Abstract
Open reading frame 29 (ORF29) of varicella-zoster virus (VZV) encodes a 120-kDa single-stranded DNA binding protein (ORF29p) that is not packaged in the virion and is expressed during latency. During lytic infection, ORF29p is localized primarily to infected cell nuclei. In contrast, ORF29p is found exclusively in the cytoplasm in neurons of the dorsal root ganglia obtained at autopsy from seropositive latently infected patients. ORF29p accumulates in the nuclei of neurons in dorsal root ganglia obtained at autopsy from patients with active zoster. The localization of this protein is, therefore, tightly correlated with the proposed VZV lytic/latent switch. In this report, we have investigated the nuclear import mechanism of ORF29p. We identified a novel nuclear targeting domain bounded by amino acids 9 to 154 of ORF29p that functions independent of other VZV-encoded factors. In vitro import assays in digitonin-permeabilized HeLa cells reveal that ORF29p is transported into the nucleus by a Ran-, karyopherin α- and β-dependent mechanism. These data are further supported by the demonstration that a glutathione S-transferase-karyopherin α fusion interacts with ORF29p, but not with a protein containing a point mutation in its nuclear localization signal (NLS). Therefore, the region of ORF29p responsible for its nuclear targeting is also involved in the association with karyopherin α. As a result of this interaction, this noncanonical NLS appears to hijack the classical cellular nuclear import machinery. Elucidation of the mechanisms governing ORF29p nuclear targeting could shed light on the VZV reactivation process.
Varicella-zoster virus (VZV) is an alphaherpesvirus that during primary lytic infection invades the dermis and epidermis to cause chicken pox (varicella). During this lytic infection, all classes of viral genes are expressed, viral DNA is replicated, and infectious virions are assembled. Following primary infection, the virus may move into the dorsal root ganglia (DRG), infecting both satellite sensory cells and neurons, or enteric ganglia (EG) to establish latency (5, 6, 13, 45, 70). During latency, viral DNA replication, late protein expression, and virion production cease. However, a few immediate-early and early genes, including open reading frames (ORFs) 4, 21, 29, 62, 63, and 66, are expressed, and the proteins they encode aberrantly localize to the cytoplasm of latently infected neurons obtained at autopsy (15-18, 22, 28, 36, 46, 47). Later during the host's life the virus may reactivate and reenter the lytic cycle, resulting in invasion of the dermatome innervated by the infected neuron to cause shingles, or zoster (6).
VZV ORF29 encodes a single-stranded DNA binding protein (ORF29p) with a predicted molecular mass of 120 kDa (38). This protein is not packaged in the virion but is expressed during latency (37, 65). Immunohistochemical analyses of epidermal cells from varicella lesions as well as VZV infection of fibroblasts in tissue culture demonstrates that the bulk of ORF29p localizes to the nucleus, with some diffuse cytoplasmic staining (38, 46). Previously, we examined neurons from DRG of VZV-seropositive individuals without clinical evidence of zoster and demonstrated that they contained ORF29p exclusively in the cytoplasm (46). This is also observed in cultured guinea pig enteric neurons infected with cell-free VZV (13). Upon VZV reactivation, ORF29p is found in the nucleus and cytoplasm of infected neurons (46). The failure of ORF29p and the other latency-associated viral proteins (LAPs) to accumulate in the nucleus suggests a tight correlation between their nuclear import/exclusion and a lytic/latent switch. Elucidation of the mechanisms governing ORF29p nuclear targeting may, therefore, shed light on the VZV reactivation process.
All nuclear transport occurs through the nuclear pore complex (NPC), which is 9 nm in diameter and allows diffusion of ions, metabolites, and small proteins under 30 to 60 kDa in size (9, 10, 58). Larger molecules must be actively transported. This involves soluble carriers termed importins binding the cargo molecule in the cytoplasm, escorting it through the NPC, and releasing it into the nucleus. The classical signal-mediated transport system works through the interaction of a canonical sequence containing a single or bipartite cluster of basic residues and the soluble, heterodimeric carrier consisting of karyopherin α and karyopherin β (kaps α and β, respectively) (34, 43, 64). The importins shuttle between the nucleus and cytoplasm through the NPC in a Ran-dependent manner (48, 49). This translocation is also often regulated by the phosphorylation state of the imported cargo (51, 55, 57).
Reports of “nonclassical” nuclear transport pathways are increasing. It is now clear that the karyopherin α/β complex is not the only transporter and that the nuclear import interaction motifs vary significantly in size and charge (21, 23, 40, 54). There are also several instances of structure-dependent import signals, where the three-dimensional structure of the protein may comprise the interaction domain or allow accessibility of the nuclear localization signal (NLS) to the import machinery (57). In addition, many proteins enter the nucleus by piggybacking onto other proteins that harbor an NLS. This is the case with herpes simplex virus 1 (HSV-1) UL37, which interacts in the cytoplasm of infected cells with the HSV-1 ORF29p homologue ICP8. As a result of this interaction, UL37 is transported into the nucleus with ICP8 via a classical nuclear import signal in the C terminus of ICP8 (4, 69). The diversity of nuclear import mechanisms continues to increase.
The mechanism of ORF29p nuclear import during VZV lytic infection is unknown. The amino acid sequence contains no classical NLSs, no sequence homology to the import signal found in HSV-1 ICP8, nor any homology to other known VZV proteins' transport sequences (38, 66, 71). Unlike many other transported molecules, the protein is not phosphorylated during lytic infection, despite the presence of many putative phosphorylation motifs (65). The state of ORF29p modification during latency has not been investigated.
We have analyzed the cellular localization patterns of ORF29p during lytic VZV infection and when it is expressed in the absence of other viral proteins. Transfection experiments demonstrate that nuclear import of ORF29p occurs independent of other VZV-encoded factors. Genetic analysis of ORF29 reveals that amino acids (aa) 9 to 154 encode a novel nuclear targeting domain that is both necessary for ORF29p nuclear import and sufficient to target green fluorescent protein (GFP) fusion molecules to the nucleus. This noncanonical targeting region appears to hijack the classical cellular transport machinery as in vitro import assays show that ORF29p is transported into the nucleus by a Ran, karyopherin α- and β-dependent mechanism. These data are further supported by the demonstration that karyopherin α2 interacts with ORF29p, but not with an NLS mutant of this protein. A better understanding of the nuclear import of ORF29p could unravel what controls the switch between VZV lytic infection and latency.
MATERIALS AND METHODS
Mammalian cells.
Human 293T fibroblast and HeLa cells were maintained as monolayer cultures in Dulbecco's modified Eagle medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (Gibco-BRL) at 37°C in a 5% CO2 atmosphere. Twenty-four hours prior to infection, transfection, and in vitro import assays, the cells were seeded onto glass coverslips for fluorescence microscopy assays or into six-well tissue culture dishes for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting.
Transfections.
All transfections into 293T cells were performed using Lipofectamine PLUS in Opti-MEM media (Invitrogen, Carlsbad, CA). Forty-eight hours following transfection, cells were processed for analysis by fluorescence microscopy.
Virus.
Jones VZV, a wild-type clinical isolate, was propagated in MeWo cell monolayers by serial passage of infected cells onto uninfected cells as described previously (29). Cell-free virus was obtained by infecting MeWo cells in 175-cm2 tissue culture flasks until cytopathic effect was present. Infected cells were scraped in the maintenance medium pelleted at 577 × g for 10 min at 4°C in a Beckman CS-6KR refrigerated centrifuge. The cells were washed twice in cold phosphate-buffered saline (PBS; 1 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, 2.7 mM KCl, pH 7.4) before being resuspended in 1 ml per flask of Hanks balanced salt solution and sonicating for two 30-s intervals. Cell debris was removed by centrifuging the lysed cells at 25 × g for 5 min at 4°C in a Tomy MX-160 high-speed refrigerated microcentrifuge. Cell-free virus stocks were stored in 0.5-ml aliquots at −80°C.
During virus infection, 293T cells were maintained in Eagle's minimal essential medium supplemented with 2% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin.
Antibodies.
Rabbit polyclonal antibodies against amino acids (aa) 1086 to 1201 of ORF29p were previously described (46). A mouse monoclonal antibody to VZV gE was purchased from ViroStat (Portland, Maine). Alexa Fluor 488-conjugated goat anti-mouse and Alexa Fluor 546 anti-rabbit antibodies were purchased from Molecular Probes (Carlsbad, CA). Goat anti-rabbit antibody conjugated to horseradish peroxidase for immunoblotting was purchased from KPL (Gaithersburg, MD).
Fluorescence microscopy. (i) Indirect IF microscopy.
Indirect immunofluorescence (IF) microscopy was performed as follows. Cells on glass coverslips were washed once with PBS, fixed for 25 min with 3.7% formaldehyde in PBS, washed two more times in PBS, and permeabilized with ice-cold acetone at −20°C for 10 min. Cells were washed twice more in PBS and blocked with 10% normal goat serum (Roche, Indianapolis, IN) in PBS-T (PBS plus 0.1% Tween 20 [Sigma, St. Louis, MO]) for 30 min. Cells were incubated with a 1:100 dilution of the appropriate primary antibody in 1% normal goat serum in PBS-T for 1 h and then washed three times for 5 min each in PBS-T. Cells were then incubated for 1 h with the appropriate Alexa Fluor-conjugated secondary antibody diluted 1:1,000 in 1% normal goat serum in PBS-T, washed once for 5 min with PBS-T, once for 10 min with PBS-T plus 0.5 μg/ml Hoechst 33258 (Sigma), and once again for 5 min in PBS-T. The coverslips were mounted with GEL/MOUNT (Biomeda, Foster City, CA).
(ii) Direct fluorescence microscopy.
293T cells on coverslips were transfected with plasmids expressing GFP fusion proteins, and the cells were washed with PBS and fixed for 25 min with 3.7% formaldehyde in PBS. The cells were then washed once with PBS, washed for 10 min with PBS plus 0.5 μg/ml Hoechst 33258, washed once more for 5 min with PBS, and then mounted on glass slides with GEL/MOUNT.
All samples were visualized with a Zeiss Axiovert 200 M inverted microscope, and images were acquired with a Zeiss Axiocam (Carl Zeiss Microimaging Inc., Thornwood, NY) using Openlab 3.1 software (Improvision, Lexington, MA). Images were assembled with Adobe Photoshop and Illustrator (Adobe Systems Inc., San Jose, CA.).
Plasmids and cloning. (i) Amplification of ORF29 from wild-type VZV.
Full-length ORF29 was amplified by PCR using the oligonucleotides ORF29ES5′PRM.ECO (5′-GGGAATTCGATGGAAAATACTCAGAAGACTG-3′) and ORF29ES3′PRM.NOT (5′-CCCGCGGCCGCCATTTCCATTGTAATGTTCCC-3′) (Gibco-BRL), the Pfu Turbo DNA polymerase (Stratagene, La Jolla, CA) and VZV strain Jones DNA as a template. Pfu Turbo DNA polymerase was used for all PCRs in this report unless otherwise noted, and all other primers used were manufactured by Proligo LLC (Boulder, Co). Viral DNA was prepared by lysing infected MeWo cells in proteinase K cocktail (100 mM NaCl, 10 mM EDTA, 50 mM Tris-HCl, pH 7.5, 0.5% SDS, 200 μg/ml proteinase K) at 50°C for 3 h. Saturated NaCl was added to the cell lysates to 35% of the total volume, and the mixture was incubated at 4°C for 10 min. Cell debris was pelleted at 22,500 × g for 10 min at 4°C in a Tomy MX-160 high-speed refrigerated microcentrifuge. VZV DNA was precipitated from the lysates with 0.7 volume of isopropanol, washed with 70% ethanol, and resuspended in Tris-EDTA (TE; 10 mM Tris-HCl, 1 mM EDTA, pH 7.9). The ORF29 PCR product was cloned into the pZErO-2.1 vector (Invitrogen) at the EcoRV site to construct pZErO29. ORF29 was then excised with EcoRI and NotI from pZErO29 and subcloned into the EcoRI and NotI sites of the pTriEx-1 vector (Novagen, Madison, WI) to generate p29-12, a chicken actin promoter-driven ORF29p expression construct.
(ii) ORF29 GFP fusion constructs.
A vector expressing the wild-type ORF29p fused to GFP, p29GFP, was generated by amplifying p29-12 with ORF29ES5′PRM.ECO and 29BamHI3′ (5′-GGGGGATCCATTTCCATTGTAATGTTCCCATGTTT-3′), cleaving with EcoRI and BamHI, and subcloning into linearized pEGFP-N1 (Clontech, Palo Alto, CA). A deletion in ORF29 of nucleotides (nt) 459 to 821 was made by digesting pZErO29 with ScaI and SspI and religating the backbone to yield pZErO29ΔNLS. The deletion mutant was then subcloned into pTriEX-1 (p29-12ΔNLS) at the EcoRI and NotI sites. As done with the wild- type ORF29, p29-12ΔNLS was amplified with oligonucleotides ORF29ES5′PRM.ECO and 29BamHI3′ and subsequently cloned into pEGFPN1 (p29ΔNLSGFP) using the same restriction enzyme sites. p1-152GFP was constructed by excising the corresponding nucleotides from p29-12 with EcoRI and SspI and subcloning the fragment into the EcoRI and PmlI sites of pEGFP-N1. p346-1203GFP, p155-1203GFP, p10-1023GFP, and p9-1203GFP were created by amplifying the corresponding nucleotides from p29-12 with 29nt1036EcoRI (5′-GGGGAATTCATGATCAAAGAATGGCCAATGTTTATAGGC-3′), 29nt463EcoRI (5′-GGGGAATTCATGTACTTGGTGGTCACGGCATTGTTTAAG-3′), 29nt28EcoRI (5′-CCGAATTCATGGTGCCCACGGGTCCCCTGG-3′), or 29nt25EcoRI (5′-CCGAATTCATGACAGTGCCCACGGGTCCCCT-3′), respectively, and oligonucleotide ORF29ES3′PRM.NOT. Note that for each of the preceding constructs an in-frame ATG is encoded within the 5′ oligonucleotide used to generate the GFP fusion constructs. The amplification products were each digested with EcoRI and NotI for cloning into pTriEx. The ORF29 fragments were then released with EcoRI and PmlI and subcloned in pEGFP-N1 at the EcoRI and SmaI sites. To construct p1-345GFP and p1-154GFP, ORF29 nucleotides 1 to 1035 and 1 to 462 were amplified from the p29-12 template by PCR using the primers ORF29ES5′PRM.ECO and either 29nt1036BamHI (5′-GGGGGATCCTATCTGTTGGAGTTTCTTCGTAAAT-3′) or 29nt463BamHI (5′-GGGGGATCCATAATATTGTATTTTCTGGCTCTAA-3′), respectively. ORF29 DNA for cloning p9-154GFP and p10-154GFP was amplified from p29-12 with primers 29nt463BamHI and either 29nt25EcoRI or 29nt28EcoRI, respectively. The PCR products were digested with EcoRI and BamHI and cloned into pEGFP-N1 at the corresponding restriction enzyme sites in the vector.
(iii) pET-21b and pGEX6p3 clones.
pET29 was constructed for in vitro transcription translation experiments by subcloning the ORF29 EcoRI-NotI fragment released from p29-12 into the pET-21b vector at the same restriction enzyme sites (Novagen, San Diego, CA). pET291-345 and pET291-345A35P were constructed by amplification of p29-12 or p29A35P, respectively, with oligonucleotides ORF29ES5′PRM.ECO and 29nt1035SalI (5′-GGGGTCGACATCTGTTGGAGTTTCTTCGTAAATT-3′). The DNA products were cleaved with EcoRI and SalI for cloning into pET-21b.
pGST29GFP was constructed by cleaving p29GFP with EcoRI and NotI and ligating the released fragment into the same sites in pGEX-6P-3 (Amersham Biosciences, Piscataway, NJ). pGST29GFP was cleaved with SmaI and NotI, and the fragment containing ORF29 nt 2366 to 3612 and the enhanced GFP sequence was subcloned into the SmaI and NotI sites of pGEX-6P-3 to construct pGST29ΔNLSGFP. All vector inserts were verified by DNA sequencing.
Point mutagenesis.
To generate a DNA pool containing random point mutations within ORF29 nucleotides 1 to 1035, 60 ng of p29-12 was used as the template in a 50-μl amplification reaction containing 3 U Taq polymerase, a depleted level of 2 nucleotides (for example, 1 mM dATP, 1 mM dGTP, 0.2 mM dCTP, and 0.2 mM dTTP), 3 mM MgCl2, 0.5 mM MnCl2, 5 μl 10× ThermoPol buffer (NEB, Beverly, MA), 2 μM oligonucleotide 29EcoRV5′ (5′-GGGCAAAGGAGATATACCATGGCGATA-3′), and 2 μM 29EcoRV3′ (5′-GGGTGGCCATTCTTTGATATCTGTTGG-3′). Seventeen PCR cycles were performed before loading the PCR product onto a 1% agarose gel for purification. The low fidelity of the Taq polymerase under these conditions yielded a pool of point mutants. The mixed pool of amplified DNAs was digested with EcoRV and cloned back into p29-12 at this restriction enzyme site. After ligation, vector DNA was transformed into TOP10 F− cells and colonies were picked to purify and isolate individual clones. The orientation of 300 potential ORF29 point mutants was tested by restriction fragment length polymorphism analysis with the SacII restriction enzyme. Plasmids containing nucleotides 1 to 1035 in the proper orientation were transfected into 293T cells and screened for subcellular distribution of ORF29p by indirect immunofluorescence. All plasmids that expressed cytoplasmic ORF29p and some expressing ORF29p in the nucleus were analyzed by DNA sequencing.
Site-directed mutagenesis was utilized to revert the mutation in p29A35P back to the wild-type sequence. PCR was performed on p29A35P with either A35Ptop (5′-GGGTTTTTGGCCGCTCGTAGCACG-3′) and 29EcoRV3′ or 29EcoRV5′ and A35Pbot (5′-GGGCGTGCTACGAGCGGCCAAAAA-3′) primers. The resulting DNA fragments were combined in a third PCR with oligonucleotides 29EcoRV5′ and 29EcoRV3′. The 1,035-nucleotide product from this amplification reaction was digested with EcoRV, subcloned into p29-12 at the corresponding restriction enzyme site, and confirmed by restriction enzyme digestion and sequencing to contain the ORF29 wild-type sequence.
ORF29p aa 1 to 345 regions containing the A35P and C236S point mutations were expressed as GFP fusions after amplification of p29A35P and p29C236S, respectively, with primers ORF29ES5′PRM.ECO and 29nt1036BamHI. The PCR products were digested with EcoRI and BamHI and cloned into pEGFP-N1.
SDS-PAGE and Western blotting.
Each well of a six-well culture dish of infected or transfected 293T cells was harvested, washed twice with PBS, and resuspended in 100 μl of radioimmunoprecipitation lysis buffer (10 mM Na2HPO4, pH 7.2, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) plus Complete protease inhibitor cocktail (Roche, Mannheim, Germany) and incubated on ice for 30 min. Cell extracts were clarified by centrifugation at 22,500 × g for 5 min at 4°C in a Tomy MX-160 high-speed refrigerated microcentrifuge. Total protein concentration was measured using the Bio-Rad protein assay (Bio-Rad, Hercules, CA). Twenty microliters of 6× SDS sample buffer (300 mM Tris-Cl, pH 6.8, 12% SDS, 0.6% bromophenol blue, 60% glycerol, 600 mM β-mercaptoethanol) was added to the samples prior to boiling them for 10 min and subjecting them to SDS-PAGE. The proteins were transferred to a nitrocellulose membrane with a Bio-Rad Trans-Blot Semi-Dry apparatus before Western blotting. After blocking the membrane in 4% nonfat milk in PBS-T, immobilized proteins were reacted with the ORF29p antibody at a 1:1,000 dilution in blocking solution. The membrane was washed three times in PBS-T, incubated with an anti-rabbit antibody conjugated to horseradish peroxidase, and washed again, and ORF29p was visualized by addition of the LumiGLO substrate (KPL) and exposure to X-ray film.
Preparation of recombinant human nuclear import factors.
Plasmids expressing GST fusions to human kap α2 and human kap β1 were kindly provided by Tarik Solomon and the Gunter Blobel laboratory at Rockefeller University, New York, NY (8, 14, 44, 52, 72). GST-tagged kap α2, kap β1, an ORF29p-GFP fusion (from pGST29GFP), an ORF29p NLS mutant fused to GFP (from pGST29ΔNLSGFP), and GST alone (from pGEX-6P-3) were expressed in Escherichia coli BL21 (DE3) for 4 h at 30°C after induction with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The soluble GST-tagged proteins were purified in their native state on glutathione agarose after incubation with GST lysis buffer (1 mM phenylmethylsulfonyl fluoride, 10 mM β-mercaptoethanol, 0.2 mg/ml Lysozyme in PBS) and sonication for 1 min to break open the bacteria. Purified ORF29p-GFP and ORF29pΔNLSGFP were obtained by site-specific cleavage with PreScission protease according to the manufacturer's instructions (Amersham Biosciences). Protein concentration was measured by the Bio-Rad Protein assay. The cleaved proteins were then concentrated and desalted using a Microcon centrifugal filter device (Millipore, Bedford, MA). All of the proteins were analyzed for purity and size by SDS-PAGE and Coomassie blue staining.
In vitro translation and interaction with GST fusion proteins.
[35S]Met-labeled mouse kap α2, ORF29p, ORF29p aa1-345, and ORF29p aa1-345A35P were synthesized by coupled in vitro transcription and translation (Promega, Madison, WI) with pET-21b-kapα2, pET29, pET291-345, and pET291-345A35P as templates, respectively (pET-21b-kapα2 described in references 44, 52, and 72). Five microliters of [35S]Met-labeled product was incubated with 40 μl of either GST alone, GST-tagged human kap α2, or human kap β1 bound to glutathione agarose beads for 1 h at 4°C in 100 μl of binding buffer (20 mM Tris-HCl, pH 7.5, 50 mM KCl, 0.5 mM EDTA, 1% NP40, 1 mM dithiothreitol [DTT], 3% bovine serum albumin). The bead-associated proteins were washed four times with binding buffer supplemented with 150 mM KCl. The bound products were resuspended in 50 μl PBS and 10 μl 6× SDS sample buffer, boiled for 10 min, and separated by SDS-PAGE before being exposed to film.
In vitro nuclear import assay.
In vitro nuclear import assays in digitonin-permeabilized HeLa cells were carried out as previously described (2, 3, 8, 50, 56). Briefly, HeLa cells were plated onto glass coverslips 1 day before being permeabilized with 35 μg/ml digitonin in transport buffer (20 mM HEPES, pH 7.4, 110 mM KOAc, 2 mM MgCl2, 1 mM DTT, 1× Complete protease inhibitor cocktail) for 5 min on ice. The permeabilized cells were washed three times with transport buffer and incubated for 30 min at room temperature with 2 μM of either recombinant ORF29p-GFP, ORF29pΔNLSGFP, or GST-SV40-NLS-GFP in a mixture of nuclear import factors. The import mixture contained one of the following combinations: (i) HeLa cytosol, 1 mg/ml BSA, 0.5 mM GTP, an energy-regenerating system (2 mM ATP, 25 mM phosphocreatine, 30 U/ml creatine phosphokinase), and 8 μM Ran; (ii) BSA, GTP, an energy-regenerating system, Ran, 2 μM kap α2, and 2 μM kap β1; (iii) BSA, kap α2, and kap β1; (iv) BSA; or (v) BSA, GTP, an energy-regenerating system, Ran, kap α2, kap β1, and 500 μg/ml wheat germ agglutinin (WGA). The final reaction volume was adjusted to 20 μl with transport buffer. Direct fluorescence microscopy was used to visualize protein nuclear import. HeLa cytosol and recombinant GST-SV40-NLS-GFP, Ran, kap α2, and kap β1 were kindly provided by Tarik Solomon and the Gunter Blobel laboratory (Rockefeller University, New York, NY).
RESULTS
Localization of ORF29p in infected and transfected cells.
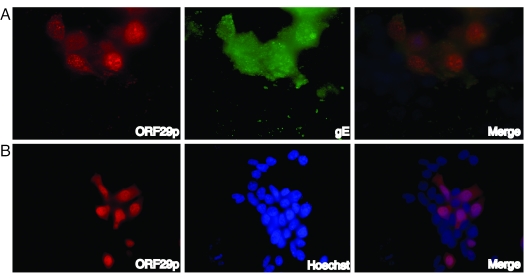
Previous studies have demonstrated that in VZV-infected fibroblasts ORF29p is present primarily in the nucleus, with diffuse cytoplasmic staining (38). To confirm these data, 293T cells were infected with cell-free VZV at a multiplicity of infection of 1. The intracellular localization of ORF29p was examined at 48 h postinfection by IF microscopy using a rabbit antiserum prepared against the C-terminal 115 amino acids of ORF29p (Fig. 1A). The nuclei in these and all subsequent localization experiments were visualized by counterstaining with Hoechst 33258. As previously described, ORF29p staining was predominantly nuclear with some cytoplasmic staining. The nuclear staining was punctate, which is similar to the expression pattern of ICP8, the HSV-1 homologue of ORF29p. These punctate regions in infected cells are probably areas of viral DNA replication and transcription as other replication and regulatory proteins colocalize there during HSV-1 infection (12, 24, 39, 59, 60, 62, 63, 73). These data show that 293T cells are permissive for VZV replication by demonstrating the expression of the early protein ORF29p and the late protein gE. We next asked where ORF29p accumulates when expressed in the absence of other virus-specified proteins. Figure 1 shows that ORF29p localizes predominantly to the nucleus when 293T cells are transfected with a plasmid DNA that expresses this protein (p29-12) under a chicken actin promoter. In contrast to infected cells, when autonomously expressed, the nuclear staining in transfected cells was more diffuse. This probably results from the absence of VZV replication complexes. These data demonstrate that expression of other VZV-specified proteins is not required for the nuclear import of ORF29p.
FIG. 1.
Cellular localization of ORF29p. 293T cells were (A) infected with VZV at an MOI of 1 or (B) were transiently transfected with p29-12, a DNA construct expressing ORF29p from a chicken actin promoter. Two days postinfection or posttransfection, cells were fixed and the localization of ORF29p and gE were examined by indirect immunofluorescence microscopy. Nuclei were counterstained with Hoechst 33258. Images were collected as described in Materials and Methods.
Western blot analysis confirmed that the ORF29p species derived from cell cultures infected with virus or transfected with plasmid DNA encoding ORF29p had identical electrophoretic mobility (data not shown).
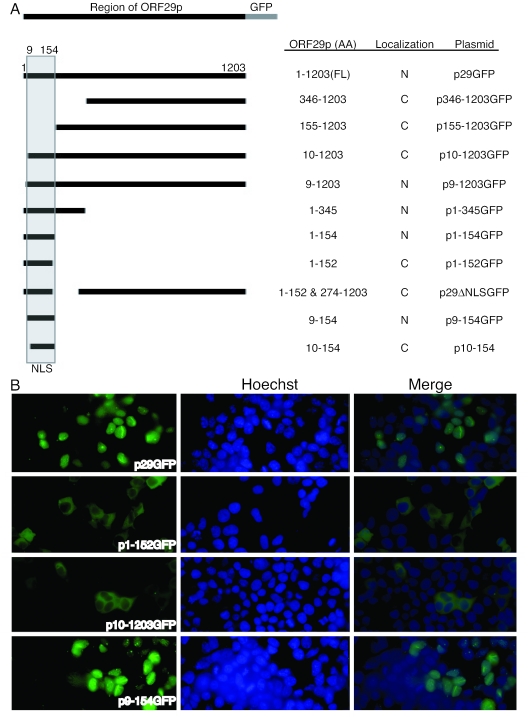
Amino acids 9 to 154 of VZV ORF29p are necessary for nuclear import and are sufficient for targeting GFP to the nucleus.
Sequence analysis of ORF29p using the Predict Protein database (http://cubic.bioc.columbia.edu/predictprotein) suggests that the protein lacks a classical NLS or any homology to the amino acid sequences that are used by ICP8 for nuclear targeting (66, 71). To determine the region(s) of ORF29p necessary for nuclear import, deletions were made in the ORF29 coding sequence using either PCR amplification or by cleavage and ligation at convenient restriction endonuclease sites. These mutant genes were expressed as GFP fusions after transfecting 293T cells (Fig. 2A). All fusions were to the C terminus of ORF29p because N-terminal fusions yielded irregular vesicular staining, possibly due to instability and degradation of the chimeric protein. Plasmid DNAs encoding the various deletion mutants of ORF29 were transfected into 293T cells, and the subcellular localization of the GFP fusion proteins was determined by direct fluorescence microscopy. Figure 2A summarizes the deletion mutants and identifies the subcellular localization of the truncated ORF29 proteins that accumulate in transfected 293T cells. Our analyses demonstrate that while full-length ORF29p targets GFP to the nuclei of transfected 293T cells, deletion of amino acids 9 to 154 abrogated the nuclear import of ORF29p (Fig. 2B). In support of this conclusion, we demonstrated that a DNA construct encoding amino acids 9 to 154 was able to target GFP to the nuclei of transfected cells whereas a construct expressing amino acids 10 to 154 fused to GFP was not (Fig. 2). This result suggests a requirement for the Thr moiety at position 9 for nuclear transport. SDS-PAGE and Western blotting were used to confirm the size and abundance of the fusion proteins in the transfected cells (data not shown). The deletion mutation analysis reveals that amino acids 9 to 154 contain a nuclear targeting sequence that is both necessary for ORF29p nuclear import and sufficient for targeting a GFP fusion protein to the nucleus. The size of this region is typical of either bipartite or structurally dependent nuclear import signals (33, 57).
FIG. 2.
Deletion mutagenesis of ORF29. Truncations of the sequence encoding ORF29p were fused to GFP at their C termini. (A) The schematic illustrates the amino acids (AA) of ORF29p that were fused to the GFP tag and the subcellular localization of the fusion protein: N for nuclear and C for cytoplasmic. (B) To determine the subcellular location, deletion mutant constructs were transiently transfected into 293T cells and the intracellular localization of GFP was visualized by direct fluorescence microscopy at 48 h posttransfection. Nuclei were stained with Hoechst 333258.
Point and insertion mutations between amino acids 35 and 245 interfere with ORF29p NLS activity in transfected 293T cells.
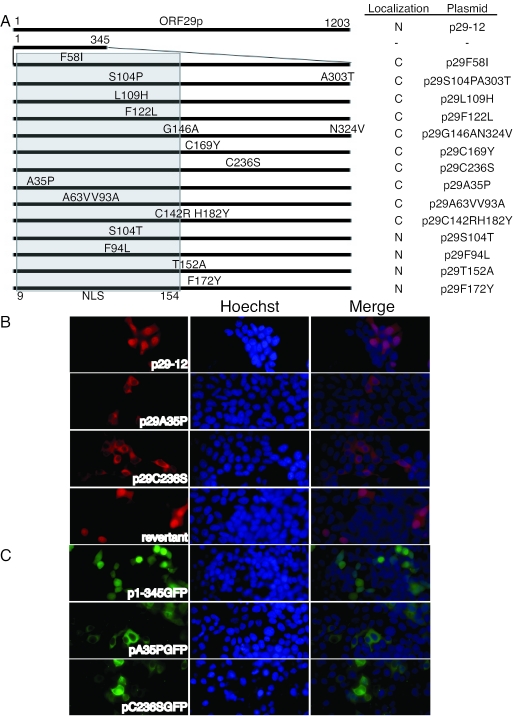
Random point mutagenesis of ORF29 (1 to 1,035 bp) was performed to identify the regions and residues that are necessary for nuclear import. This region of ORF29 was amplified using error-prone PCR as described in Materials and Methods. The low fidelity of the Taq polymerase under these conditions yielded a pool of point mutants. This DNA pool was then cloned back into a cassette containing the rest of the ORF29 to reconstruct the full-length ORF29 gene. Individual clones were isolated, transfected into 293T cells, and screened for the cellular localization of ORF29p by indirect IF. All clones that encoded protein which localized to the cytoplasm were sequenced along with some that encoded proteins that localized to the nucleus. Figure 3A identifies the residues that were altered in each clone and summarizes the intracellular distribution of the protein they encode. Single-point mutations A35P and C236S restricted ORF29p to the cytoplasm of transfected cells (Fig. 3B). The point mutations that abolished ORF29p nuclear localization resided both within and downstream of the nuclear targeting domain that was characterized by deletion mapping (Fig. 3A).
FIG. 3.
Point mutagenesis of the ORF29 NLS. (A) ORF29 nucleotides 1 to 1035 were amplified using error-prone PCR and Taq polymerase to generate a pool of random point mutants. Mutated fragments were cloned so as to reconstruct the full-length ORF29 gene. The schematic illustrates the amino acid changes in each construct and the subcellular localization of the full-length protein: N for nuclear and C for cytoplasmic. (B) To determine the subcellular location, plasmid DNAs encoding the point mutants were transiently transfected into 293T cells and protein localization was visualized by indirect immunofluorescence microscopy with an ORF29p-specific antiserum at 48 h posttransfection. The point mutant A35P was reverted to wild type by site-directed mutagenesis. Localization of the protein encoded by the revertant was analyzed by indirect immunofluorescence. (C) Individual mutated DNAs were also subcloned into a C-terminal GFP fusion construct and transfected into 293T cells to visualize localization by direct fluorescence microscopy. All nuclei were stained with Hoechst 333258.
To verify that the A35P mutation was responsible for inhibiting nuclear import of ORF29p and that this did not result from a secondary mutation or cloning error, this mutant was repaired and the cellular localization of the protein it encoded was determined by indirect IF after transfection of 293T cells. The protein encoded by the revertant targeted to the nuclei of transfected cells just like wild-type ORF29p (Fig. 3A). This proves that a single-residue change at amino acid 35 is sufficient to abrogate nuclear transport.
The ORF29p fragments containing the A35P and C236S amino acid substitutions were fused at their C termini to a GFP tag to see if these alterations affected nuclear import. The DNA constructs encoding these chimeric proteins were transfected into 293T cells, and the cellular distribution of their proteins was observed by direct fluorescence microscopy (Fig. 3B). The GFP signal from the fusion proteins containing mutant NLSs was cytoplasmic. Thus, point mutations within and adjacent to the ORF29p nuclear targeting domain inhibit ORF29p nuclear import and abolish the NLS activity when fused to GFP.
Transposon-based linker insertional mutagenesis of the 5′ 1,035 nucleotides of ORF29 was also performed to identify other critical residues involved in ORF29p nuclear import. The resulting mutated DNA fragments were cloned into a cassette containing the rest of the ORF29 gene to reconstruct the full-length coding sequence. Clones containing insertions were then analyzed for the subcellular localization of ORF29p. All insertions that fell within the region containing the point mutations (aa 35 to 236) studied previously resulted in cytoplasmic localization (data not shown). Western blot analysis was performed on the products encoded by the mutant genes to confirm that proteins of the predicted molecular weight were being made (data not shown).
The results from the point and insertion mutagenesis analyses support our finding from deletion mapping studies that amino acids 9 to 154 of ORF29p contain a functional NLS. The size of this nuclear targeting region and the sensitivity of its import activity to sequence changes within and adjacent to amino acids 9 to 154 suggest that the ORF29p NLS is either bipartite or a large structurally dependent domain (33, 57).
Karyopherins α2 and β1 are sufficient and necessary for ORF29p in vitro nuclear import.
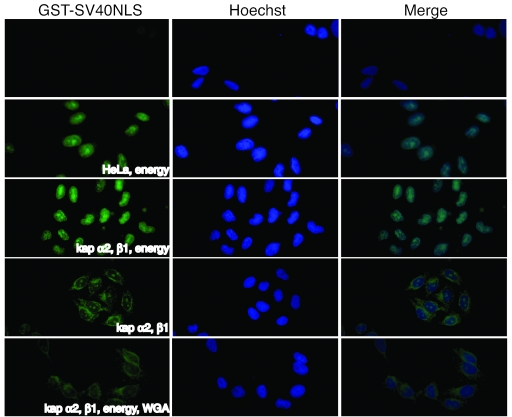
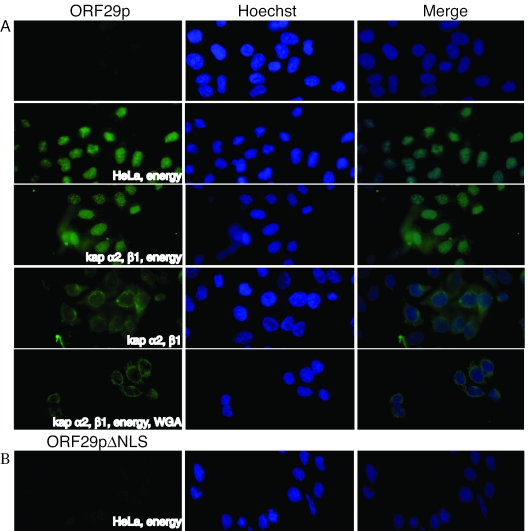
Although ORF29p encodes a nonclassical NLS, we wanted to determine if it used the karyopherins to enter the nucleus, possibly through novel interactions or by bridging. To test whether kaps α2 and β1 were sufficient and necessary for ORF29p nuclear translocation, nuclear import assays in digitonin-permeabilized HeLa cells were performed. This assay, in which digitonin permeabilizes the plasma membrane but leaves the nuclear envelope intact, has been previously used to investigate nuclear transport pathways (1, 8). Digitonin-treated HeLa cells retain import-competent nuclei but are depleted of most cytosolic factors; therefore, various transport molecules may be added to determine if they are involved in a particular nuclear import pathway. Permeabilized cells were incubated with either ORF29p-GFP or SV40-NLS-GFP in the presence of an energy-regenerating system, GTP, Ran, and either HeLa cytosol extract or recombinant mouse kap α2 and human kap β1. The mouse kap α2 is 98% identical at the amino acid level to its human homologue and has been shown to recognize the same substrates (26, 42, 72). The localizations of SV40-NLS-GFP and ORF29p-GFP were determined by direct fluorescence microscopy (Fig. 4 and 5A). As expected, the molecule harboring the canonical SV40-NLS was transported into the nucleus using the classical import machinery. Likewise, ORF29p-GFP was transported into the nuclei of permeabilized cells in the presence of total cytosolic protein and also when combined with only kap α2, kap β1, and Ran.
FIG. 4.
In vitro nuclear import of an SV40-NLS-GFP fusion protein. HeLa cells grown on glass coverslips were permeabilized with digitonin and then incubated for 30 min with 2 μM of GFP fused to GST-SV40-NLS in the presence or absence of 2 μM kap α2, 2 μM kap β1, HeLa cytosolic extract, 500 μg/ml WGA, and an energy reconstitution system (500 μM GTP, 8 mM Ran, 2 μM ATP, 25 mM phosphocreatine, 30 U/ml creatine phosphokinase) as indicated. The permeabilized HeLa cells were fixed, and fusion proteins were visualized by direct fluorescence microscopy. Nuclei were stained with Hoechst 333258.
FIG. 5.
In vitro nuclear import of ORF29p-GFP fusion proteins. HeLa cells grown on coverslips were permeabilized with digitonin and then incubated for 30 min with 2 μM of GFP fused to either (A) ORF29p or (B) ORF29p aa 789 to 1203 (ORF29pΔNLS) in the presence or absence of 2 μM kap α2, 2 μM kap β1, HeLa cytosolic extract, 500 μg/ml WGA, and an energy reconstitution system (500 μM GTP, 8 μM Ran, 2 mM ATP, 25 mM phosphocreatine, 30 U/ml creatine phosphokinase) as indicated. Permeabilized HeLa cells were fixed, and fusion proteins were visualized by direct fluorescence microscopy. Nuclei were stained with Hoechst 333258.
To test the specificity of ORF29p nuclear translocation, the in vitro import assays were repeated in the absence of the energy reconstitution system or in the presence of wheat germ agglutinin (WGA) (Fig. 4 and 5A). WGA inhibits nuclear transport by binding to the nucleoporins. The kaps α2 and β1 retain the ability to bind the nucleoporins during energy depletion or WGA treatment but will not translocate through the pore complex (74). Therefore, in the presence of WGA, the importins and any binding partners accumulate at the periphery of the nuclear membrane and exhibit a nuclear rim-staining pattern. Under either of these inhibitory conditions, both SV40-NLS-GFP and ORF29p-GFP accumulate at the nuclear membrane. This observation supports the hypothesis that kaps α2 and β1 are involved in targeting ORF29p to the nucleus in a Ran-dependent manner.
To confirm that the ORF29p NLS we identified in the mutagenesis studies is responsible for protein translocation during the in vitro import assays, the experiments with digitonin-permeabilized HeLa cells were repeated using an ORF29 NLS deletion mutant encoding amino acids 789 to 1203 fused to GFP (Fig. 5B). Under all conditions, the NLS mutant was unable to be translocated to the nucleus. The absence of any fluorescent signal above background indicated that the NLS mutant was not only defective for nuclear import but probably also for binding the importins.
Our results with ORF29p mimic what is seen when an SV40-NLS-GFP fusion molecule is tested in similar in vitro import assays (Fig. 4 and 5). However, the genetic and computational analyses indicate that ORF29p nuclear transport utilizes a nonclassical NLS. Therefore, ORF29p may be hijacking the same cellular importin system that is utilized by canonical nuclear targeting motifs through a novel NLS interaction.
ORF29p amino acids 1 to 345 interact with human karyopherin α2.
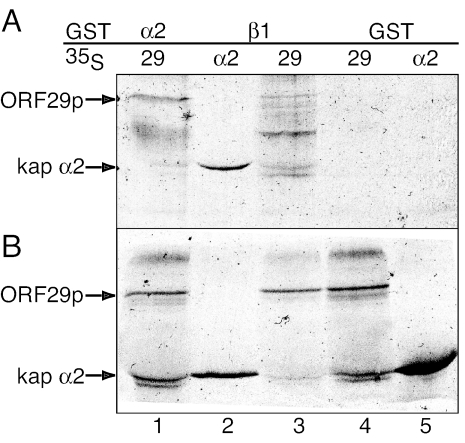
The ability of ORF29p to be targeted to the nuclei of digitonin-permeabilized HeLa cells by only kaps α2 and β1, Ran, and an energy-regenerating system suggests that ORF29p is interacting either directly or indirectly with one or both of the importins. Nuclear rim localization of ORF29p following energy depletion or WGA treatment supports the hypothesis that ORF29p is interacting with the karyopherins and is thus being sequestered at the nuclear pore under these conditions. We therefore asked if ORF29p interacted with the karyopherins by using in vitro binding assays. GST-kap α2, GST-kap β1, and GST immobilized on glutathione Sepharose were incubated with radiolabeled ORF29p or kap α2 generated from coupled in vitro transcription/translation reactions. The bound proteins were subjected to SDS-PAGE, and the gel was exposed to film (Fig. 6A). Ten percent of the unbound protein volume was also analyzed by SDS-PAGE to confirm the presence of labeled protein product in each sample (Fig. 6B). Kap α2 and kap β1 are known to interact, and the interaction of [35S]Met-labeled kap α2 with GST-kap β1 served as a positive control in this experiment. Figure 6A demonstrates that ORF29p interacted with kap α2, but not with GST-kap β1 or GST alone.
FIG. 6.
Interaction of karyopherins with in vitro-synthesized ORF29p. (Lanes 1, 3, and 4) ORF29p and (lanes 2 and 5) kap α2 were synthesized in a rabbit reticulocyte system in the presence of [35S]Met and mixed with either (lanes 4 and 5) GST alone or (lanes 1 to 3) GST-kap fusion proteins that were expressed in E. coli BL21 cells and bound to glutathione beads. (A) Bound proteins and (B) in vitro-translated protein inputs were subjected to SDS-PAGE analysis on a 4 to 12% gradient gel and exposed to film.
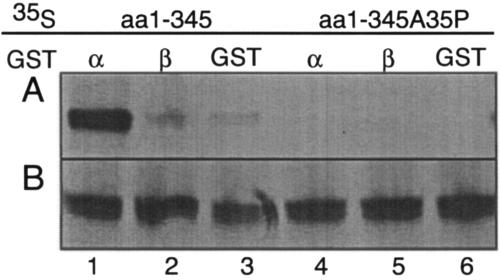
We then asked if kap α2 interacted with the putative NLS of ORF29p. GST-kap α2, GST-kap β1, and GST immobilized on glutathione Sepharose were incubated with radiolabeled ORF29p amino acids 1 to 345. The bound proteins were subjected to SDS-PAGE and visualized by autoradiography (Fig. 7A). As seen with the full-length protein, ORF29p amino acids 1 to 345 interacted with GST-kap α2, but not GST-kap β1 or GST. The interaction of kap α with the full-length ORF29p as well as with aa 1 to 345 was stable to washes with salt concentrations as high as 250 μM. To confirm the specificity of this interaction and its requirement for the ORF29p nuclear targeting domain, the NLS point mutant A35P was tested for interaction with GST-kap α2. This protein failed to interact (Fig. 7A). Figure 7B shows that ORF29p was present in all samples. These data strongly suggest that the novel ORF29p NLS functions by interacting with karyopherin α and that the residues involved in nuclear import are also required for this interaction.
FIG. 7.
Interaction of the ORF29p NLS with karyopherins. (Lanes 1 to 3) ORF29p aa1-345 and (lanes 4 to 6) ORF29p aa1-345A35P were synthesized in a rabbit reticulocyte system in the presence of [35S]Met and mixed with either (lanes 3 and 6) GST alone or (lanes 1 and 2 and 4 and 5) GST-karyopherin fusion proteins that were expressed in BL21 cells and bound to glutathione beads. (A) Bound proteins and (B) in vitro-translated protein inputs were subjected to SDS-PAGE analysis on a 4 to 12% gradient gel and exposed to film.
DISCUSSION
ORF29p localizes predominantly to the nucleus during productive infection (38, 46). During latent infection of the DRG and EG, ORF29p is found exclusively in the cytoplasm, and following VZV reactivation, the protein accumulates in the nucleus (13, 46). Therefore, the subcellular distribution of ORF29p correlates with the type of VZV infection. Identification of the pathways involved in this protein's differential localization is hampered by the absence of canonical nuclear import, export, and shuttling motifs within the ORF29p sequence. ORF29p also shows no similarity to the nuclear targeting signal of its homologue, HSV-1 ICP8 (38, 71). The possibility that the mechanisms involved in ORF29p and ICP8 subcellular segregation have diverged is not surprising as their expression and localization patterns differ throughout their respective infectious cycles. Both proteins are primarily nuclear during productive infection, but in contrast to ORF29p, ICP8 is not expressed during latency (11, 19, 38, 61, 65). The binding partners of these homologues have deviated as well. Whereas ICP8 interacts with UL37 and plays a role in UL37's nuclear import, ORF29p does not associate with the UL37 VZV homologue, ORF21p (4, 19, 69). Our study was designed to map the ORF29p nuclear import signal and to determine how it works in an effort to elucidate what controls the switch between VZV lytic infection and latency.
In this report, we have identified a novel nuclear targeting domain within the amino acid sequence of ORF29p and investigated the possible pathways involved. We show that ORF29p nuclear import occurs in the absence of other viral proteins. Because all facilitated nuclear import involves protein interactions with either importins or bridge molecules that in turn bind the cellular importins, ORF29p must associate with a cell-encoded factor. There are, however, no previous accounts of ORF29p interactions with host proteins.
The ORF29p NLS maps between amino acids 9 and 154, and mutational analysis has demonstrated this sequence to be both sufficient and necessary for nuclear targeting. The size of this domain and its sensitivity to mutations throughout the region demonstrate that this NLS is not comprised of a simple linear array of amino acids; rather it is characteristic of a bipartite signal or a signal that is assembled as a result of protein folding (33, 57). Deletion, point, and insertion mutations both within and adjacent to amino acids 9 to 154 are able to abrogate the nuclear targeting activity. This is additional evidence that this signaling sequence arises in response to protein folding, as has been demonstrated for NF-κB (7, 30, 75). The NLS may constitute a specific structure, or the conformation of the residues surrounding the interaction domain may affect the accessibility of the targeting sequence (32, 68).
To determine if ORF29p amino acids 1 to 345 encoded a structure that previously was implicated in nuclear import, the sequence was submitted for analysis to the Predict Protein database to examine the predicted secondary structure of the NLS and its adjacent residues (66). The output for the N-terminal 345 amino acids was aligned with the characterized point mutants, and no obvious correlation between secondary structure, location of the mutations, and known protein structures involved in nuclear import was detected (data not shown). This lack of correlation is, however, inconclusive without knowing the actual structure of ORF29p.
In vitro import assays in permeabilized HeLa cells revealed that mouse karyopherin α2 and human karyopherin β1 are sufficient and necessary for ORF29p nuclear targeting. The pathway involved is Ran dependent and sensitive to wheat germ agglutinin. These characteristics are all biochemical hallmarks of the classical nuclear import system where karyopherin α binds the basic residues in a traditional NLS via its central area and karyopherin β with its N terminus. Karyopherin β promotes the interaction of the resultant heterotrimer with the NPC and subsequent nuclear translocation (20, 25, 27, 31, 53). The ORF29p NLS described here is noncanonical, and it appears to hijack a conserved system within the host cell. Translocation in vitro and in vivo requires the ORF29p NLS in a functional form, as NLS mutants were not able to target to the nuclear compartment. Although this karyopherin α/β-dependent import occurred in the absence of other cytosolic factors, it remains possible that other nuclear/cytoplasmic shuttling molecules are available for ORF29p interaction and bridging to the importins. Moreover, it is conceivable that in the context of a productive infection other virus-specified proteins could act as charons to ferry ORF29p to the nucleus.
The involvement of karyopherin α in ORF29p nuclear import was further supported by the in vitro interaction of a GST-human karyopherin α2 fusion and [35S]Met-labeled ORF29p. These experiments still do not distinguish between ORF29p interacting directly or indirectly with karyopherin α or with bridge molecules in the rabbit reticulocyte extract. A region harboring the ORF29p NLS, amino acids 1 to 345, also bound karyopherin α, while a point mutant capable of abrogating the NLS activity did not. These data demonstrate that the inability of the NLS mutant to translocate coincides with its failure to associate with the karyopherins. This conclusion is supported by in vitro import assays that reveal that the ORF29p NLS mutant fails to accumulate at the nuclear pore with karyopherin α and β following energy depletion and WGA.
The VZV infectious cycle is cell type specific in that invasion of the epidermis and dermis results in a lytic infection, while infection of neurons and satellite cells of the sensory ganglia leads to latency (5, 13, 45, 70, 76). These different stages also correlate with the localization of ORF29p, suggesting that compartmentalization of this protein is cell type dependent. Upon reactivation, however, ORF29p is able to accumulate in the nuclei of the sensory ganglia cells, implying that the process of ORF29p nuclear import is conserved among all permissive human cell types (28, 46). Our studies revealed that ORF29p amino acids 9 to 154 harbor a novel NLS, which functions in a karyopherin α/β-dependent manner. At least six isoforms of human karyopherin α, grouped into three subfamilies based on sequence homology, have been described (21, 23, 40, 54, 67). The assays described here used the human and mouse karyopherin α2 homologues, which are highly conserved members of the same subfamily (35, 40, 72). It is, therefore, not surprising that these homologues exhibit a similar binding affinity to ORF29p. In general, karyopherin α2 appears to be expressed ubiquitously in mammalian cells, although the protein levels may be adjusted in some cell types during differentiation and proliferation (35, 41, 42, 72). If the nuclear import mechanism of ORF29p is conserved in both fibroblasts and sensory neurons, this indicates that another mechanism must exist to confer the differential localization of ORF29p during lytic and latent infection.
The regulation of the distribution of ORF29p and the other VZV LAPs may be critical to the ability of VZV to establish a lifelong infection within its host. We have defined a novel NLS that is responsible for ORF29p nuclear targeting and its interactions with members of a ubiquitous, classical cellular transport pathway. During latency, however, ORF29p is selectively maintained in the cytoplasm. Determination of the molecular basis for retention of this protein and other LAPs may further our understanding of how latency is maintained in neurons.
Acknowledgments
These studies were supported by a grant from the Public Health Service, AI-024021, to S.J.S.
REFERENCES
- 1.Adam, E. J., and S. A. Adam. 1994. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol. 125:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, S. A., R. S. Marr, and L. Gerace. 1990. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol. 111:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam, S. A., R. Sterne-Marr, and L. Gerace. 1991. In vitro nuclear protein import using permeabilized mammalian cells. Methods Cell Biol. 35:469-482. [DOI] [PubMed] [Google Scholar]
- 4.Albright, A. G., and F. J. Jenkins. 1993. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J. Virol. 67:4842-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arvin, A. M. 1996. Varicella-zoster virus. Clin. Microbiol. Rev. 9:361-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arvin, A. M. 1996. Varicella-zoster virus: overview and clinical manifestations. Semin. Dermatol. 15:4-7. [PubMed] [Google Scholar]
- 7.Blank, V., P. Kourilsky, and A. Israel. 1991. Cytoplasmic retention, DNA binding and processing of the NF-kappa B p50 precursor are controlled by a small region in its C-terminus. EMBO J. 10:4159-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonifaci, N., J. Moroianu, A. Radu, and G. Blobel. 1997. Karyopherin beta2 mediates nuclear import of a mRNA binding protein. Proc. Natl. Acad. Sci. USA 94:5055-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bonner, W. M. 1975. Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J. Cell Biol. 64:421-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonner, W. M. 1975. Protein migration into nuclei. II. Frog oocyte nuclei accumulate a class of microinjected oocyte nuclear proteins and exclude a class of microinjected oocyte cytoplasmic proteins. J. Cell Biol. 64:431-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boucaud, D., H. Yoshitake, J. Hay, and W. Ruyechan. 1998. The varicella-zoster virus (VZV) open-reading frame 29 protein acts as a modulator of a late VZV gene promoter. J. Infect. Dis. 178(Suppl. 1):S34-S38. [DOI] [PubMed] [Google Scholar]
- 12.Burkham, J., D. M. Coen, C. B. C. Hwang, and S. K. Weller. 2001. Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75:2353-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, J. J., A. A. Gershon, Z. S. Li, O. Lungu, and M. D. Gershon. 2003. Latent and lytic infection of isolated guinea pig enteric ganglia by varicella zoster virus. J. Med. Virol. 70(Suppl. 1):S71-S78. [DOI] [PubMed] [Google Scholar]
- 14.Chi, N. C., E. J. Adam, and S. A. Adam. 1995. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J. Cell Biol. 130:265-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohrs, R. J., M. Barbour, and D. H. Gilden. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J. Virol. 70:2789-2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohrs, R. J., M. B. Barbour, R. Mahalingam, M. Wellish, and D. H. Gilden. 1995. Varicella-zoster virus (VZV) transcription during latency in human ganglia: prevalence of VZV gene 21 transcripts in latently infected human ganglia. J. Virol. 69:2674-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohrs, R. J., D. H. Gilden, P. R. Kinchington, E. Grinfeld, and P. G. Kennedy. 2003. Varicella-zoster virus gene 66 transcription and translation in latently infected human ganglia. J. Virol. 77:6660-6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohrs, R. J., K. Srock, M. B. Barbour, G. Owens, R. Mahalingam, M. E. Devlin, M. Wellish, and D. H. Gilden. 1994. Varicella-zoster virus (VZV) transcription during latency in human ganglia: construction of a cDNA library from latently infected human trigeminal ganglia and detection of a VZV transcript. J. Virol. 68:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohrs, R. J., J. Wischer, C. Essman, and D. H. Gilden. 2002. Characterization of varicella-zoster virus gene 21 and 29 proteins in infected cells. J. Virol. 76:7228-7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 21.Cortes, P., Z. S. Ye, and D. Baltimore. 1994. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc. Natl. Acad. Sci. USA 91:7633-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Croen, K. D., J. M. Ostrove, L. J. Dragovic, and S. E. Straus. 1988. Patterns of gene expression and sites of latency in human nerve ganglia are different for varicella-zoster and herpes simplex viruses. Proc. Natl. Acad. Sci. USA 85:9773-9777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuomo, C. A., S. A. Kirch, J. Gyuris, R. Brent, and M. A. Oettinger. 1994. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc. Natl. Acad. Sci. USA 91:6156-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 25.Gorlich, D., P. Henklein, R. A. Laskey, and E. Hartmann. 1996. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 15:1810-1817. [PMC free article] [PubMed] [Google Scholar]
- 26.Gorlich, D., S. Kostka, R. Kraft, C. Dingwall, R. A. Laskey, E. Hartmann, and S. Prehn. 1995. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 5:383-392. [DOI] [PubMed] [Google Scholar]
- 27.Gorlich, D., S. Prehn, R. A. Laskey, and E. Hartmann. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79:767-778. [DOI] [PubMed] [Google Scholar]
- 28.Grinfeld, E., and P. G. Kennedy. 2004. Translation of varicella-zoster virus genes during human ganglionic latency. Virus Genes 29:317-319. [DOI] [PubMed] [Google Scholar]
- 29.Grose, C., and P. A. Brunel. 1978. Varicella-zoster virus: isolation and propagation in human melanoma cells at 36 and 32°C. Infect. Immun. 19:199-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henkel, T., U. Zabel, K. van Zee, J. M. Muller, E. Fanning, and P. A. Baeuerle. 1992. Intramolecular masking of the nuclear location signal and dimerization domain in the precursor for the p50 NF-kappa B subunit. Cell 68:1121-1133. [DOI] [PubMed] [Google Scholar]
- 31.Herold, A., R. Truant, H. Wiegand, and B. R. Cullen. 1998. Determination of the functional domain organization of the importin alpha nuclear import factor. J. Cell Biol. 143:309-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jans, D. A. 1995. The regulation of protein transport to the nucleus by phosphorylation. Biochem. J. 311:705-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jans, D. A., and S. Hubner. 1996. Regulation of protein transport to the nucleus: central role of phosphorylation. Physiol. Rev. 76:651-685. [DOI] [PubMed] [Google Scholar]
- 34.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 35.Kamei, Y., S. Yuba, T. Nakayama, and Y. Yoneda. 1999. Three distinct classes of the alpha-subunit of the nuclear pore-targeting complex (importin-alpha) are differentially expressed in adult mouse tissues. J. Histochem. Cytochem. 47:363-372. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy, P. G. E., E. Grinfeld, and J. E. Bell. 2000. Varicella-zoster virus gene expression in latently infected and explanted human ganglia. J. Virol. 74:11893-11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinchington, P. R., D. Bookey, and S. E. Turse. 1995. The transcriptional regulatory proteins encoded by varicella-zoster virus open reading frames (ORFs) 4 and 63, but not ORF 61, are associated with purified virus particles. J. Virol. 69:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kinchington, P. R., G. Inchauspe, J. H. Subak-Sharpe, F. Robey, J. Hay, and W. T. Ruyechan. 1988. Identification and characterization of a varicella-zoster virus DNA-binding protein by using antisera directed against a predicted synthetic oligopeptide. J. Virol. 62:802-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kohler, M., S. Ansieau, S. Prehn, A. Leutz, H. Haller, and E. Hartmann. 1997. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 417:104-108. [DOI] [PubMed] [Google Scholar]
- 41.Kohler, M., A. Fiebeler, M. Hartwig, S. Thiel, S. Prehn, R. Kettritz, F. C. Luft, and E. Hartmann. 2002. Differential expression of classical nuclear transport factors during cellular proliferation and differentiation. Cell Physiol. Biochem. 12:335-344. [DOI] [PubMed] [Google Scholar]
- 42.Köhler, M., C. Speck, M. Christiansen, F. R. Bischoff, S. Prehn, H. Haller, D. Görlich, and E. Hartmann. 1999. Evidence for distinct substrate specificities of importin α family members in nuclear protein import. Mol. Cell. Biol. 19:7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lanford, R. E., and J. S. Butel. 1984. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell 37:801-813. [DOI] [PubMed] [Google Scholar]
- 44.Le Roux, L. G., and J. Moroianu. 2003. Nuclear entry of high-risk human papillomavirus type 16 E6 oncoprotein occurs via several pathways. J. Virol. 77:2330-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lungu, O., P. W. Annunziato, A. Gershon, S. M. Staugaitis, D. Josefson, P. LaRussa, and S. J. Silverstein. 1995. Reactivated and latent varicella-zoster virus in human dorsal root ganglia. Proc. Natl. Acad. Sci. USA 92:10980-10984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lungu, O., C. A. Panagiotidis, P. W. Annunziato, A. A. Gershon, and S. J. Silverstein. 1998. Aberrant intracellular localization of varicella-zoster virus regulatory proteins during latency. Proc. Natl. Acad. Sci. USA 95:7080-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meier, J. L., R. P. Holman, K. D. Croen, J. E. Smialek, and S. E. Straus. 1993. Varicella-zoster virus transcription in human trigeminal ganglia. Virology 193:193-200. [DOI] [PubMed] [Google Scholar]
- 48.Melchior, F., B. Paschal, J. Evans, and L. Gerace. 1993. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 123:1649-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore, M. S., and G. Blobel. 1993. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature 365:661-663. [DOI] [PubMed] [Google Scholar]
- 50.Moore, M. S., and G. Blobel. 1992. The two steps of nuclear import, targeting to the nuclear envelope and translocation through the nuclear pore, require different cytosolic factors. Cell 69:939-950. [DOI] [PubMed] [Google Scholar]
- 51.Moroianu, J. 1999. Nuclear import and export pathways. J. Cell Biochem. Suppl. 32-33:76-83. [DOI] [PubMed] [Google Scholar]
- 52.Moroianu, J., G. Blobel, and A. Radu. 1997. RanGTP-mediated nuclear export of karyopherin alpha involves its interaction with the nucleoporin Nup153. Proc. Natl. Acad. Sci. USA 94:9699-9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moroianu, J., M. Hijikata, G. Blobel, and A. Radu. 1995. Mammalian karyopherin alpha 1 beta and alpha 2 beta heterodimers: alpha 1 or alpha 2 subunit binds nuclear localization signal and beta subunit interacts with peptide repeat-containing nucleoporins. Proc. Natl. Acad. Sci. USA 92:6532-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nachury, M. V., U. W. Ryder, A. I. Lamond, and K. Weis. 1998. Cloning and characterization of hSRP1 gamma, a tissue-specific nuclear transport factor. Proc. Natl. Acad. Sci. USA 95:582-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 56.Nelson, L. M., R. C. Rose, and J. Moroianu. 2002. Nuclear import strategies of high risk HPV16 L1 major capsid protein. J. Biol. Chem. 277:23958-23964. [DOI] [PubMed] [Google Scholar]
- 57.Nigg, E. A. 1997. Nucleocytoplasmic transport: signals, mechanisms and regulation. Nature 386:779-787. [DOI] [PubMed] [Google Scholar]
- 58.Paine, P. L., L. C. Moore, and S. B. Horowitz. 1975. Nuclear envelope permeability. Nature 254:109-114. [DOI] [PubMed] [Google Scholar]
- 59.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 61.Quinlan, M. P., and D. M. Knipe. 1983. Nuclear localization of herpesvirus proteins: potential role for the cellular framework. Mol. Cell. Biol. 3:315-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 63.Rice, S. A., M. C. Long, V. Lam, and C. A. Spencer. 1994. RNA polymerase II is aberrantly phosphorylated and localized to viral replication compartments following herpes simplex virus infection. J. Virol. 68:988-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 65.Roberts, C. R., A. C. Weir, J. Hay, S. E. Straus, and W. T. Ruyechan. 1985. DNA-binding proteins present in varicella-zoster virus-infected cells. J. Virol. 55:45-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rost, B., and J. Liu. 2003. The PredictProtein server. Nucleic Acids Res. 31:3300-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seki, T., S. Tada, T. Katada, and T. Enomoto. 1997. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem. Biophys. Res. Commun. 234:48-53. [DOI] [PubMed] [Google Scholar]
- 68.Sheldon, L. A., and R. E. Kingston. 1993. Hydrophobic coiled-coil domains regulate the subcellular localization of human heat shock factor 2. Genes Dev. 7:1549-1558. [DOI] [PubMed] [Google Scholar]
- 69.Shelton, L. S. G., A. G. Albright, W. T. Ruyechan, and F. J. Jenkins. 1994. Retention of the herpes simplex virus type 1 (HSV-1) UL37 protein on single-stranded DNA columns requires the HSV-1 ICP8 protein. J. Virol. 68:521-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Straus, S. E. 1993. Shingles. Sorrows, salves, and solutions. JAMA 269:1836-1839. [DOI] [PubMed] [Google Scholar]
- 71.Taylor, T. J., and D. M. Knipe. 2003. C-terminal region of herpes simplex virus ICP8 protein needed for intranuclear localization. Virology 309:219-231. [DOI] [PubMed] [Google Scholar]
- 72.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 73.Wilkinson, D. E., and S. K. Weller. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yoneda, Y., N. Imamoto-Sonobe, M. Yamaizumi, and T. Uchida. 1987. Reversible inhibition of protein import into the nucleus by wheat germ agglutinin injected into cultured cells. Exp. Cell Res. 173:586-595. [DOI] [PubMed] [Google Scholar]
- 75.Zabel, U., T. Henkel, M. S. Silva, and P. A. Baeuerle. 1993. Nuclear uptake control of NF-kappa B by MAD-3, an I kappa B protein present in the nucleus. EMBO J. 12:201-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zerboni, L., C. C. Ku, C. D. Jones, J. L. Zehnder, and A. M. Arvin. 2005. Varicella-zoster virus infection of human dorsal root ganglia in vivo. Proc. Natl. Acad. Sci. USA 102:6490-6495. (First published 25 April 2005; 10.1073/pnas.0501045102.) [DOI] [PMC free article] [PubMed] [Google Scholar]