCAGCACTTAGCCAACCACCCCGCCGCG3′; 16st32, 5′-CCAAAGCCGTAGCCTTAGGCACCG-3′; 16st67, 5′-ACTCAGTAGACAGTGCTCCAATCCTGACTGCAT-3′; 16st84, 5′-AAGGACGGATCCACTGTAATAGTA-3′; PSS/WT, 5′-AAGTATCCTCTCCCGAAATCATCAGGCAGCACTTGGCCAACC-3′; and PSS/20, 5′-AAGTATCCTCTCCCGAAATCATCAGGCAGCACTTAGCCAACC-3′.
Cell culture.
Epithelial cells, NIKS cells (1), and W12E clone 20863 (31) were maintained at subconfluence on mitomycin C-treated m1 3T3 feeder cells in F medium with all supplements as previously described (19, 21, 23). Human foreskin fibroblasts, used in the formation of dermal equivalents in organotypic raft cultures, were cultured in Ham's F12 medium (Invitrogen-Gibco, Carlsbad, CA) containing 10% fetal bovine serum (FBS), penicillin, and streptomycin (100 μg/ml) prior to raft culture. 293T cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% FBS, penicillin, and streptomycin.
Generation of NIKS cell populations harboring wild-type or E4 mutant HPV16 genomes.
Recircularized HPV 16 DNAs were prepared as previously described (26). Briefly, per transfection, 5 μg of plasmid containing HPV16 DNA was digested with BamHI to release the viral genome from the bacterial vector. BamHI-digested DNA was recircularized by intramolecular ligation, and recircularized DNA was recovered with a QIAGEN spin column (QIAprep Spin Miniprep kit; QIAGEN, Valencia CA). The recovered DNA was cotransfected with 1.2 μg of pEGFP-N1 (BD Bioscience Clontech, Palo Alto, CA) into 3 × 105 NIKS cells that were prepared in low Ca2+ F medium without feeder cells as previously described (19, 23). G418 selection was performed for 4 days as follows: 100 μg/ml for 2 days and then 50 μg/ml for next 2 days. Cells were then cultured in the absence of G418 to expand the G418-resistant transfectants, and the resulting colonies were pooled and expanded for Southern analyses. During G418 selection and subsequent expansion, feeder cells were freshly prepared and added to NIKS cells every other day. In each transfection experiment, 5 to 10 independent transfections were carried out per HPV16 genome type (the wild type [WT]or E4 mutant) to derive an equal number of independent transfected populations.
Southern blotting.
Total genomic DNAs were isolated as previously described (26). A total of 1 ng of pGEM3Z grown up in bacteria (Promega, Madison, WI) was added to each total genomic DNA preparation to ensure complete digestion with DpnI. Total genomic DNAs isolated from each population were digested with DpnI and either BamHI (20 μg) or HindIII (30 μg) for at least 8 h at 37°C and electrophoresed on a 0.8% agarose gel. Total genomic DNA isolated from untransfected NIKS cells was used as a negative control. Total genomic DNA isolated from W12E cells, containing extrachromosomal HPV16 DNA (31), was used as a positive control (data not shown). Parental pEFHPV-16W12E DNA was digested with BamHI, and appropriate serial dilutions were loaded onto the gel as copy number standards. Following electrophoresis, DNAs were transferred to a nylon membrane (Hybond-N; Amersham, Piscatway, NJ) and hybridized to a [α-32P]dCTP-radiolabeled DNA probe for full-length HPV16 prepared with a random primer kit (Rediprime II; Amersham). Radioactive bands were detected using a Storm imaging system (Amersham).
Organotypic raft cultures.
NIKS cells harboring the wild-type or E4 mutant HPV16 genomes were grown in raft cultures to induce the three-dimensional architecture of the stratified epithelium, as described previously, but with the following modifications (23, 26). NIKS cells transfected with wild-type or E4 mutant HPV16 genomes were transferred onto the dermal equivalents in transwell inserts (Costar no. 3450; Corning, Inc., distributed by Fisher Scientific Co., Pittsburgh, PA) that were submerged in Deep Well plates (BIOCOAT; Becton Dickinson Labware, Bedford, MA) with the keratinocyte plating medium (F medium containing 0.5% fetal bovine serum [FBS], the Ca2+ concentration increased to 1.88 mM, and all supplements except for epidermal growth factor [EGF]) for 4 days. At day 4 postplating, the transwell inserts were raised by placing three or four 2.5 cm- by 2.5-cm square cotton pads (catalog no. 740-E; Schleicher & Schuell BioScience, Inc., Keene, NH) underneath them, thereby exposing the epithelial cells to the air-liquid interface. The rafts were fed from below the transwell inserts with the cornification medium (F medium containing 5% FBS, the Ca2+ concentration increased to 1.88 mM, and all supplements except for EGF), which was replaced every other day until harvesting at day 11 postlifting. To estimate cellular DNA synthesis, the rafts were incubated in cornification medium containing 10 μM bromodeoxyyuridine (BrdU; Sigma, Saint Louis, MO) for 8 h prior to harvest. In the time course experiment, the rafts were incubated with BrdU for 8 h from 60 h to 52 h prior to harvest; then, the raft cultures were washed with PBS three times before being placed onto new cotton pads and allowed to continue to grow in the cornification medium without BrdU. Rafts were removed from transwell inserts and fixed as previously described (23). Fixed rafts were paraffin embedded, and 5-μm sections were prepared on plus-coated histology slides.
Western blotting.
Cell populations harboring the wild-type or E4 mutant HPV16 genomes were harvested by trypsin digestion, pelleted, resuspended in protein lysis buffer (50 mM Tris [pH 7.4], 150 ml NaCl, 1 mM EDTA, 1 mM phenylmethysulfonyl, 1 μg/ml leupeptin, 1 μg/ml pepstain, 1 mM NaF, 1% NP-40), incubated on ice for 30 min, and subjected to centrifugation at 14,000 rpm for 15 min at 4°C (Eppendorf model 5415C microcentrifuge) to separate the detergent-soluble (supernatant) from the insoluble (pellet) fraction. The pellet was homogenized in an equal volume (i.e., the same volume as the volume of protein lysis solution used originally to lyse the cells) of Laemmli sodium dodecyl sulfate (SDS) buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS) without reducing reagent by repeated pipetting before being recentrifuged at 14,000 rpm for 10 min at room temperature (Eppendorf model 5415C microcentrifuge). The resulting supernatant was used as the detergent-insoluble fraction. To isolate proteins from rafts, the stratified epithelium was peeled off from the dermal equivalent and lysed in the protein lysis buffer with a homogenizer. The cell lysate was incubated on ice for 30 min and separated into the detergent-soluble or -insoluble fraction as described above. The concentration of protein in the detergent-soluble fraction was measured with the DC protein assay kit according to the manufacturer's protocol (Bio-Rad, Hercules, CA). The volume of detergent-soluble fraction containing the amount of protein indicated for each experiment (see figure legends) was mixed with an appropriate volume of 6× SDS loading buffer (1× buffer was 62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% SDS, and 0.02% bromophenol blue) containing 100 mM dithiothreitol and boiled for 5 min. In the cases where the detergent-insoluble fraction was analyzed (i.e., for E1∧E4 analyses), the same volume of detergent-insoluble fraction as the volume of the detergent-soluble fraction was likewise mixed with 6× SDS loading buffer. Protein samples were electrophoresed through an SDS-15% polyacrylamide gel (ready-made gel; Bio-Rad) and transferred onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA). The membrane was blocked with 5% skimmed milk powder-PBS-T (phosphate-buffered saline [PBS]containing 0.1% Tween 20) for 1 h at room temperature and incubated overnight at 4°C with primary antibody diluted in blocking solution. The membrane was washed with PBS-T three times for 10 min each at room temperature and incubated for 1 h at room temperature with secondary antibody (peroxidase-conjugated anti-mouse immunoglobulin G; Jackson ImmunoResearch, West Grove, PA) diluted 1:10,000 in blocking solution. The membrane was again washed with PBS-T three times for 10 min each time at room temperature, and antibody conjugates were detected with an ECL kit (Amersham) according to the manufacturer's instructions. β-Actin was used as a loading control. Primary antibodies used included E7 antibodies, a mixture of two antibodies (1:200 dilution of ED7 [Santa Cruz, Santa Cruz, CA]; and 1:150 of 8C9 [Zymed, San Francisco, CA]), β-actin antibody (1:10,000 dilution of AC15 [Sigma, Saint Louis, MO]), human filaggrin antibody (1:200 dilution of BT-576 [Biomedical Techonologies, Inc., Stoughton, MA]), and E1∧E4 antibodies (1:20 dilution of supernatant from hybridoma culture of TGV402 monoclonal antibody) (14). For detection of E4 in monolayer culture, biotinylated anti-mouse antibody (1:2,000 dilution; DAKO Cytomation, Carpinteria, CA) was used as a secondary antibody. After incubation with the secondary antibody, the membrane was incubated with horseradish peroxidase-conjugated avidin (DAKO Cytomation) diluted 1:2,000 in PBS-T for 30 min at room temperature.
Transient DNA replication assay.
pHPV16E1E2TTL is a full-length clone of the HPV16 genome that contains translational termination linkers inserted in both the E1 and E2 ORFs. The E2S206T expression vector, pCMV4E2S206T, was generated by oligonucleotide-directed mutagenesis using the pCMV4 HPV16E2 (54) plasmid as a template. A total of 0.5 μg of pHPV16E1E2TTL was cotransfected with or without 1 μg of either the pCMV4 E2 wild type or pCMV4 E2S206T into 293T cells with Lipofectamine 2000 (Invitrogen, Carlsbad, CA) as instructed by the manufacturer. 293T cells were prepared in DMEM containing 10% FBS and no antibiotics in a 6-cm-diameter tissue culture dish at the day before transfection. A total of 0.5 μg of pEGFP-NI was also cotransfected to monitor that the efficiency of each transfection was roughly equal. Various amounts (0, 1, 3, or 5 μg) of E1-expressing vector pCMV4 HPV16E1 wild type (54) were cotransfected with these plasmids. An empty vector, pCMV4, was added to keep the total amount of DNA for each transfection equal as needed. At 72 h after transfection, low-molecular-weight Hirt DNAs were isolated from each sample as previously described (23). Hirt DNA was digested with DpnI and BamHI for at least 8 h at 37°C and electrophoresed on a 0.8% agarose gel. Southern blotting was performed as described above. A radiolabeled probe was prepared with a PCR-amplified long control region fragment as a template.
Luciferase assay.
A total of 0.5 μg of either pCMV4 E2 wild type or pCMV4 E2S206T was cotransfected with 0.25 μg of a luciferase reporter plasmid, either pBS1073 or pBS1013 (33), and 0.25 μg of pGFP-N1 into 293T cells using Lipofectamine 2000 as instructed by the manufacturer. 293T cells were prepared in DMEM containing 10% FBS with no antibiotics in 12-well tissue culture plates at the day before transfection. At 48 h after transfection, cells were lysed in a luciferase lysis buffer and subjected to a luciferase assay as instructed by the manufacturer (Promega, Madison, WI). Chemiluminescence per second for luciferase activity was measured with a microplate reader. Protein concentration was measured by the Protein Dc kit (Bio-Rad) to ensure that protein concentrations in each sample were roughly equal.
Immunostaining.
Cross sections of rafts on plus-coated slides were sequentially deparaffinized, rehydrated, and then treated by antigen-unmasking techniques before being immunostained as previously described (19, 23). E1∧E4 positive cells were detected with fluorescein isothiocyanate (FITC)-conjugated anti-E1∧E4 TGV402 as previously described (23). BrdU immnohistocheminal staining (see Fig. 4) was carried out primarily using anti-BrdU antibody (Ab-2; Calbiochem, San Diego, CA), and BrdU-positive cells were detected with the Vecstain ABC kit (Vector Laboratories, Burlingame, CA) as previously described (19, 23). For BrdU fluorescence, sections were incubated with biotinylated anti-BrdU antibody (Zymed, San Francisco, CA) diluted 1:100 in blocking solution (PBS containing 5% horse serum) overnight at 4°C and then washed with PBS twice for 3 min at room temperature. BrdU-positive cells were detected with a 1:100 dilution of Texas red-conjugated streptavidin (Vetor) in PBS after incubation for 30 min at room temperature. For keratin-10 (K10), sections were blocked with PBS containing 5% horse serum (Vector Laboratories) and then incubated with primary antibody, anti-K10 (CK8.60; Sigma, Saint Louis, MO) diluted 1:200 in blocking solution for 2 h at room temperature. These sections were washed with PBS twice for 3 min at room temperature and incubated with Texas red-conjugated anti-mouse secondary antibody (Vector Laboratories) diluted 1:100 in PBS for 30 min at room temperature. The stained sections were washed with PBS twice for 3 min and mounted with fluorescence mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole) (Vector). To costain with E4, primary antibodies, such as anti-BrdU or anti-K10, were mixed and incubated together with anti-E4 antibody overnight at 4°C. The percentage of BrdU-positive cells in rafts was evaluated by counting immunohistochemically stained BrdU-positive cells and total nuclei in the basal and suprabasal compartments of the rafts under a microscope. Ten visual fields at a magnification of ×400 were randomly chosen for quantification in each raft.
FIG. 4.
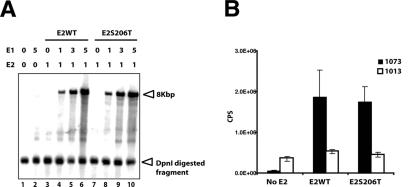
Transient viral DNA replication assay and transcriptional transactivation assay. (A) A total of 1 μg of either pCMV4E2 wild type or pCMV4E2S206T-expressing vector was cotransfected with 0.5 μg of pHPV16E1E2TTL and various amounts of pCMV4E1 into 293T cells. After 72 h, low-molecular-weight Hirt DNAs were isolated and subjected to DpnI and BamHI digestion. DpnI-resistant DNAs were detected by Southern blotting with a radiolabeled probe. The amounts of pCMV4E2 or pCMV4E1 in each transfection are indicated above the panel. DpnI-resistant pHPV16E1E2TTL DNA were detected only in the presence of both E1 and E2 (lanes 4 to 6 and 8 to 10). A DpnI-digested fragment of pHPV16E1E2TTL (indicated by an arrowhead) was detected equally in all samples, indicating a similar efficiency of transfection for each sample. (B) A total of 0.5 μg of either wild-type pCMV4E2 or pCMV4E2S206T was cotransfected with 0.25 μg of a luciferase reporter plasmid, pBS1073 (black columns) or pBS1013 (white columns), into 293T cells. After 48 h, cells were harvested and subjected to analysis to measure luciferase activity as instructed by the manufacturer (Promega, Madison, WI). The y axis indicates chemiluminescence per second (CPS). Three independent transfections were performed.
FISH.
Fluorescence in situ hybridization (FISH) was performed to detect amplified HPV16 DNA in rafts as previously described (26). pEFHPV-16W12E was used as a template to generate a digoxigenin (DIG)-labeled probe for HPV16 DNA. To detect amplified HPV16 DNA and E1∧E4 simultaneously, sections were blocked with PBS containing 5% horse serum for 30 min at room temperature and incubated overnight with FITC-conjugated anti-E1∧E4 antibody diluted 1:500 in blocking buffer at 4°C after being hybridized and then washed. Rhodamine-conjugated anti-DIG antibody (Roche Applied Science, Indianapolis, IN) diluted 1:200 in PBS containing 5% horse serum was used to detect amplified HPV16 DNA. Sections were mounted with fluorescence mounting medium containing DAPI. To quantify the frequency of FISH-positive cells in rafts, FISH-positive cells were counted under a fluorescence microscope in the context of an entire raft and were divided by the total number of nuclei. The total number of nuclei per raft was estimated by multiplying the average number of total nuclei per field by the number of fields per raft. The average number of nuclei per field was obtained by counting the number of DAPI-positive nuclei in three representative fields of ×400 magnification in each raft.
RESULTS
Generation of E4 mutant HPV16 genomes.
To identify a role for the E1∧E4 protein in the HPV16 viral life cycle, we generated a panel of E4 mutant HPV16 genomes (Fig. 1) by introducing nucleotide substitutions and analyzed the behavior of these E4 mutant HPV16 genomes in the viral life cycle. The E4 mutant HPV16 genomes (16st10, 16st15, 16st20, 16st32, 16st67, and 16st84) were designed to express C-terminally truncated forms of the E1∧E4 protein by the introduction of translation termination codons at various position within the E4 ORF as indicated in Fig. 1. 16st10, 16st15, 16st20, and 16st84 have single-nucleotide substitutions at nucleotide (nt) 3370, T to A (codon 10 of E1∧E4, TAT to TAA); nt 3384, T to A (codon 15, TTA to TAA); nt 3399, G to A (codon 20, TGG to TAG); and nt 3591, T to A (codon 84, TTA to TAA), respectively. 16st32 has two nucleotide substitutions, one at nt 3435, C to A (codon 32, TCG to TAG), and the other at nt 3441, G to A (codon 34, TGG to TAG), both of which introduce translational termination codons. Likewise, in the 16st67 genome, two translational termination codons were introduced by nucleotide substitutions at nt 3540, G to A (codon 67, TGG to TAG), and nt 3558, C to G (codon 73, TCA to TGA). All mutations in these E4 mutant HPV16 genomes were silent in the overlapping E2 ORF, with exception of the 16st10 genome. The mutation in the 16st10 genome resulted in an amino acid substitution at codon 206 (serine to threonine) in the overlapping E2 ORF. Protein sequence alignments revealed that serine 206 was not conserved among papillomavirus E2 proteins; for instance, bovine papillomavirus type 1 (BPV-1) E2 contains a valine at the codon equivalent to 206 of the HPV16 E2 protein. For this reason, the particular amino acid substitution in the E2 protein (S206T) encoded by 16st10 was predicted not to affect E2 protein function. This proved to be the case (see below). In addition to these E4 truncation mutants, we also made amino acid substitutions in the E4 ORF of both the wild-type HPV16 genome, designated PSS/WT, and the 16st20 genome, designated PSS/20, in which the leucine-rich motif of the E1∧E4 protein (Fig. 1) was changed from LLKLL to LPKSS by three nucleotide substitutions at nt 3378, T to C (codon 13, CTG to CCG); nt 3384, T to A (codon 15,TTA to TCA); and nt 3387, T to C (codon 16,TTA to TCA). These three mutations were silent in the overlapping E2 ORF.
Gross truncation of the E1∧E4 protein or disruption of the leucine-rich motif leads to a defect in viral plasmid DNA replication in proliferating, poorly differentiated human keratinocytes that express E1∧E4 at low levels.
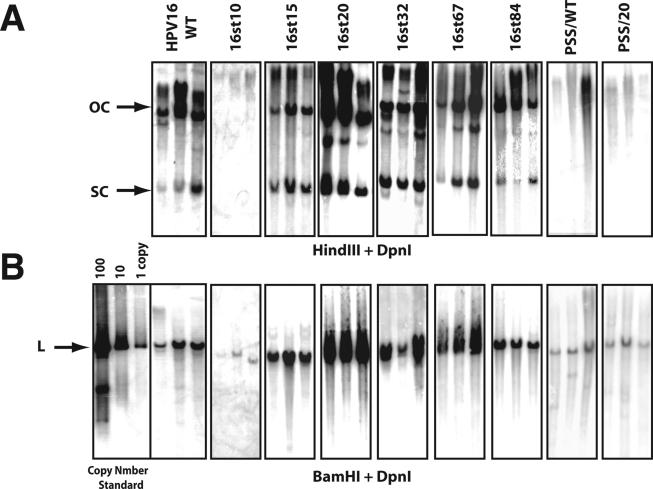
To assess the role of E1∧E4 in the viral life cycle, the E4 wild-type and mutant HPV16 genomes were released from the bacterial plasmid vector, recircularized, and cotransfected with a plasmid conferring neomycin resistance into NIKS cells, an immortalized keratinocyte cell line that supports the papillomavirus life cycle (20). Following G418 selection, cell populations were established and expanded under growth conditions that maintain poorly differentiated or basal-cell-like properties. Under these culture conditions, we can assess the properties of HPV16 during the early stage of the viral life cycle. Total genomic DNAs were isolated from these cell populations and digested with DpnI, to remove any bacterially synthesized input DNA and either HindIII (for which no restriction sites is present in the HPV16 genome) or BamHI (for which one site is present in HPV16). The presence and the extrachromosomal status of the HPV16 viral DNA were analyzed by Southern blotting. To measure the viral DNA copy number of wild-type or E4 mutant HPV16 genomes present in NIKS populations, appropriate amounts of recombinant HPV16 plasmid DNA equal to 1, 10, and 100 copies per cell equivalents were electrophoresed along with total genomic DNAs; hybridization signals to these copy number standards were quantified by phosphorimager analysis. Figure 2 shows representative results of the Southern blots carried out with equal amounts of total genomic DNA isolated from multiple cell populations transfected with the wild-type or E4 mutant HPV16 genomes. Extrachromosomal viral DNA genomes (referred to herein as replicons) were present in all NIKS populations transfected with the wild-type genome, as shown by the presence of two bands that corresponded to open circular (OC) and supercoiled (SC) DNA (Fig. 2A). In addition to the OC or SC form of viral DNAs, viral DNAs were occasionally detected in higher-molecular-weight factions in some of NIKS cell populations. The viral DNA in higher-molecular-weight fractions could be integrated or multimeric viral DNA. The copy number of the wild-type HPV16 genome, quantified by measuring the intensity of hybridization of the linearized genome compared to that of copy number standards (generated by digestion with BamHI) (Fig. 2B), ranged from 5 to 12 copies per cell (Table 1). We failed to detect OC or SC forms of extrachromosomal viral DNA in any NIKS populations transfected with the 16st10 genome (Fig. 2A), despite sensitivity of the Southern blots down to 0.1 copy equivalents of linear HPV genome per cell (data not shown). Instead, we only found hybridization in the high-molecular-weight fraction of the HindIII-restricted samples, suggestive of viral DNA integration. The presence of integrated concatameric 16st10 HPV16 genomes was supported by the fact that we could detect linear 16st10 HPV16 DNA, albeit at a low copy number (Fig. 2B and Table 1), when the total genomic DNA was digested with BamHI, which cleaves HPV16 once. Extrachromosomal viral DNA replicons were detected in each NIKS population transfected with the E4 mutant HPV16 genomes 16st15, 16st20, 16st32, 16st67, and 16st84. The viral DNA copy numbers of 16st15, 16st67, and 16st84 were in a range that was statistically similar to that of the wild-type genome, whereas the viral DNA copy numbers of 16st20 and 16st32 were significantly increased (Table 1). The viral DNA copy numbers of 16st20 or 16st32 genomes were approximately 100 copies per cell or 50 copies per cell, respectively, compared to approximately 10 copies per cell for the wild-type HPV16 DNA. We also performed transfections of either the wild-type or E4 mutant HPV16 genomes in early-passage human foreskin keratinocytes. Southern analyses of these populations demonstrated that the replicative properties of the E4 mutant HPV16 genomes were consistent with those observed with NIKS cell populations, indicating that the properties of E4 mutant HPV16 genomes for viral DNA replication were not influenced by immortalization (data not shown). Because the human foreskin keratinocyte populations did not efficiently immortalize in our hands, we performed our subsequent analyses on the NIKS cell populations.
FIG. 2.
Southern analysis of total genomic DNA isolated from NIKS cell populations transfected with wild-type or E4 mutant HPV16 genomes. Shown are Storm phosphorimages of hybridization patterns from Southern analyses carried out to assess the presence of extrachromosomal HPV16 genomes (A) and the copy number of the HPV16 genomes (B) present in NIKS populations transfected with the wild-type or E4 mutant HPV16 genomes. Total genomic DNAs were extracted from NIKS cell populations at passage 1, following G418 selection. Bacterially synthesized DNA was added in each extraction as a spike DNA to ensure complete digestion of DpnI (see Materials and Methods); Southern blots for DpnI-undigested DNA are not shown. Equal amounts of total genomic DNA from each population were digested with DpnI and HindIII (no site in HPV16 genome) (A) or BamHI (one site in HPV16 genome) (B) and subjected to Southern blot analysis using a radiolabeled DNA probe for HPV16 DNA. Arrows indicate OC, SC, and linear (L) HPV16 genomes. BamHI-digested parental pEFHPV16-W12E was used as a copy number standard.
TABLE 1.
Viral DNA copy number of wild type or E4 mutant HPV16 genomes
| Genome | Presence of extrachromosomal viral DNAa | Plasmid copy no.b,d | Relative plasmid copy no.c,d |
|---|---|---|---|
| WT | Y | 8.4 ± 3.5 | 1 |
| 16st10 | N | 1.0 ± 0.2* | 0.1* |
| 16st15 | Y | 11 ± 2.5 | 1.3 |
| 16st20 | Y | 80 ± 21* | 7.4* |
| 16st32 | Y | 31 ± 14* | 4.9* |
| 16st67 | Y | 4.4 ± 2.3 | 0.5 |
| 16st84 | Y | 9.6 ± 1.5 | 1.2 |
| PSS/WT | N | 1.3 ± 0.3* | 0.2* |
| PSS/20 | N | 1.6 ± 0.3* | 0.2* |
Y, extrachromosomal viral DNA was detected; N, extrachromosomal DNA was not detected (see Fig. 2).
Viral DNA copy number in each NIKS cell population was estimated based on the intensity of copy number standard. Each value is the average copy number of the wild-type or E4 mutant HPV16 genome, based upon analyses of multiple (n > 10 per genotype) independent cell populations generated from a representative set of transfections.
The copy number for wild-type HPV16 populations was set at 1.
*, copy number values for E4 mutant HPV16 genomes that were statistically significant from that of the wild-type HPV16 genome, based on the two-sided Wilcoxon rank sum test.
Based upon the results described above (see also Fig. 1 and Table 1), mutations engineered into the E4 ORF had varied consequences for viral DNA replication. We considered two alternative hypotheses to account for these findings. The first hypothesis is that E1∧E4 itself plays a role in modulating the replication properties of the viral DNA genome. A necessary corollary to this hypothesis is that an intact leucine-rich motif is necessary and sufficient for plasmid replication, because only 16st10 lacks this motif entirely; 16st15 retains most of it, and 16st20, which displays a high-copy-number phenotype, retains all of this motif (Fig. 1). To test this hypothesis, we generated and analyzed amino acid substitution mutants, PSS/WT and PSS/20. These mutants have amino acid substitutions within the leucine-rich motif, replacing three leucine residues in the context of either the otherwise-wild-type HPV16 genome or the 16st20 genome, respectively (Fig. 1). The PSS mutations do not alter the amino acid sequence of the overlapping E2 ORF. If the leucine-rich motif is indeed required to confer the replication competent phenotype, then these PSS mutants should be defective for plasmid replication. This was indeed the case. Similar to 16st10, the PSS/WT and PSS/20 mutants were grossly deficient in their ability to replicate extrachromosomally, as judged by the absence of detectable OC or SC DNA, the presence of non-unit-length genomes after digestion with BamHI with unit-length genomes (Fig. 2), and their very low copy numbers (Table 1). These results were consistent over 15 cell populations established from multiple independent sets of transfections. The inability of the PSS mutations to replicate as nuclear plasmids is consistent with the hypothesis that an intact leucine-rich motif in E1∧E4 is required for plasmid DNA replication.
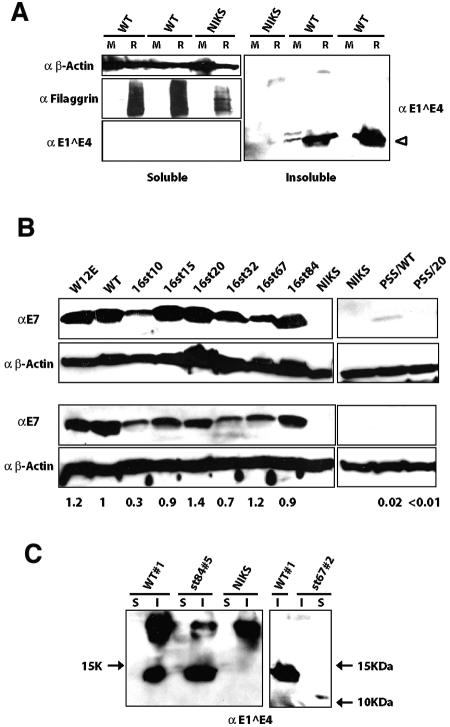
If E1∧E4 contributes to viral plasmid replication in poorly differentiated, proliferating keratinocytes (as our data suggest), then it must be expressed in these cells. When the E1∧E4 protein is overexpressed in cultured keratinocytes, it is predominantly found in the detergent-insoluble fraction (51). To determine if the E1∧E4 protein is expressed in poorly differentiated keratinocytes, Western blot analyses were performed on detergent-soluble and detergent-insoluble protein fractions harvested from NIKS cells harboring wild-type HPV16 genomes extrachromosomally that were maintained either in monolayer culture or in organotypic (raft) culture (Fig. 3A). It is known that keratinocytes in monolayer culture, even on feeder cells, can initiate differentiation when they become confluent. To prevent this from occurring, we carefully maintained NIKS cells in a subconfluent state. E1∧E4 protein was reproducibly detected in the detergent-insoluble fraction of monolayer at low levels and in the same fraction of raft cultures at much higher levels. We also performed Western blot analyses for filaggrin, a differentiation marker of the stratified epithelium, to assess the differentiation state of the cells. Filaggrin was chosen because the filaggrin-positive cell layer overlaps the cell layer where HPV16 E1∧E4 protein is found following immunodetection in raft culture (16, 40). As expected, filaggrin was detected only in cell lysates harvested from raft cultures and was not apparent in monolayer cultures, verifying that the cells had not undergone terminal differentiation (Fig. 3A, left). Thus, the E1∧E4 protein is expressed at low but detectable levels in poorly differentiated keratinocytes harboring the wild-type HPV16 genome and, as expected, is upregulated in its expression in a differentiation-dependent manner.
FIG. 3.
Western blot analysis for E1∧E4 and E7 expression in NIKS cells harboring the wild-type or E4 mutant HPV16 genomes. (A) E1∧E4 expression in monolayer (M) and raft (R) cultures. A total of 100 μg of detergent-soluble protein and an equivalent volume of the detergent-insoluble fraction of cellular proteins isolated from parental NIKS cells (NIKS) or NIKS cell populations harboring wild-type HPV16 genomes (WT) were subjected to E4-specific immunoblot analysis as described in Materials and Methods. Filaggrin (38 kDa), which was used as a marker for differentiation, was detected mainly in the detergent-soluble fractions isolated from raft cultures but not from monolayer cultures, verifying that the cells maintained in monolayers had not undergone terminally differentiation. β-Actin (50 kDa) was detected in the detergent-soluble fraction as a loading control. (B) E7 expression in monolayer NIKS cell populations. A total of 80 μg of protein from the detergent-soluble fraction extracted from NIKS cell populations harboring the wild-type or E4 mutant HPV16 genomes wereas subjected to E7-specific immunoblot analysis as described in Materials and Methods. β-Actin was detected as a loading control. W12E cells were used as a positive control cell line. Western blotting for E7 was repeated at least three times (shown are two representative populations), and levels of E7 in multiple, independently derived cell populations harboring wild-type or E4 mutant HPV16 genomes were quantified with NIH Image software. The average levels of E7 in cells harboring E4 mutant genomes relative to that in cells harboring the wild type are shown below the panel. (C) E1∧E4 expression in raft cultures harboring E4 mutant HPV16 genomes. E1∧E4 was detected in the detergent-insoluble fraction (I) of rafts harboring the wild type as well as 16st84, whereas it was detected in the detergent-soluble fraction (S) of rafts harboring 16st67 (approximately 10 kDa). Note that the E1∧E4 antibody used (TVG402) recognizes an epitope that maps within the central portion of E1∧E4. Arrows indicate molecular mass markers.
Absence of evidence to indicate pleiotropic effects of E4 mutations on functions other than E4.
An alternative, second hypothesis that we felt necessary to address is that E4 mutations may have pleiotropic effects on the functionality of the HPV16 genome that are not restricted to the disruption of the E4 ORF. This was certainly a possibility for 16st10, which has a serine-to-threonine amino acid substitution at position 206 in E2. To test whether this mutant E2 protein (E2S206T) encoded in the 16st10 genome is altered for its function, we performed transient DNA replication and transcriptional transactivation assays (Fig. 4A and B). The nucleotide substitution (codon 206 of E2, TCC to ACC) was introduced into a wild-type E2 expressing vector, the pCMV4 HPV16E2 wild type (54), by site-directed mutagenesis. To analyze E2's ability to support viral DNA replication, a transient DNA replication assay was performed. Either the pCMV4E2 wild type or pCMV4E2S206T was cotransfected into 293T cells with the pCMV4E1 E1 expression vector and the HPV16 genomic replicon pHPV16E1E2TTL, which is disrupted for expression of E1 and E2, by the presence of translational termination linkers in E1, as well as in E2, ORFs. After 72 h, low-molecular-weight Hirt DNAs were isolated from each transfection and then subjected to BamHI and DpnI digestion. Southern blotting was performed to detect DpnI-resistant pHPV16E1E2TTL DNA. Since viral DNA replication is dependent on the ratio between E1 and E2, various amounts of the E1 expression vector, pCMV4HPV16E1 (54), were cotransfected with either the pCMV4E2 wild-type or pCMV4E2S206T E2 expression vector. As shown in Fig. 4A, Dpn I-resistant viral DNA was detected only in DNAs isolated from transfections in the presence of both E1 and E2 (Fig. 4A, lanes 4 to 6 and 8 to 10), indicating that pHPV16E1E2TTL does not replicate in the absence of exogenous E1 and E2 expression. Wild-type E2, as well as E2S206T, was able to support transient viral DNA replication equally well in cooperation with E1 in an E1 dose-dependent manner. Dpn I-digested fragments of pHPV16E1E2TTL DNA were detected equally in all lanes (Fig. 4A, lanes 1 to 10), indicating that the efficiency of transfections was roughly equal. To analyze E2S206T's ability to function as a transcriptional activator, a promoter activity assay was performed using a luciferase reporter plasmid, pBS1073, which contains four E2 binding sites upstream of a thymidine kinase promoter (33). A parental luciferase reporter plasmid, pBS1013, which does not contain any E2 binding sites, was used as a negative control. Either the pCMV4E2 wild type or pCMV4E2S206T was cotransfected with pBS1073 or pBS1013 into 293T cells. A green fluorescent protein expression vector was also cotransfected to ensure that efficiency of transfection was similar between E2 wild-type and E2S206T transfections. As shown in Fig. 4B, expression of wild-type E2 protein transcriptionally activated luciferase activity from pBS1073 but not from pBS1013. Similar to wild-type E2, E2S206T transcriptionally activated luciferase activity from pBS1073 but not pBS1013. Taken together with the transient replication data, these analyses indicated that the E2S206T encoded in 16st10 mutant HPV16 genomes is functionally indistinct from wild-type E2.
The mutation in the 16st20 genome does not alter the E2 ORF, and therefore changes in E2 function cannot account for its elevated plasmid copy number. However, we did consider the possibility that this mutation alters some uncharacterized cis element, leading to a cis-dominant phenotype. One possible cis element would be a splice enhancer or suppressor that modulates the utilization of the nearby 3′ splice site at nt 3358. It has been reported that the 3′ splice site at nt 3325 in BPV-1, which is equivalent to the site at nt 3358 in HPV16, is suboptimal and may be regulated by additional cis elements. Immediately downstream of the 3′ splice site at nt 3325 in the BPV-1 genome is a bipartite splicing regulatory element, consisting of purine-rich exonic splicing enhancers 1 and 2, followed immediately by a pyrimidine-rich exonic splicing suppressor. These sequences constitute potential binding sites for the serine- and arginine-rich proteins ASF/SF2, which are involved in splicing (67, 68). Such cis regulatory elements have not yet been identified in the HPV16 genome, and none of the mutations introduced into the E4 ORF in the context of this study overlap any potential binding sites for the serine- and arginine-rich protein; indeed, they were chosen with this in mind. Nevertheless, we addressed this concern by measuring the relative utilization of the nt 880 to nt 2709 versus nt 880 to nt 3358 splice events in cells harboring the wild-type versus 16st20 mutant HPV16 replicons. Total RNAs were isolated from NIKS cell populations that harbor wild-type or E4 mutant (16st15, 16st20, and 16st84) HPV16 genomes maintained in monolayers and then subjected to reverse transcription with random hexamers. Subsequent PCR was done using a 5′ primer complementary to sequences upstream of the 5′ splice site at nt 880 (nt 823 to 46), together with two 3′ primers complementary to sequences downstream of the 3′ splice sites at nt 2709 (nt 2853 to 2876) or nt 3358 (nt 3538 to 3561). The relative abundance of the two PCR products arising from the spliced mRNAs at nt 880 to nt 2709 (224 bp) and nt 880 to nt 3358 (260-bp) reflected the relative utilization of these two splice events. We found no gross changes in the relative abundance of the two PCR products for E4 mutant genomes compared to that of the wild type, suggesting that the splicing of these E4 mutant HPV16 genomes remained intact overall (data not shown). Further evidence arguing against a cis-acting, dominant mutation in 16st20 is the observation that the introduction of the PSS mutation in the 16st20 background led to the replication-defective phenotype, just as it did on an otherwise wild-type background.
As an additional means of monitoring viral gene expression, we quantified the level of E7 expression by Western blotting. To do this, protein lysates were prepared from monolayer cultures of several NIKS cell populations harboring the wild-type or E4 mutant HPV16 genomes and subjected to Western blotting to detect the E7 protein. Figure 3B shows E7 Western blots for two representative cell populations harboring wild-type or E4 mutant HPV16 genomes. Average levels of E7 expression in cells harboring E4 mutant HPV16 genome relative to that in cells harboring wild-type HPV16 genome were quantified with NIH Image software (results are shown below the bottom panel of Fig. 3B). In multiple, independently derived populations of cells, the level of E7 expression in cells harboring the E4 mutants (16st15, 16st20, 16st32, 16st67, and 16st84) was generally similar to that observed with cells harboring the wild-type HPV16 genome. We predicted that in those cells harboring the 16st10, PSS/WT, and PSS/20 genomes, viral early gene expression would be significantly altered because the genomes were replication defective. As expected, the level of E7 expression in cells harboring the three low-copy-number mutants was significantly lower than that observed with the wild-type HPV16 genomes (i.e., for 16st10 versus the wild type; P = 0.02 for six populations harboring each genome that was tested) or could barely be detected (PSS/WT and PSS/20). We found no gross or reproducible alteration in the levels of E7 protein when cells harboring the 16st20 mutant were compared with cells containing the wild-type HPV16 replicons. The level of E7 expression in W12E cells, which contained >500 copies of extrachromosomal HPV16 DNA genomes per cell, was likewise similar to that in NIKS cells harboring approximately 10 copies of wild-type viral DNA per cell. These data suggest that the number of extrachromosomal viral DNA genomes does not directly correlate with levels of early gene expression. Taken together, these data indicated that the mutations we engineered into the E4 ORF did not influence early gene expression as long as the genomes were maintained as nuclear plasmids.
To further assess the underlying cause for the replication and transcription defects of the 16st10, PSS/WT, and PSS/20 genomes, we performed transient replication assays with 293T cells and found, based upon the levels of DpnI-resistant DNA, that each of these mutant genomes was able to replicate transiently at levels commensurate with that of wild-type HPV16 (data not shown). This would suggest that neither cis nor trans replication factors (e.g., E1 and E2) per se are disrupted in these genomes and that the long-term replication defects (and consequent reductions in viral transcription) reflect some defect manifest at the level of maintenance of plasmid DNA once it becomes established in the cell.
Analysis of E1∧E4's role in the productive stage of the viral life cycle.
Because expression of the E1∧E4 protein is most abundant in the terminally differentiating cells of the stratified epithelium (16, 47, 59), E1∧E4 has been postulated to play a role in the late stage of the viral life cycle. To access the role of the E1∧E4 protein in the late stage of the HPV16 viral life cycle, we generated raft cultures of NIKS cell populations transfected with either the wild-type or the E4 mutant HPV16 genomes and analyzed the abilities of these genomes to execute the late stage of viral life cycle. Organotypic raft culture recapitulates the three-dimensional architecture of the stratified epithelium and supports the complete HPV life cycle (20, 39). The hallmarks of late stage of the viral life cycle that we analyzed included induction of cellular DNA synthesis, viral DNA amplification, and late gene expression in the terminally differentiated layers of the stratified epithelium.
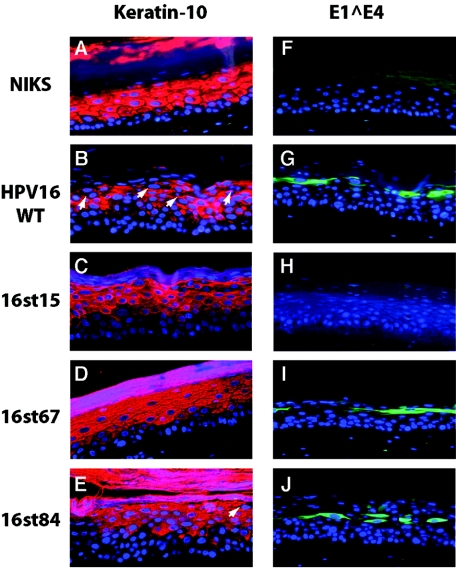
Initially, we monitored levels of late gene expression. E1∧E4 expression was monitored using an E1∧E4-specific monoclonal antibody, TGV402, which recognizes the central portion of the E1∧E4 protein (14). Immunofluorescence analysis revealed that in raft cultures harboring 16st67 or 16st84, E1∧E4 expression was detected in the terminally differentiating cells in a manner similar to that seen with raft cultures harboring the wild-type HPV16 replicon (Fig. 5). As expected, E1∧E4 was not detected in raft cultures of NIKS populations harboring 16st15 (Fig. 5), 16st20, and 16st32 replicons using this monoclonal E1∧E4 antibody (data not shown), because the epitope recognized by TGV402 was not present in the E1∧E4 gene products predicted to be expressed from these mutant genomes. In the raft cultures of NIKS populations transfected with the replication-defective mutant, PSS/WT, we were not able to detect E1∧E4 protein, even though the epitope should be retained in the predicted E1∧E4 gene product. This latter finding likely reflects the very low levels of viral gene expression from the replication-defective mutants, as shown by the E7 Western analysis (Fig. 3B). We also performed Western blots with the same E1∧E4 antibody (TGV402) and found that, consistent with immunofluorescence analysis, E1∧E4 products were detected in raft cultures harboring 16st67 or 16st84, as well as the wild type (Fig. 3C), but were not detected in raft cultures harboring other E4 mutants (data not shown). Previous studies have shown that C-terminal truncations remove E1∧E4 from the detergent-insoluble faction to the detergent-soluble fraction (51). As expected, E1∧E4 was detected in the detergent-soluble fraction of raft cultures harboring the 16st67 genome. In contrast, E1∧E4 was detected in the detergent-insoluble fraction of raft cultures harboring the 16st84 genome in a manner similar to that of rafts harboring the wild-type HPV16 replicons. This result indicates that the C-terminally (Δ84-92) truncated form of E1∧E4 expressed from the 16st84 mutant genome remained in the detergent-insoluble fraction. This contrasts with results from previous studies carried out under conditions in which E1∧E4 was overexpressed in cells; under these conditions, truncation of the C-terminal five amino acids was sufficient to remove E1∧E4 from the detergent-insoluble fraction (51, 65).
FIG. 5.
Analysis of E1∧E4 expression and cellular differentiation in raft cultures. Cross sections of raft cultures were subjected to immunofluorescence with antibodies to keratin-10 (A to E) (red) or E1∧E4 antibody (F to J) (green) and counterstained with DAPI to indicate nuclei (blue), as indicated in Materials and Methods. Arrows indicate the K10-negative cells in the more superficial spinous and granular layers. Note that monoclonal antibodies used to detect E1∧E4 recognize an epitope not present in the 16st15 E1∧E4 gene product. Shown are panels from rafts of NIKS cells alone (A and F), NIKS cells harboring the wild-type genome (B and G), or NIKS cells harboring the indicated E4 mutant HPV16 genomes (C to E and H to J).
HPV16 E1∧E4 protein enhances the ability of terminally differentiating cells to support suprabasal DNA synthesis during the late stage of the viral life cycle.
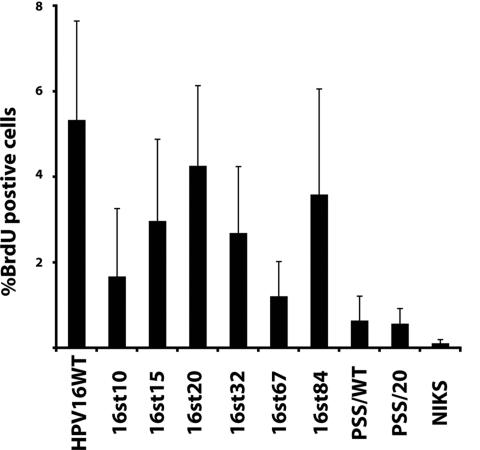
The HPV16 E7 protein, in the context of the full-length viral genome or when expressed alone, reprograms suprabasal, terminally differentiating cells to support DNA synthesis (7, 19). This suprabasal DNA synthesis is thought to contribute to the ability of these terminally differentiating cells to support viral DNA amplification during the late stage of the viral life cycle. The HPV16 E5 protein quantitatively contributes to this suprabasal DNA synthesis in the context of viral life cycle (23). To address whether E1∧E4 contributes to suprabasal DNA synthesis, raft cultures were incubated in medium containing the nucleotide analog BrdU prior to harvest. Cross sections from raft cultures were subjected to BrdU-specific immunohistochemistry (Fig. 6). The percentages of BrdU-positive cells in the suprabasal compartment of raft cultures were scored for multiple independent NIKS populations (n > 5) that had been transfected with either the wild-type or E4 mutant genomes (Fig. 7). As expected, the percentage of BrdU-positive cells in the suprabasal compartment was significantly increased in raft cultures of cells harboring the wild-type HPV16 replicon, compared to raft cultures of cells harboring no HPV DNA (P < 0.01). The percentages of suprabasal BrdU-positive cells were significantly increased in raft cultures of cells containing the E4 mutant HPV16 genomes 16st10, 16st15, 16st20, 16st32, 16st67, and 16st84 compared to raft cultures of cells not harboring HPV DNA (P ≤ 0.01). The fact that suprabasal DNA synthesis was observed in raft cultures expressing truncated forms of E1∧E4 is in agreement with previous studies indicating that the E7 protein is sufficient to reprogram terminally differentiating cells to undergo DNA synthesis (7). Indeed, E7 expression was detected in all NIKS cell populations harboring these E4 mutant genomes that were capable of inducing suprabasal DNA synthesis (Fig. 3B). Furthermore, the two E4 mutants that did not display suprabasal DNA synthesis at levels significantly greater than that observed in NIKS alone, PSS/WT (P = 0.07) and PPS/20 (P = 0.38), expressed E7 protein at barely detectable (2% that of the wild type) or undetectable (<1% that of the wild type) levels (Fig. 3B).
FIG. 6.
Analysis of DNA synthesis and viral DNA amplification in organotypic raft cultures of NIKS cells alone or NIKS cells harboring the wild-type or E4 mutant HPV16 genomes. To identify cells supporting DNA synthesis, histological sections (A to E and K to O) from raft cultures that were incubated in medium containing BrdU for 8 h prior to harvest were immunohistochemically stained with anti-BrdU antibody, and the nuclei were counterstained with hematoxylin (blue). In cells that had incorporated BrdU, nuclei were stained brown. To detect cells in which the HPV16 genome is amplified, serial histological sections (F to J and P to T) were subjected to HPV16-specific FISH. Cells containing amplified HPV16 DNA were detected with FITC-conjugated anti-DIG antibody (Roche; green) and counterstained with DAPI (Vector) to indicate nuclei (blue). The arrows in panel G indicate punctate, small dots that are found only in rafts harboring the 16st10 genome, which show a different pattern of signal from rafts harboring the wild-type or other E4 mutant HPV16 genomes. Raft cultures of NIKS cells alone (A and P) and NIKS cells harboring the wild-type HPV16 genome (A and F), E4 mutant HPV16 genome 16st10 (B and G), 16st15 (C and H), 16st20 (D and I), 16st32 (E and J), 16st67(L and Q), 16st84 (M and R), PSS/WT (N and S), and PSS/20 (O and T) are shown.
FIG. 7.
Quantification of BrdU-positive cells in raft cultures of NIKS cells alone or NIKS cells harboring the wild-type or E4 mutant HPV16 genomes. BrdU-positive cells in raft cultures (as shown in Fig. 6) were scored for multiple independently derived NIKS cell populations (n > 5) harboring wild-type or E4 mutant HPV16 genomes. The percentages of BrdU-positive cells in cells within the suprabasal compartment of raft culture are shown.
While suprabasal DNA synthesis was found to arise in raft cultures harboring E4 mutant replicons, the level of this DNA synthesis was generally reduced compared to that observed with raft cultures of cells harboring the wild-type HPV16 replicons. Specifically, the level of suprabasal DNA synthesis was significantly reduced for 16st15 (P = 0.06), 16st32 (P = 0.05), and 16st67 (P = 0.02), compared to that of the wild-type HPV16 genome. These results indicate that E1∧E4, while not required for suprabasal DNA synthesis, contributes quantitatively to this phenotype, akin to that observed with E5 mutant HPV16 genomes (23). No significant difference in the level of suprabasal DNA synthesis was observed in raft cultures harboring 16st20 (P = 0.6) or 16st84 (P = 0.5) replicons compared to raft cultures harboring wild-type HPV16 replicons. We interpret the 16st84 data to indicate that the last nine amino acids of E1∧E4 are dispensable for its quantitative contribution to suprabasal DNA synthesis. The high copy number of the 16st20 genome may compensate for the quantitative requirement for E1∧E4, although this does not appear to be a consequence of increased levels of E7 (Fig. 3B). Alternatively, 16st20 may display a gain of function phenotype, consistent with its ability to induce heightened levels of viral DNA amplification (see below). We noted that 16st20 did display significant reduction in the frequency of BrdU-positive cells, compared to that of wild-type HPV16 genome (P < 0.05) when BrdU-positive cells were scored in the context of all layers, including basal cells. These data suggest that a gross truncation of E4 protein in HPV16 genome leads to a reduction in the ability of the HPV16 genome to increase cellular DNA synthesis.
The E1∧E4 protein significantly contributes to viral DNA amplification in the productive stage of the HPV16 viral life cycle.
In the stratified epithelium infected with HPV16, high-level expression of E1∧E4 correlates with the onset of viral DNA amplification (16, 40). This has led to the hypothesis that the E1∧E4 protein contributes to viral DNA amplification. Consistent with this hypothesis for HPV16 E1∧E4, Peh et al. reported that E4 mutant CRPV genomes are defective for viral DNA amplification (46). To determine whether or not the E1∧E4 protein of HPV16 contributes to viral DNA amplification, we monitored viral DNA amplification in raft cultures of NIKS populations transfected with wild-type or E4 mutant HPV16 genomes using HPV16-specific FISH. Evidence for viral DNA amplification, as indicated by brightly fluorescent nuclei, was detected in terminally differentiated cells of raft cultures harboring the wild-type HPV16 genome (Fig. 6). Table 2 summarizes the frequency of FISH-positive cells for raft cultures of multiple independent NIKS populations transfected with wild-type or E4 mutant HPV16 genomes. As expected, there was an absence of brightly fluorescent nuclei in raft cultures containing the E4 mutant HPV16 genomes (16st10, PSS/WT, and PSS/20) (Fig. 6; Table 2) that were defective for viral plasmid DNA replication, indicating an absence of detectable viral DNA amplification. In the case of 16st10, a few nuclei were found with weak, punctate fluorescent signals that were distinct from that the uniformly bright fluorescent nuclei observed in rafts harboring wild-type HPV16 replicons. We currently have no understanding of the basis of these signals, which were not seen in other populations.
TABLE 2.
Frequency of FISH-positive cells in raft cultures harboring wild-type or E4 mutant HPV16 genomes
| Genome | Presence of extrachromo- somal viral DNAa | No. of rafts with FISH- positive cells/ all rafts testedf | Frequency of FISH- positive cellsb,e | Relative frequency of FISH-positive cellsc,e |
|---|---|---|---|---|
| WT | Y | 13/14 | 0.38 ± 0.44 | 1 |
| 16st10 | N | (4)/6 | 0.05 ± 0.05d* | 0.05d* |
| 16st15 | Y | 10/16 | 0.21 ± 0.34 | 0.6 |
| 16st20 | Y | 10/10 | 5.54 ± 4.4* | 14* |
| 16st32 | Y | 5/6 | 0.62 ± 0.79 | 1.6 |
| 16st67 | Y | 1/8 | <0.01* | <0.01* |
| 16st84 | Y | 2/8 | 0.18 ± 0.47* | 0.5* |
| PSS/WT | N | 0/3 | 0* | 0* |
| PSS/20 | N | 0/3 | 0* | 0* |
Y, extrachromosomal viral DNA was detected; N, extrachromosomal DNA was not detected (see Fig. 2).
The percentage of FISH-positive cells in total number of cells was scored for multiple independently generated rafts harboring the wild-type or E4 mutant HPV16 genomes, as described in Materials and Methods. Raft cultures that do not contain FISH-positive cells were included in the calculation.
The average frequency of FISH-positive cells for wild-type HPV16 populations was set at 1.
Frequency of cells containing small punctate signal (see Fig. 6 and Results).
Asterisks indicate values for E4 mutant HPV16 genomes that were statistically significant from that of the wild-type HPV16 genome, based on the two-sided Wilcoxon rank sum test.
Value in parentheses reflects the number of rafts containing punctate small signal (see arrows in Fig. 6G).
Analysis of viral amplification for E4 mutants that can replicate as nuclear plasmids provided some unexpected and disparate results. The frequency of FISH-positive cells was significantly reduced in raft cultures of cells harboring 16st84 (P < 0.01) or 16st67 (P < 0.001) replicons, indicating a gross reduction in the efficiency of viral DNA amplification. These results indicate that full-length HPV16 E1∧E4 contributes to viral DNA amplification. However, the frequency of FISH-positive cells was not significantly decreased in raft cultures of cells harboring 16st15 (P = 0.08), 16st20, or 16st32 (P = 0.56) replicons compared to the raft cultures of cells harboring the wild-type HPV16 replicon. The amplification-competence of these further truncated E1∧E4 mutants indicate that full-length E4 protein is not required for viral DNA amplification. One hypothesis that may account for these seemingly contradictory results is that the C-terminal nine amino acids of E1∧E4 that are absent in both 16st84 and 16st67 modulate an amplification-suppression activity that is encoded by the central region of E1∧E4 between amino acids 32 and 67 (i.e., the region absent in the more truncated E1∧E4 mutants 16st15, 16st20, and 16st32). Were this hypothesis true, then full-length E1∧E4 would have to be interpreted as not contributing to the amplification of the viral genome. This hypothesis is not consistent with the prior findings made with CRPV, however (46). An alternative hypothesis is that the 16st15, 16st20, and 16st32 gene products display a gain-of-function phenotype. If this were the case, then one would conclude that full-length E1∧E4 does contribute to viral DNA amplification, as indicated by the amplification defect seen with 16st67 and 16st84, consistent with the CRPV data (46). We favor this hypothesis because 16st20 actually displayed a level of viral DNA amplification that was significantly greater (P < 0.001) than that observed with the wild-type HPV16 genome. It would be difficult to account for this apparent gain-of-function phenotype based upon the first hypothesis.
E1∧E4 protein is required for the appearance of differentiation-defective cells in the more superficial spinous and granular layers.
HPV infection induces aberrant differentiation in the stratified epithelium (18, 41, 61). Similarly, organotypic raft cultures harboring the wild-type HPV16 replicons display an altered epithelial differentiation program, shown by a delay in the onset of expression of keratin-10 and involucrin in the lower spinous layer and the presence of cells in the more superficial spinous and the granular layers that are deficient for both early (keratin-10 and involucrin) and late (filaggrin and loricrin) markers for differentiation (20). The two major viral oncoproteins, E6 and E7, have been shown to contribute to the delay in the initial onset of the differentiation program in early-passage human keratinocytes (38, 66), NIKS cells (19), and mouse epidermis (3, 24, 58) but do not alter differentiation in the more superficial cellular compartment. The HPV E1∧E4 protein has been shown to associate with the keratin IF network in monolayer cultures of keratinocytes, and overexpression of HPV16 and -18 E1∧E4 in these cells results in the collapse of keratin bundles, although overexpression of HPV1 E1∧E4 does not (15, 50). Previous studies have also shown that HPV16 E1∧E4 colocalizes with the keratin IF network in the suprabasal compartment of stratified epithelium infected with HPV16 (59) and that the expression of differentiation-dependent keratins is selectively absent in cells expressing HPV11 and 16 E1∧E4 in vivo (6, 16). Consistent with these observations, evidence of keratin reorganization caused by HPV16 in lesions has been reported (65). However, whether E1∧E4 is required for aberrant differentiation has not been assessed. To address E1∧E4's contribution to aberrant differentiation, we analyzed expression of epithelial differentiation markers in raft cultures harboring the wild-type or the E4 mutant HPV16 replicons. Cross sections of raft cultures were subjected to immunofluorescence staining for the epithelial differentiation markers, keratin-10 (Fig. 5), involucrin, filaggrin, and loricrin (data not shown). K10 expression was detected uniformly throughout the suprabasal compartments of raft cultures of cells not harboring HPV16. In contrast, disruption of K10 expression was observed in raft cultures harboring the wild-type HPV16 genome, manifest both in a delay in the onset of K10 expression in the lower spinous layer and in the appearance of K10-negative cells in the more superficial spinous and granular layers (Fig. 5). In the raft cultures harboring E4 mutant HPV16 replicons, we again observed the delay in the expression of K10 in the lower spinous layer, indicating that E1∧E4 protein does not contribute to this delay in the onset of K10 expression. In contrast, we failed to detect K10-negative cells in the more superficial spinous and granular layers in raft cultures harboring E4 mutant HPV16 replicons, except for 16st84 (Fig. 5). The majority of the granular layer cells in rafts of cells harboring 16st84 genome remained positive for K10 and involucrin, as was observed with rafts of cells harboring the other E4 mutant HPV16 replicons; however, we did find a few K10-negative cells in the granular layer of the rafts of cells harboring the 16st84 genome. This result suggests that the truncated E1∧E4 product from 16st84 (Δ84-92) retains some attenuated ability to induce K10-negative cells in granular cells. It has been shown that C-terminally truncated HPV16 E1∧E4 proteins that lose their ability to induce keratin collapse correspondingly lose their detergent-insoluble property in cultured cells (51). We found that Δ84-92 retains the detergent-insoluble property (Fig. 3C), consistent with it retaining some ability, albeit attenuated, to induce K10-negative cells in the granular layer. When taken together, these studies indicate that the E1∧E4 gene product is specifically required for the appearance of differentiation-defective cells in the more superficial compartment of the rafts.
Temporal and spatial patterns of E1∧E4 expression as it relates to suprabasal DNA synthesis, viral DNA amplification, and disturbances in terminal differentiation.
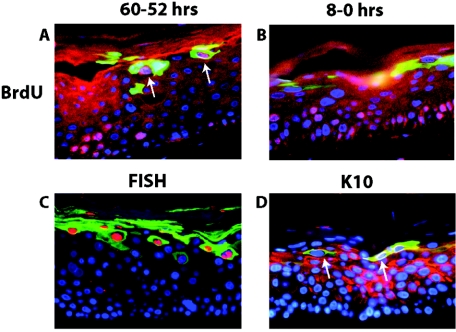
Our mutational analysis indicated that E1∧E4 contributes to multiple parameters of the viral life cycle. We therefore wanted to assess the temporal and spatial relationships between the expression of E1∧E4 and these different parameters. To this end, we carried out double-immunofluorescence studies on raft cultures harboring the wild-type HPV16 replicons (Fig. 8). It is important to recognize, in interpreting the results of these experiments, that our sensitivity in detecting E1∧E4 by immunofluorescence was limited, as shown by the fact that we could not detect E1∧E4 in the basal compartment of raft cultures by immunofluorescence (Fig. 5 and Fig. 8), even though it was detected by Western analysis in monolayer cultures maintained in a poorly differentiated, basal-like state (Fig. 3A). To assess the relationship between suprabasal DNA synthesis and E1∧E4 expression, we performed BrdU-E1∧E4 double immunofluorescence (Fig. 8A and B) on raft cultures that were incubated with BrdU for 8 h either 2 days or immediately prior to their being harvested. In the culture labeled for 8 h immediately prior to its harvesting, BrdU-positive cells were found in the intermediate cell layers far below those cells that stained positively for E1∧E4, which were exclusively located in the more superficial layers. In contrast, double-positive cells could be detected within the more superficial layers in the raft culture labeled with BrdU 2 days prior to being harvested. These results indicate not only that the onset of suprabasal DNA synthesis precedes the onset of the differentiation-dependent, high-level expression of E1∧E4, but also that cells expressing high levels of E1∧E4 protein no longer support the levels of cellular DNA synthesis that are necessary to permit its detection by the BrdU-labeling technique. A very different result was observed when we carried out double immunofluorescence for E1∧E4 and viral DNA amplification (the latter with FISH). Here, we found that nearly all FISH-positive cells were E1∧E4 positive (Fig. 8C). These double-positive cells were exclusively found in the more superficial layers of the raft cultures, far above the position of cells that were actively supporting DNA synthesis at the time of harvest as detected by BrdU incorporation (Fig. 8B). These results, when taken together, indicate that viral DNA amplification arises in cells expressing high levels of E1∧E4 temporally following the onset and presence of detectable suprabasal DNA synthesis. Because we had determined a role of E1∧E4 in perturbing terminal differentiation, we carried out E1∧E4/K10 double immunofluorescence (Fig. 8D). We found that majority of E1∧E4-positive cells in the more superficial compartment of the rafts were deficient for K10 staining, although a few double-positive cells were detected. These data are consistent with its role in perturbing differentiation.
FIG. 8.
Colocalization analysis of E1∧E4 with hallmarks of the productive stage of the viral life cycle. Shown are representative cross sections of raft cultures harboring the wild-type HPV16 genome that have been subjected to double immunofluorescence to detect the relative position of E1∧E4-positive cells in relation to cells supporting DNA synthesis (A and B), viral DNA amplification (C), and aberrant differentiation (D). (A and B) Raft cultures harboring wild-type HPV16 genomes were incubated in medium containing BrdU from 60 h to 52 h prior to harvest and then cultured in medium without BrdU until harvesting (A) or from 8 h to 0 h prior to harvest (B) (see Materials and Methods). Histological cross sections of rafts were subjected to double immunofluorescence using antibodies to BrdU (red) and E1∧E4 (green) and counterstained with DAPI (blue). (A) Arrows indicate BrdU-E1∧E4 double-positive cells. (C) Histological cross sections of rafts harboring the wild-type HPV16 genome were subjected to HPV16-specific FISH. Following hybridization to the probe, sections were incubated with FITC-conjugated anti-E1∧E4 antibody (green), and the amplified HPV16 genomes were detected by rhodamine-conjugated anti-DIG antibody (red). (D) Histological cross sections of rafts harboring wild-type HPV16 were subjected to double immunofluorescence using antibodies to K10 (red) and E1∧E4 (green). K10-negative cells in the superficial spinous and granular layers are indicated by arrows.
DISCUSSION
The HPV E1∧E4 protein has long been predicted to play a role in the late stage of the viral life cycle, primarily because it can be immunohistochemically detected in the terminally differentiated compartment of stratified epithelium in warts (16, 59) and in organotypic cultures of epithelial cell populations harboring HPV replicons (Fig. 5). Our results indicate that E1∧E4 does indeed play critical roles in the late stage of the viral life cycle, affecting the efficiency of induction of suprabasal DNA synthesis and viral DNA amplification, as well as in perturbing terminal differentiation. Surprisingly, we also discovered that mutations in the E4 ORF of HPV16 alter the plasmid replication state of the viral genome in human keratinocytes that were cultured to maintain their basal-like state (Fig. 2 and Table 1) and that the E1∧E4 protein is expressed in these basal-like cells, albeit at lower levels than that observed with raft cultures which are composed primarily of differentiated cells (Fig. 3). This latter finding is intriguing, given that the E1∧E4-spliced mRNA is present in early viral transcripts and widely used as a means of detecting the earliest signs of a viral infection. Our results provide the first detailed assessment of E1∧E4's role not only in the late stage but also in the early stage of the viral life cycle.
E1∧E4's role in plasmid DNA replication.
Recent advances in our understanding of the biological properties of E1∧E4 provide potential insights into the mechanism by which E1∧E4 contributes to plasmid DNA replication in the context of the nonproductive infective state. First, E1∧E4 can stimulate the activity of the papillomaviral early protein E2. E2 modulates viral transcription, contributes to E1-dependent replication of the viral genome, and mediates stable plasmid maintenance in dividing cell populations. E1∧E4 has been shown to stimulate the first two properties of E2 (R. Sorathia and J. Doorbar, unpublished data; T. Nakahara and H. Sakai, unpublished data). An effect of E1∧E4 on E2's role in plasmid maintenance has not been assessed. Furthermore, E1∧E4 has been found to bind directly to E2 protein (Sorathia and Doorbar, unpublished), providing a possible mechanism by which E1∧E4 could directly alter E2's function. Another potentially relevant property of E1∧E4 is its association with promyelocytic leukemia protein (PML), a cellular protein that is an essential component of nuclear subdomains called PML oncogenic domains (PODs), or nuclear domain 10 (ND10) (designated PODs/ND10). Roberts et al. have suggested that HPV1 E1∧E4 relocates ND10s via its association with PML (52). PODs/ND10 have been proposed to play a role in multiple steps in the papillomavirus life cycle, including efficient establishment of the viral genome in the nucleus upon infection (11), viral DNA replication (62), viral gene expression (25), and virus assembly (12, 22). Although E1∧E4's association with PML has not been characterized for HPV16 E1∧E4, it is of particular relevance that viral DNA replication is thought to occur at or near ND10. Swindle et al. demonstrated that the E1 and E2 proteins of HPV11 are colocalized near to PODs/ND10, together with the plasmid containing the replication origin of HPV11 in transiently transfected cells (62). Thus, E1∧E4 may modulate viral DNA replication by altering the activity of E2 directly as a result of E2 binding or indirectly by affecting the location of PODs/ND10. To date, mutational analyses defining the domains of E1∧E4 that are necessary for its association with either E2 or PML have not been described; therefore, we cannot assess whether our results using various mutations in the E4 ORF do or do not correlate with E1∧E4's association with E2 or PML. It is interesting to note however, that the leucine-rich motif of E1∧E4, which we found to be essential for E1∧E4s role in plasmid DNA replication, is predicted to contribute to E1∧E4's function in the context of its association with both E2 and effects on PODs/ND10. Specifically, E4 can sequester E2 to the keratin IF network (J. Doorbar, unpublished data) and this association of E4 with the keratin IF network is mediated by the leucine-rich motif (49, 51). In addition, Roberts et al. have suggested that the association between the leucine-rich motif of E1∧E4 and the keratin IF network is involved in E4-mediated relocation of ND10 (52).
E1∧E4 contributes to suprabasal DNA synthesis.
In the late stage of the viral life cycle, most E4 mutant HPV16 replicons were quantitatively reduced in their ability to induce suprabasal DNA synthesis to levels that were significantly lower than those seen with raft cultures harboring the wild-type HPV16 genome (Fig. 6 and 7). This result indicates that E1∧E4 contributes to the ability of HPV16 to induce suprabasal cells to support DNA synthesis. A requirement for E1∧E4 in inducing suprabasal DNA synthesis clearly is not absolute, as all replication-competent E4 mutant genomes retained a capacity for inducing suprabasal DNA at levels significantly higher than those seen with cell populations harboring no HPV16 genomes. This quantitative requirement for E1∧E4 is similar to that observed for the HPV16 and HPV31 E5 genes (17, 23) but contrasts with the absolute requirement for the HPV16 E7 oncogene (19). We found that the onset of DNA synthesis in suprabasal cells clearly preceded the differentiation-dependent upregulation of E1∧E4 as detected by immunofluorescence (Fig. 8). The level of E1∧E4 needed for its detection by immunofluorescence is high, since E1∧E4 is expressed in basal cells based upon our Western analysis (Fig. 3B) yet cannot be detected in these cells by immunofluorescence (Fig. 8). Therefore, we interpret the double-immunofluorescence results to indicate that low levels of E1∧E4 can contribute quantitatively to suprabasal DNA synthesis.
A possible mechanism by which E1∧E4 contributes to suprabasal DNA synthesis is its ability to induce G2/M arrest (10, 43). Overexpression of E1∧E4 leads to an accumulation of cells with 4N content, consistent with the cells being in G2 or M phase. Interestingly, these cells kept synthesizing DNA over time, as shown by the appearance of cells with a more than 4N DNA content, suggesting that E1∧E4 inhibits mitosis but not DNA synthesis (35, 43). By and large, the E1∧E4 requirements for its quantitative contribution to suprabasal DNA synthesis correlate with its requirements for inducing G2/M arrest. For instance, the E4 mutant HPV16 genomes that are predicted to express E1∧E4 peptides that are incapable of inducing G2/M arrest, such as 16st10, 16st15, and 16st32, were reduced in their ability to induce suprabasal DNA synthesis; whereas 16st84, which is predicted to retain an ability to induce G2/M arrest, induced suprabasal DNA synthesis to levels commensurate with those of wild-type HPV16. The only possible outlier to this correlation is 16st67, which was reduced in its ability to induce suprabasal DNA synthesis in human keratinocyte raft cultures (Fig. 7). A similar truncation mutant in HPV18 E4 (Δ60-92) also was reduced in its ability to induce G2/M arrest in HeLa cells (43); however, the HPV16 E4 mutant (Δ51-92) when expressed in Schizosaccharomyces pombe, could induce G2/M arrest (10). The defectiveness of the 16st67 mutant genome may simply reflect the relative instability of the mutant E1∧E4 protein, as shown by its low steady-state level compared to that of the wild-type E1∧E4 protein (Fig. 3C), and is similar to what was observed for the HPV18 E4 (Δ60-92) mutant protein in HeLa cells (43). Of potential relevance in this regard, the 16st67 protein was found in the detergent-soluble fraction (Fig. 3C). Prior studies have argued that the stability of full-length E1∧E4 protein is related to its migration into the detergent-insoluble state (65). Chien et al. showed that a majority of cells undergoing suprabasal DNA synthesis did not exhibit a prolonged S phase in rafts expressing E7 alone, suggesting that E7 by itself is sufficient to reprogram cells to initiate suprabasal DNA synthesis but is not sufficient to maintain active for DNA synthesis (endoreduplication) (8). E1∧E4 may facilitate endoreduplication by inhibiting mitosis but not DNA synthesis in the context of the viral life cycle.
E1∧E4's contribution to amplification is separable from its contribution to the induction of suprabasal DNA synthesis but may relate to its role in plasmid DNA replication in basal cells.
Our study demonstrates a role of E1∧E4 in viral DNA amplification that is consistent with prior E4 mutational analyses in the context of CRPV (46). The induction of suprabasal DNA synthesis is thought to be a prerequisite for viral DNA amplification in the productive stage of the viral life cycle. In rafts harboring the E7-null HPV16 genome, suprabasal DNA synthesis was not detected; correspondingly, no viral DNA amplification occurred (19). Interestingly, the genetic requirements for E1∧E4 in supporting viral DNA amplification are separable from its requirements for inducing suprabasal DNA synthesis. The E4 mutant HPV16 genome, 16st84, was able to induce suprabasal DNA synthesis but not viral DNA amplification (Fig. 7 and Table 2). The direct involvement of E1∧E4 in viral DNA amplification is consistent with the observation that viral DNA amplification arises in E1∧E4-positive cells that are spatially distinct from those cells that support suprabasal DNA synthesis (Fig. 8). The distinct genetic requirement of E1∧E4 is further shown by the fact that the E4 mutant genomes, 16st15 and 16st32, displayed a reduction in suprabasal DNA synthesis but not in viral DNA amplification. The mechanism by which E1∧E4 contributes to viral DNA amplification in the context of the late stage of the viral life cycle is unclear; however, it may relate to its role in plasmid DNA replication in the context of the nonproductive, early stage of the viral life cycle. Supportive of such a relationship is the fact that for many of the mutants analyzed (16st10, 16st15, 16st20, 16st32, PSS/20, and PSS/WT) the presence of the leucine-rich motif of E1∧E4 correlates with the competence of the mutant genome to replicate as nuclear plasmids, as well as to undergo viral DNA amplification.
E1∧E4 contributes to the perturbation of differentiation specifically in the more superficial layers of stratified squamous epithelia.
HPV16 has two effects on differentiation. It delays the onset of differentiation in the lower layers of the stratum spinosum and perturbs differentiation within the upper layers of the stratum spinosum and stratum granulosum. Our studies indicate that E1∧E4 is necessary to perturb differentiation in the more superficial layers of the epidermis but show that it does not contribute to the delayed onset of differentiation in the lower layers of the stratum spinosum. This specific effect of E1∧E4 on differentiation correlated with its spatial pattern of high-level expression in the raft cultures (Fig. 8D). How E1∧E4 causes the perturbation of differentiation is unclear, but we suspect that it is mediated through its capacity to induce keratin IF reorganization, a property that correlates with E1∧E4's detergent-insoluble characteristic (51). Interestingly, E7 is necessary and sufficient to cause the delay in differentiation in the lower layers of the stratum spinosum (19, 24; A. Collins et al., submitted for publication). Thus, in the context of the HPV16 life cycle, E7 and E1∧E4 alter cell differentiation in a spatially distinct manner.
An integrated model for the role of E1∧E4 and other viral proteins in the HPV life cycle.
Based on these findings and other findings made in the context of like experimental model systems, we propose a model for the role of E1∧E4 and other viral genes in the nonproductive versus productive stages of the HPV life cycle. A hallmark of the early stage of the viral life cycle, which takes place in the basal compartment, is the establishment of the viral genome as low-copy-number nuclear plasmid DNA. Four viral early gene products appear to contribute to this process for HPVs: E1 (28), E2 (29, 60), E4 (this study), and E6 (45, 63) (E. Flores and P. F. Lambert, unpublished data). While a direct role has been established for E1 and E2 in stable plasmid DNA replication, it remains unclear exactly as to how E4 and E6 contribute to this process. Upon basal cell division, one of the daughter cells migrates into the suprabasal compartment and initiates a program of terminal differentiation that includes the ordered expression of differentiation specific factors, as well as exit from the cell cycle. In these suprabasal cells where HPVs initiate the productive stage of their life cycle to produce progeny viruses, the cellular differentiation program is perturbed in multiple ways. The first observable perturbation is a delay in the onset of differentiation, as shown by a paucity of K10 staining in the lowermost layers of the stratum spinosum. E7 is necessary for this delay (19), and its requirement correlates with its ability to bind and degrade pRb and the related pocket proteins p107 and p130 (Collins et al., submitted). E6 also can delay the onset of differentiation in human keratinocytes (30, 57), which suggests that E6 may enhance the delay in the onset of differentiation, together with E7. A second aspect of the differentiation process that is perturbed by HPVs is the withdrawal of suprabasal cells from the cell cycle. Normally, suprabasal cells are unable to initiate new rounds of DNA synthesis. HPVs reprogram these cells to stay and/or reenter the cell cycle, as shown by the capacity of these cells to incorporate nucleotide analogs (e.g., BrdU) into newly synthesized DNA. While E4 (this study) and E5 (17, 23) contribute quantitatively to this process in the context of the viral life cycle, E7 is absolutely required; this requirement correlates with E7's binding to pRb (Collins et al., submitted). E5 may facilitate these differentiating cells to stay in and/or to reenter the cell cycle through its activation of EGF signaling pathways. In contrast, E4 may promote these cells to undergo endoreduplication by its association with the cyclin B/Cdk1 complex (9). In these latter studies, in which individual viral genes are expressed out of context of the viral genome, the DNA synthesis measured is necessarily limited to cellular DNA, as no viral genomes are present. Likewise, in the context of viral life cycle studies in cells harboring the full-length viral genome, BrdU incorporation most probably represents cellular DNA synthesis, not viral DNA synthesis, as cells incorporating BrdU are located in lower strata within the raft cultures than the cells that are detected by FISH to have amplified the viral DNA (Fig. 8B versus 8C). A spatial separation of cells expressing cellular S-phase markers and cells supporting viral DNA amplification in HPV-infected tissues has been documented (47). We interpret the spatial separation of BrdU and FISH positivity to indicate that as cells migrate to the more superficial strata, they are programmed by the virus to undergo a switch that directs the cellular replication machinery to amplify viral DNA, at which time cellular DNA synthesis no longer occurs or occurs inefficiently. We postulate that E4 is critical for this switch. As was previously found with CRPV (46), some E4 mutant HPV16 genomes are defective for DNA amplification (Fig. 6; Table 2); importantly, in the case of one such mutant (16st84), this defect was separable from defects in suprabasal DNA synthesis. The second observation is that high-level expression of E1∧E4, as detected by immunofluorescence, correlates with viral DNA amplification (Fig. 8C). The precise mechanism by which E4 contributes to this hypothesized switch remains unknown. But it is interesting that the same superficial strata of the raft cultures in which we found a correlation between high-level expression of E1∧E4 and viral DNA amplification is where we also detected E4-dependent defects in cellular differentiation (Fig. 8). Whether this effect of E4 on differentiation contributes to the switch remains unclear. Alternatively, or in addition to any effect on viral DNA amplification, the effect we observed on K10 staining may relate to E4's argued role in promoting virus egress from cells by altering the cornified cell envelope (6). Based upon staining patterns for the major capsid protein, L1 (5, 16, 59), progeny viruses are thought to accumulate in cells in the granular layers of the stratified epithelium where high-level E1∧E4 expression correlates with K10-negative status. Also in these cells, L2 is expressed and facilitates encapsidation of the viral genome (26).
It is relevant to consider the role of E1∧E4 in the context of cervical cancer in light of our findings. Loss of E1∧E4 expression in cervical lesions infected with HPV16 correlates with increased severity of those lesions and the appearance of frank cancer (40, 44, 56). This loss of E4 expression, like that of E2, is believed to be caused by integration of the viral genome into host chromosomes, which can lead to disruption of the E4 ORF (2, 32, 53) and/or destabilization of mRNAs encoding E4 (32). It has been argued that the absence of E1∧E4 is conducive to tumorigenesis, perhaps because of its capacity to induce G2/M arrest (10, 43) or its newly discovered role in inducing cellular apoptosis (48). Given our findings, It is also possible that E1∧E4 contributes to replication of the viral genome as a nuclear plasmid and that loss of E4 expression, similar to that of E2, may result in progressive integration events of the viral genome, contributing to tumorigenesis in HPV-associated cancers.
ADDENDUM
Following submission of the manuscript, a similar study analyzing E4 mutant HPV31 genomes in context of the viral life cycle was published (R. Wilson, F. Fehrmann, and L. A. Laimins, J. Virol., 79:6732-6740, 2005). Some phenotypes described for E4 mutant HPV31 genomes were similar to those described for our cognate E4 mutant HPV16 genomes, as follows. (i) The E4M4 mutant, which is equivalent to 16st10, was defective for viral DNA replication in human keratinocytes, while E4M9, which is equivalent to 16st15, was competent for viral DNA replication. (ii) Suprabasal BrdU-positive cells were decreased in raft cultures harboring E4M9, compared to the raft culture-harboring wild-type HPV31 genome.
Acknowledgments
We thank members of the Lambert and Doorbar laboratories for helpful discussions and comments. We thank Karl Munger for human foreskin keratinocytes and for the Western protocol for E7 protein.
This work was supported by NIH grants CA22443, CA07175, and AI52049 and by the United Kingdom's Medical Research Council.
REFERENCES
- 1.Allen-Hoffmann, B. L., S. J. Schlosser, C. A. Ivarie, C. A. Sattler, L. F. Meisner, and S. L. O'Connor. 2000. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J. Investig. Dermatol 114:444-455. [DOI] [PubMed] [Google Scholar]
- 2.Baker, C. C., W. C. Phelps, V. Lindgren, M. J. Braun, M. A. Gonda, and P. M. Howley. 1987. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J. Virol. 61:962-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balsitis, S. J., J. Sage, S. Duensing, K. Munger, T. Jacks, and P. F. Lambert. 2003. Recapitulation of the effects of the human papillomavirus type 16 E7 oncogene on mouse epithelium by somatic Rb deletion and detection of pRb-independent effects of E7 in vivo. Mol. Cell. Biol. 23:9094-9103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bouvard, V., G. Matlashewski, Z. M. Gu, A. Storey, and L. Banks. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Brown, D. R., J. T. Bryan, L. Pratt, V. Handy, K. H. Fife, and M. H. Stoler. 1995. Human papillomavirus type 11 E1-E4 and L1 proteins colocalize in the mouse xenograft system at multiple time points. Virology 214:259-263. [DOI] [PubMed] [Google Scholar]
- 6.Bryan, J. T., and D. R. Brown. 2000. Association of the human papillomavirus type 11 E1∧E4 protein with cornified cell envelopes derived from infected genital epithelium. Virology 277:262-269. [DOI] [PubMed] [Google Scholar]
- 7.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 8.Chien, W. M., F. Noya, H. M. Benedict-Hamilton, T. R. Broker, and L. T. Chow. 2002. Alternative fates of keratinocytes transduced by human papillomavirus type 18 E7 during squamous differentiation. J. Virol. 76:2964-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davy, C. E., D. J. Jackson, K. Raj, W. L. Peh, S. A. Southern, P. Das, R. Sorathia, P. Laskey, K. Middleton, T. Nakahara, Q. Wang, P. J. Masterson, P. F. Lambert, S. Cuthill, J. B. A. Millar, and J. Doorbar. 2005. Human papillomavirus type 16 E1∧E4-induced G2 arrest is associated with cytoplasmic retention of active Cdk1/cyclin B1 complexes. J. Virol. 79:3998-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davy, C. E., D. J. Jackson, Q. Wang, K. Raj, P. J. Masterson, N. F. Fenner, S. Southern, S. Cuthill, J. B. A. Millar, and J. Doorbar. 2002. Identification of a G2 arrest domain in the E1∧E4 protein of human papillomavirus type 16. J. Virol. 76:9806-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Day, P. M., C. C. Baker, D. R. Lowy, and J. T. Schiller. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. USA 101:14252-14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Day, P. M., R. B. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doorbar, J., R. C. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. M. Griffin, P. Masterson, S. Stacey, Y. Mengistu, and J. Dunlop. 2000. The E1E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 74:10081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doorbar, J., S. Ely, N. Coleman, M. Hibma, D. H. Davies, and L. Crawford. 1992. Epitope-mapped monoclonal antibodies against the HPV16E1-E4 protein. Virology 187:353-359. [DOI] [PubMed] [Google Scholar]
- 15.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV-16 E1-E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 16.Doorbar, J., C. Foo, N. Coleman, L. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterization of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 17.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fletcher, S., and M. Norval. 1983. On the nature of the deep cellular disturbances in human-papilloma-virus infection of the squamous cervical epithelium. Lancet ii:546-549. [DOI] [PubMed] [Google Scholar]
- 19.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, C. A. Sattler, and P. F. Lambert. 1999. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology 262:344-354. [DOI] [PubMed] [Google Scholar]
- 21.Flores, E. R., and P. F. Lambert. 1997. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J. Virol. 71:7167-7179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florin, L., F. Schafer, K. Sotlar, R. E. Streeck, and M. Sapp. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology 295:97-107. [DOI] [PubMed] [Google Scholar]
- 23.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gulliver, G. A., R. L. Herber, A. Liem, and P. F. Lambert. 1997. Both conserved region 1 (CR1) and CR2 of the human papillomavirus type 16 E7 oncogene are required for induction of epidermal hyperplasia and tumor formation in transgenic mice. J. Virol. 71:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heino, P., J. Zhou, and P. F. Lambert. 2000. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology 276:304-314. [DOI] [PubMed] [Google Scholar]
- 26.Holmgren, S. C., N. A. Patterson, M. A. Ozbun, and P. F. Lambert. 2005. The minor capsid protein L2 contributes to multiple steps in the papillomaviral life cycle. J. Virol. 79:3938-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 28.Hubert, W. G. 2005. Variant upstream regulatory region sequences differentially regulate human papillomavirus type 16 DNA replication throughout the viral life cycle. J. Virol. 79:5914-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubert, W. G., and L. A. Laimins. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hudson, J. B., M. A. Bedell, D. J. McCance, and L. A. Laimins. 1990. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type 18. J. Virol. 64:519-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeon, S., B. L. Allen-Hoffmann, and P. F. Lambert. 1995. Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69:2989-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeon, S., and P. F. Lambert. 1995. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: implications for cervical carcinogenesis. Proc. Natl. Acad. Sci. USA 92:1654-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim, K., P. A. Garner-Hamrick, C. Fisher, D. Lee, and P. F. Lambert. 2003. Methylation patterns of papillomavirus DNA, its influence on E2 function, and implications in viral infection. J. Virol. 77:12450-12459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Knight, G. L., J. R. Grainger, P. H. Gallimore, and S. Roberts. 2004. Cooperation between different forms of the human papillomavirus type 1 E4 protein to block cell cycle progression and cellular DNA synthesis. J. Virol. 78:13920-13933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lambert, P. F. 1991. Papillomavirus DNA replication. J. Virol. 65:3417-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leechanachai, P., L. Banks, F. Moreau, and G. Matlashewski. 1992. The E5 gene from human papillomavirus type 16 is an oncogene which enhances growth factor-mediated signal transduction to the nucleus. Oncogene 7:19-25. [PubMed] [Google Scholar]
- 38.McCance, D. J., R. Kopan, E. Fuchs, and L. A. Laimins. 1988. Human papillomavirus type 16 alters human epithelial cell differentiation in vitro. Proc. Natl. Acad. Sci. USA 85:7169-7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meyers, C., M. G. Frattini, J. B. Hudson, and L. A. Laimins. 1992. Biosynthesis of human papillomavirus from a continuous cell line upon epithelial differentiation. Science 257:971-973. [DOI] [PubMed] [Google Scholar]
- 40.Middleton, K., W. Peh, S. Southern, H. Griffin, K. Sotlar, T. Nakahara, A. El-Sherif, L. Morris, R. Seth, M. Hibma, D. Jenkins, P. Lambert, N. Coleman, and J. Doorbar. 2003. Organization of human papillomavirus productive cycle during neoplastic progression provides a basis for selection of diagnostic markers. J. Virol. 77:10186-10201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullink, H., N. M. Jiwa, J. M. Walboomers, A. Horstman, W. Vos, and C. J. Meijer. 1991. Demonstration of changes in cytokeratin expression in condylomata accuminata in relation to the presence of human papilloma virus as shown by a combination of immunohistochemistry and in situ hybridization. Am. J. Dermatopathol. 13:530-537. [DOI] [PubMed] [Google Scholar]
- 42.Munger, K., and W. C. Phelps. 1993. The human papillomavirus E7 protein as a transforming and transactivating factor. Biochim. Biophys. Acta 1155:111-123. [DOI] [PubMed] [Google Scholar]
- 43.Nakahara, T., A. Nishimura, M. Tanaka, T. Ueno, A. Ishimoto, and H. Sakai. 2002. Modulation of the cell division cycle by human papillomavirus type 18 E4. J. Virol. 76:10914-10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palefsky, J. M., B. Winkler, J. P. Rabanus, C. Clark, S. Chan, V. Nizet, and G. K. Schoolnik. 1991. Characterization of in vivo expression of the human papillomavirus type 16 E4 protein in cervical biopsy tissues. J. Clin. Investig. 87:2132-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park, R. B., and E. J. Androphy. 2002. Genetic analysis of high-risk e6 in episomal maintenance of human papillomavirus genomes in primary human keratinocytes. J. Virol. 76:11359-11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peh, W. L., J. L. Brandsma, N. D. Christensen, N. M. Cladel, X. Wu, and J. Doorbar. 2004. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. J. Virol. 78:2142-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and J. Doorbar. 2002. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 76:10401-10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raj, K., S. Berguerand, S. Southern, J. Doorbar, and P. Beard. 2004. E1 empty set E4 protein of human papillomavirus type 16 associates with mitochondria. J. Virol. 78:7199-7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roberts, S., I. Ashmole, L. J. Gibson, S. M. Rookes, G. J. Barton, and P. H. Gallimore. 1994. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J. Virol. 68:6432-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts, S., I. Ashmole, G. D. Johnson, J. W. Kreider, and P. H. Gallimore. 1993. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology 197:176-187. [DOI] [PubMed] [Google Scholar]
- 51.Roberts, S., I. Ashmole, S. M. Rookes, and P. H. Gallimore. 1997. Mutational analysis of the human papillomavirus type 16 E1-E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J. Virol. 71:3554-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts, S., M. L. Hillman, G. L. Knight, and P. H. Gallimore. 2003. The ND10 component promyelocytic leukemia protein relocates to human papillomavirus type 1 E4 intranuclear inclusion bodies in cultured keratinocytes and in warts. J. Virol. 77:673-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Romanczuk, H., and P. M. Howley. 1992. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. USA 89:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakai, H., T. Yasugi, J. D. Benson, J. J. Dowhanick, and P. M. Howley. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 70:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sedman, S. A., M. S. Barbosa, W. C. Vass, N. L. Hubbert, J. A. Haas, D. R. Lowy, and J. T. Schiller. 1991. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J. Virol. 65:4860-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sherman, L., N. Alloul, I. Golan, M. Durst, and A. Baram. 1992. Expression and splicing patterns of human papillomavirus type-16 mRNAs in pre-cancerous lesions and carcinomas of the cervix, in human keratinocytes immortalized by HPV 16, and in cell lines established from cervical cancers. Int. J. Cancer 50:356-364. [DOI] [PubMed] [Google Scholar]
- 57.Sherman, L., and R. Schlegel. 1996. Serum- and calcium-induced differentiation of human keratinocytes is inhibited by the E6 oncoprotein of human papillomavirus type 16. J. Virol. 70:3269-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, S., H. C. Pitot, and P. F. Lambert. 1999. The human papillomavirus type 16 E6 gene alone is sufficient to induce carcinomas in transgenic animals. J. Virol. 73:5887-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sterling, J. C., J. N. Skepper, and M. A. Stanley. 1993. Immunoelectron microscopical localization of human papillomavirus type 16 L1 and E4 proteins in cervical keratinocytes cultured in vivo. J. Investig. Dermatol. 100:154-158. [DOI] [PubMed] [Google Scholar]
- 60.Stubenrauch, F., A. M. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun, Q., K. Tsutsumi, M. Yokoyama, M. M. Pater, and A. Pater. 1993. In vivo cytokeratin-expression pattern of stratified squamous epithelium from human papillomavirus-type-16-immortalized ectocervical and foreskin keratinocytes. Int. J. Cancer 54:656-662. [DOI] [PubMed] [Google Scholar]
- 62.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thomas, J. T., W. G. Hubert, M. N. Ruesch, and L. A. Laimins. 1999. Human papillomavirus type 31 oncoproteins E6 and E7 are required for the maintenance of episomes during the viral life cycle in normal human keratinocytes. Proc. Natl. Acad. Sci. USA 96:8449-8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Q., H. Griffin, S. Southern, D. Jackson, A. Martin, P. McIntosh, C. Davy, P. J. Masterson, P. A. Walker, P. Laskey, M. B. Omary, and J. Doorbar. 2004. Functional analysis of the human papillomavirus type 16 E1∧E4 protein provides a mechanism for in vivo and in vitro keratin filament reorganization. J. Virol. 78:821-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woodworth, C. D., S. Cheng, S. Simpson, L. Hamacher, L. T. Chow, T. R. Broker, and J. A. DiPaolo. 1992. Recombinant retroviruses encoding human papillomavirus type 18 E6 and E7 genes stimulate proliferation and delay differentiation of human keratinocytes early after infection. Oncogene 7:619-626. [PubMed] [Google Scholar]
- 67.Zheng, Z. M., P. He, and C. C. Baker. 1996. Selection of the bovine papillomavirus type 1 nucleotide 3225 3′ splice site is regulated through an exonic splicing enhancer and its juxtaposed exonic splicing suppressor. J. Virol. 70:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng, Z. M., M. Huynen, and C. C. Baker. 1998. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc. Natl. Acad. Sci. USA 95:14088-14093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.zur Hausen, H. 1996. Papillomavirus infections—a major cause of human cancers. Biochim. Biophys. Acta 1288:F55-F78. [DOI] [PubMed] [Google Scholar]