Abstract
Cre recombinase from bacteriophage P1 recognizes a 34-bp recombination site, loxP, with exquisite sequence specificity and catalyzes the site-specific insertion, excision, or rearrangement of DNA. To better understand the molecular basis of protein–DNA recognition and generate recombinases with altered specificities, we have developed a directed evolution strategy that can be used to identify recombinases that recognize variant loxP sites. To be selected, members of a library of Cre variants produced by targeted random mutagenesis must rapidly catalyze recombination, in vivo, between two variant loxP sites that are located on a reporter plasmid. Recombination results in an altered pattern of fluorescent protein expression that can be identified by flow cytometry. Fluorescence-activated cell sorting can be used either to screen positively for recombinase variants that recognize a novel loxP site, or negatively for variants that cannot recognize the wild-type loxP site. The use of positive screening alone resulted in a relaxation of recombination site specificity, whereas a combination of positive and negative screening resulted in a switching of specificity. One of the identified recombinases selectively recombines a novel recombination site and operates at a rate identical to that of wild-type Cre. Analysis of the sequences of the resulting Cre variants provides insight into the evolution of these altered specificities. This and other systems should contribute to our understanding of protein–DNA recognition and may eventually be used to evolve custom-tailored recombinases that can be used for gene study and inactivation.
Site-specific recombinases carry out a multitude of critical functions in nature ranging from gene rearrangement to genome segregation. Cre, a 38-kDa recombinase from bacteriophage P1 that resolves genome dimers into monomers (1, 2), is one of the simplest and best understood of known recombinases. Like other members of the λ integrase family, Cre catalyzes recombination between two identical double-stranded DNA sites of a particular sequence (1–3). The enzyme requires no accessory proteins or cofactors and functions efficiently under a wide variety of cellular and noncellular conditions.
The recombination site recognized by Cre is a 34-bp double-stranded DNA sequence known as loxP (Fig. 1). The loxP site is palindromic with the exception of its eight innermost base pairs, which impart directionality to the site. Crystallographic analyses of Cre–DNA complexes (4–6) have helped to elucidate details of the catalytic mechanism of Cre-mediated site-specific recombination and identify protein–protein and protein–DNA interactions within the Cre–loxP catalytic complex. In a productive Cre–loxP complex, two loxP sites are aligned in an antiparallel orientation and are bound by four identical Cre subunits that join to form a ring in which each subunit contacts two adjacent subunits and one loxP half site.
Figure 1.
LoxP, the natural recombination site of Cre. The outermost 13-bp regions on each side (black) are inverted repeats. The middle region (gray) is asymmetric and confers directionality to the site. The base pair identities most important for Cre binding (12) are boxed. Arrows indicate sites of cleavage during recombination.
Cre can catalyze DNA integration, excision, or rearrangement, depending on the relative location and orientation of the two participating loxP sites. Because of its simplicity and versatility, Cre has found widespread use in conditional mutagenesis and gene expression, gene replacement and deletion, and chromosome engineering experiments (7, 8). Despite its extensive utility, applications of Cre are currently restricted by the requirement that targeted DNA regions contain appropriately positioned loxP sites, which effectively limits applications to targets for which sites have been artificially introduced. The ability to target for recombination a region of DNA containing an unnatural recombination site would significantly extend the uses of recombinases by allowing the replacement or deletion of precise segments of genomic DNA. Moreover, the large amount of information available regarding the structure and function of Cre makes this an excellent experimental system to study the molecular details of protein–DNA recognition.
Here we describe the development and application of a high-throughput genetic screening strategy for identifying recombinases that selectively recognize unnatural recombination sites. The strategy uses fluorescence-activated cell sorting (FACS) to identify desired recombinases within a library of Cre variants generated by targeted mutagenesis. The system makes use of two plasmids that can be can be propagated together in Escherichia coli. One plasmid bears a library of Cre variants, in which amino acid residues that are predicted to make sequence-specific contacts to loxP have been targeted for randomization; the other plasmid produces a distinctive pattern of fluorescent protein expression upon recombination. FACS is used to sort cells on the basis of the activity of the recombinases that they possess, and thereby identify Cre variants that specifically recognize unnatural recombination sites. The approach described here differs considerably from a recently reported approach to altering the specificity of Cre, in which the authors used a PCR-based selection method to identify Cre variants from a library made by global mutagenesis (9).
Materials and Methods
Construction of Plasmid pR and Cre Libraries.
Plasmid pR (Fig. 2A Left) was constructed by triple ligation of a BsiWI/BsrBI overlap PCR fragment containing the Cre gene upstream of the rrnB transcription termination region, a BsiWI/AatII overlap PCR fragment containing the araC gene and ara promoter region from the pBAD/Myc-His A plasmid (Invitrogen), and a FspI/AatII fragment from pACYC177 (New England Biolabs). Libraries of Cre variants were constructed by overlap PCR using oligonucleotides (Operon Technologies, Alameda, CA) containing NNK (n = A, T, G, or C and K = G or T) at the positions targeted for randomization. Residues Met-44, Val-48, Thr-87, Gln-90, His-91, and Tyr-283 were randomized to construct the C1 library, and residues Ile-174, Thr-258, Arg-259, Glu-262, and Glu-266 were randomized for the C2 library. Cre variants were ligated into the BsiWI/BstXI fragment of pR to generate the pR-C1 and pR-C2 library plasmids. Ligation products were transformed into DH10B competent cells (Invitrogen) to yield libraries of ≈2 × 108 and ≈1 × 108 colony-forming units (cfu) for libraries C1 and C2, respectively. Twenty individual clones from each library were mapped and sequenced to confirm library quality (data not shown). Supercoiled library plasmid DNA was amplified by maxiprep (Qiagen, Chatsworth, CA) and used to transform the E. coli selection strains pS-M5 and pS-M7 (see below).
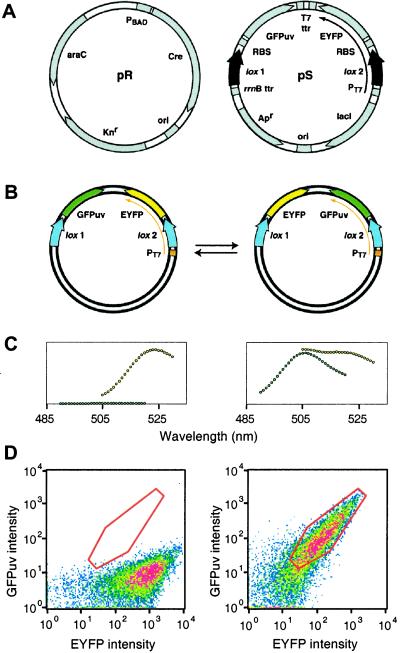
Figure 2.
Recombinase library and reporter system. (A) Plasmid system for genetic screening. The pR plasmid (Left) contains the Cre gene under control of the ara promoter (PBAD), the araC gene, which allows modulated expression from PBAD, a kanamycin selectable marker (knr), and a p15a origin of replication (ori). The pS plasmid (Right) contains, in its starting arrangement, two loxP sites (lox 1 and lox 2), the EYFP gene under control of the T7/lac promoter (PT7), GFPuv downstream of a transcription termination region (rrnB ttr), the lacI gene, which allows inducible expression from PT7, an ampicillin selectable marker (ampr), and a colE1 origin of replication (ori). The EYFP and GFPuv genes are flanked by ribosome binding sites (RBS) and T7 RNA polymerase transcription termination regions (T7 ttr). Plasmids pS-M1–M8 have identical lox M1–M8 variant sites at lox 1 and lox 2. (B) Recombination at the lox 1 and lox 2 sites within pS and pS-M plasmids results in rearrangement of the plasmid and reorientation of PT7 upstream of the GFPuv gene. Recombination is reversible. Cre recombination in vivo can be followed by fluorimetry (C) or cytometry (D) of cells. Cells containing pS alone (Left) express only EYFP, whereas cells containing pR and pS (Right) express roughly equal amounts of GFPuv and EYFP.
Construction of the Reporter Plasmids pS and pS-M1–M8.
The pS reporter plasmid (Fig. 2A Right) was constructed by triple ligation of an EcoRI/ApaLI overlap PCR fragment containing the GFPuv gene [CLONTECH; located downstream of loxP site #1 and a ribosome binding site (RBS), and in front of a T7 RNA polymerase transcription termination region], an ApaLI/XbaI overlap PCR fragment containing the EYFP gene (CLONTECH; located downstream of the T7 RNA polymerase promoter, loxP site #2, and an RBS, and upstream of a T7 RNA polymerase transcription termination region), and an EcoRI/XbaI fragment of plasmid pET-12a (Novagen). An rrnB transcription termination region was then inserted between the AatII and EcoRI sites (in front of loxP site #1) to reduce background expression from the GFPuv gene. Reporter plasmids pS-M1–M8 were constructed by ligating EcoRI/ApaLI and ApaLI/XbaI PCR fragments, made with primers allowing the replacement of loxP sites #1 and #2 with loxP variant sites, to the EcoRI/XbaI fragment of pS. Plasmids were transfected into E. coli DH10B-DE3 competent cells, prepared using a λ-DE3 lysogenization kit (Novagen), to produce the pS and pS-M1–M8 selection strains.
Fluorimetric Analysis of in Vivo Recombinase Activity.
In vivo recombinase activity was assayed by transformation of pR or individual library clones into selection strains pS or pS-M1–M8. Cells were recovered in SOC medium for 1 h at 37°C and then used to inoculate 5-ml cultures containing LB, ampicillin (100 μg/ml), kanamycin (35 μg/ml), and glucose (0.02%). Cells were grown to saturation (≈12 h), washed with LB medium, and incubated in 5 ml of LB containing ampicillin, kanamycin, and isopropyl β-D-thiogalactoside (IPTG; 100 nM) for 4 h at 37°C. IPTG-induced cells (200 μl) were pelletted, resuspended in 1 ml of PBS, transferred to a cuvette, and analyzed using a FluoroMax-2 fluorimeter. Relative expression levels of GFPuv and EYFP were assessed by the relative emission intensities measured at 505 and 523 nm with excitation at 396 and 480 nm for GFPuv and EYFP, respectively. For cells containing pS and wild type Cre, GFPuv fluorescence was ≈6-fold higher than EYFP fluorescence, which reflects differences between the two proteins in expression efficiency, extinction coefficient, and quantum yield. The relative abundance of the two reporter arrangements was therefore calculated by adjusting EYFP intensity by a factor of 6.
Library Screening Using FACS.
For initial sorts, pR-C1 and pR-C2 library plasmids were transformed into selection strains pS-M5 and pS-M7, respectively, using 1 μg of DNA in 400 μl of competent cells for each transformation. Cells were recovered in 10 ml of SOC for 1 h, transferred to 1-liter cultures containing LB, ampicillin (100 μg/ml), kanamycin (35 μg/ml), and glucose (0.02%), and grown to saturation (≈12 h). Aliquots (5 ml) were removed, washed once with 5 ml LB, resuspended in LB containing ampicillin, kanamycin, and IPTG (100 nM), and incubated for 4 h at 37°C. IPTG-induced cells (200 μl) were pelletted and resuspended in 3 ml of PBS. Cells were sorted either positively for expression of both GFPuv and EYFP, or negatively for expression of EYFP alone, using a BDIS FACVantage TSO cell sorter with a Coherent Enterprise II ion laser. Excitation wavelengths were 351 and 488 nm, and emissions were detected using 575/25 nm and 530/30 nm band pass filters for GFPuv and EYFP, respectively. Collected cells were diluted into at least 10 volumes of LB, containing ampicillin and kanamycin, and grown to saturation. Plasmid DNA was miniprepped (Qiagen) and digested with PstI and SacI to remove reporter plasmids. Digested DNA (200 ng) was used to retransform selection cells in preparation for the next round of screening.
Overexpression, Purification, and in Vitro Analysis of Recombinases.
PCR fragments of Cre and C2(±) #4 were inserted between the NcoI and HindIII sites of plasmid pBAD/Myc-His A. Ligation products were transfected into DH10B to produce protein expression strains. Plasmid clones were confirmed by restriction mapping and sequencing. Cultures (500 ml) containing LB and ampicillin were inoculated with expression strains, grown to an OD (600) of 1.0, induced with 0.02% arabinose, and incubated for 3 h at 37°C. Protein was purified using the QIAexpressionist kit (Qiagen) and dialyzed in 50% glycerol, 300 mM NaCl, 10 mM Tris (pH 7.5), and 0.1% β-mercaptoethanol (BME). Protein concentrations were determined by analysis on a 10% SDS polyacrylamide gel.
To construct substrates for in vitro recombination assays, the orientation of loxP site #1 within the pS and pS-M7 plasmids was first reversed by replacing the EcoRI/HinDIII fragment with a PCR fragment containing the appropriate lox site with the orientation of the spacer region reversed. The resulting plasmids, pSp and pS-M7p (25 μg of each), were digested with EcoNI, followed by phenol-chloroform extraction and ethanol precipitation. Digested DNA (4 μg) was dephosphorylated with calf intestinal phosphatase (CIP), phenol-chloroform extracted, and ethanol precipitated. Dephosphorylated DNA (1.5 μg, 0.33 pmol) was 32P-labeled by using [γ-32P]ATP, phenol-chloroform extracted, ethanol precipitated, and redissolved in 20 μl of water.
The dependence of recombination efficiency on enzyme concentration was analyzed using three separate sets of reactions for each enzyme/substrate combination assayed. All reactions were carried out using a final concentration of either 0.2 nM 32P-labeled or 2 nM unlabeled substrate and enzyme concentrations ranging from 2 to 2,000 nM in a reaction buffer containing 300 mM NaCl, 20 mM Tris (pH 7.5), 1 mM EDTA, and 0.1% BME, for 2 h at 37°C. Reactions were stopped as described (10) and analyzed using agarose gel electrophoresis and a Storm PhosphorImager (Molecular Dynamics). Time-course experiments were carried out using 1,000 nM recombinase and 5 nM unlabeled substrate for 1–90 min, quenched, and analyzed using agarose gel electrophoresis in the presence of 0.5 μg/ml ethidium bromide and an Eagle Eye II densitometer (Stratagene).
Results and Discussion
Strategy for Evolution of Novel Cre Variants.
A FACS-based genetic screen was developed to allow both positive screening for Cre variants that recognize novel loxP sites, and negative screening for Cre variants that cannot recognize the wild-type loxP site. The screening system consists of a recombinase plasmid, pR, and a recombination site reporter plasmid, pS (Fig. 2A) that contain distinctive compatibility and selectability markers to allow their simultaneous propagation in E. coli. Plasmid pR, a low-copy derivative of pACYC177, contains the Cre gene under the control of the arabinose promoter, allowing tightly regulated and highly variable levels of protein expression (11). Plasmid pS is a medium-copy derivative of pBR322 designed for use in a FACS-based screen to allow the rapid analysis of recombinase activity in living cells. Plasmid pS contains two loxP sites, the genes for two GFP variants, EYFP and GFPuv, and a T7/lac promoter located upstream of the EYFP gene and one of the loxP sites. Plasmid pS was designed such that recombination would result in a reversible plasmid rearrangement, reorientation of the GFPuv gene downstream of the T7/lac promoter, and a change in expression from EYFP to GFPuv (Fig. 2B). Cells containing an active recombinase should rapidly and reversibly rearrange the pS plasmid, resulting in equal expression levels of EYFP and GFPuv, whereas cells containing an inactive recombinase or pS alone should express only EYFP.
DH10B-DE3 E. coli cells expressing T7 RNA polymerase were transformed with either pS alone or with both pR and pS. Cells transformed with pS alone expressed only EYFP, whereas cells transformed with both plasmids expressed roughly equal amounts of GFPuv and EYFP. Recombinase activity could thus be assessed either fluorimetrically (Fig. 2C) or cytometrically (Fig. 2D). In vivo recombinase activity appeared to be independent of induction with arabinose, indicating that the arabinose promoter is sufficiently leaky to allow the production of Cre protein in quantities sufficient to allow rapid recombination. The rate of screening possible using FACS, up to 7 × 107 cells per hour, was adequate to allow complete coverage of the Cre variant libraries after transformation of E. coli. The rate of enrichment of cells containing both the pR and pS plasmids versus pS alone is ≈5,000-fold per round of FACS (data not shown).
Design of Unnatural LoxP Sites and Cre Variant Libraries.
Each subunit of Cre binds to a loxP half-site through a multitude of protein–DNA contacts (4–6). The majority of these interactions appear to be nonspecific, involving the DNA backbone. Direct interactions between the protein and DNA bases, although relatively few in number, are critical for Cre function, as the enzyme must exhibit high sequence specificity in binding to its recombination site. The sequence of base pairs 2–7 of loxP (see Fig. 1) appears to be particularly important for Cre binding, as demonstrated by gel-shift experiments in which the effects of single base pair changes within loxP on binding of Cre were investigated (12). The importance of the sequence of these base pairs may be attributable to the relatively large number of contacts between the protein and bases in this region.
The initial objective in the design of variant loxP sites was to identify minimal changes to loxP that would eliminate wild-type Cre activity and yet serve as substrates for Cre variants identified from mutant libraries. Previous results have demonstrated the relative importance of the identities of base pairs 2–7 in Cre binding (12). Analysis of the crystal structures indicates that base pairs two and three are contacted by residue Y283 in the minor groove and residues M44, T87, and Q90 in the major groove, whereas base pairs 5–7 are contacted by residues R259 and E262. These data suggest that changes within the region encompassing base pairs 2–7 might eliminate recognition by wild-type Cre. A series of eight loxP variant sites, lox M1–M8 (Fig. 3), were designed containing symmetrical mutations from the wild-type sequence on each side of the loxP site within base pairs 2–7. Variant lox half sites were designed to contain one, two, or three transition mutations, because the number of changes necessary to eliminate recognition by wild-type Cre was not known. The variant sites were inserted into the pS plasmid, replacing the wild-type loxP sites, and the resulting plasmids, pS-M1–M8, were individually cotransfected, along with pR, into E. coli to assess their susceptibility to recombination by wild-type Cre. The lox M1–M8 sites were recombined to different extents by wild-type Cre, as assessed by fluorimetry (Fig. 3). Five of the sites, M1, M2, M4, M6, and M8, were recombined to lower but still detectable extents by wild-type Cre (the high sensitivity of the in vivo assay allows detection of even small amounts of recombinase activity). Three of the eight sites, lox M3, M5, and M7, were not detectably recombined. The lox M5 and M7 sites were chosen as targets for directed evolution experiments.
Figure 3.
Cre recombination of loxP and loxP variant sites. Each of the eight loxP variants shown (M1–M8) were inserted into pS, replacing both of the loxP sites, and tested for recombination in vivo. All of the sites, except lox M3, M5, and M7, supported detectable levels of recombination by wild-type Cre.
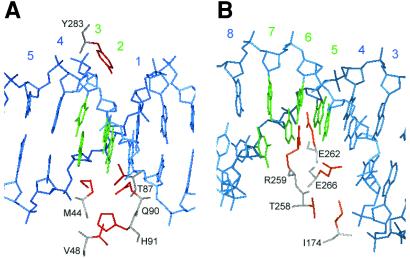
Given the limited number of library members that can be transfected into E. coli (108–109), a targeted library approach was anticipated to have a higher likelihood of success while requiring fewer rounds of selective amplification and mutagenesis compared with an approach involving random mutagenesis of the entire gene. Thus, Cre libraries were generated in which particular amino acids involved in sequence-specific contacts with loxP were randomized. The three-dimensional structures of Cre (4–6) were analyzed to identify amino acid residues located within close proximity to the mutated base pairs of the lox M5 and M7 sites. Six amino acid residues were identified within ≈10 Å of altered base pairs 2 and 3 of lox M5, including M44, T87, Q90, and Y283, which directly contact DNA bases, and residues V48 and H91, which contact residues M44, T87, and Q90 (Fig. 4A). Five amino acid residues were identified within ≈10 Å of altered base pairs 5–7 of lox M7, including R259 and E262, which directly contact DNA bases, and residues T258, E266, and I174, which contact residues R259 and E262 (Fig. 4B). Codons corresponding to the identified residues were randomized at the DNA level by overlap PCR to generate two targeted libraries, C1 and C2, designed for the lox M5 and M7 variant sites, respectively. The partially randomized genes were inserted into the pR plasmid, replacing wild-type Cre, to generate pR-C1 and pR-C2, and transformed into E. coli. Following transformation, the C1 and C2 libraries contained ≈2 × 108 and 1 × 108 members, respectively.
Figure 4.
Targeted Cre libraries. (A) The C1 library, in which residues M44, V48, T87, Q90, H91, and Y283 (red) have been randomized, was designed for use with the lox M5 site, in which base pairs 2 and 3 (green) have been altered. (B) The C2 library, in which residues I174, T258, R259, E262, and E266 (red) have been randomized, was designed for use with the lox M7 site, in which base pairs 5, 6, and 7 (green) have been altered.
Library Screening.
Two different screening methods were used to direct the evolution of Cre site specificity. One method involved the application of positive selection pressure alone, through screening for recombination of the loxP variant site, while a second method involved the application of both positive selection pressure and negative pressure against recombination of the wild-type loxP site. Supercoiled pR-C1 and pR-C2 library plasmids were transformed into DH10B-DE3 cells containing the pS-M5 and pS-M7 reporter plasmids (M5 and M7 selection strains), respectively. Following growth to saturation and induction of the T7/lac promoter, ≈108 cells from each library were screened by FACS. Cells expressing roughly equal concentrations of GFPuv and EYFP were collected and amplified by growth to saturation in LB, and their pR plasmids harvested and transfected into the corresponding selection strains in preparation for the next rounds of sorting. The C1 and C2 lines were then sorted both positively and negatively to branch the C1 line into C1(+) and C1(±) and the C2 line into C2(+) and C2(±).
The C1(+) and C2(+) lines were carried although a total of three rounds of positive screening, and the C1(±) and C2(±) lines were carried through a total of five rounds of alternating positive and negative screening. Cytometric analysis during the final rounds of sorting indicated that lines C1(+), C2(+), and C2(±) were significantly enriched in activity for their respective target loxP variant site; eight clones from each line were sequenced. The C1(±) line was not enriched in activity compared with the naive library and was not further characterized.
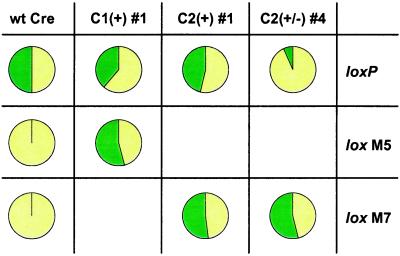
Sequence analysis of the C1(+) line identified a single clone that was represented eight times. The C1(+) #1 clone contained three mutations relative to the wild-type sequence (M44 → G, G90 → S, and H91 → A; Table 1) and recombined the pS-M5 variant reporter plasmid in vivo, as evidenced by the approximately equal expression of GFPuv and EYFP (Fig. 5). The C1(+) #1 enzyme retained the ability to recognize the wild-type loxP-containing pS plasmid, however, as demonstrated by the GFPuv to EYFP expression ratio of 38:62 that indicates an incomplete switch in substrate sequence specificity. Sequence analysis of the C2(+) and C2(±) lines revealed eight distinct clones from each line, all of which actively recombined the lox M7 plasmid (data not shown). The in vivo activity of a typical clone from the C2(+) line, C2(+) #1 (Table 1), is shown in Fig. 5. C2(+) #1 exhibits relaxed substrate specificity in vivo, causing GFPuv and EYFP to be expressed approximately equally from both the pS-M7 and pS plasmids. In contrast, several of the clones from the C2(±) line appear to recombine the lox M7 site more efficiently than the wild-type loxP site (data not shown). The clone from the C2(±) line with the largest preference for recognition of the lox M7 site is C2(±) #4 (Table 1). This clone efficiently recombines the pS-M7 plasmid, resulting in approximately equal expression of GFPuv and EYFP, but not the wild-type loxP-containing site in pS, as demonstrated by the GFPuv to EYFP expression ratio of 6:94 (Fig. 5).
Table 1.
Amino acid compositions of Cre libraries and evolved variants
| M44 | V48 | T87 | Q90 | H91 | I174 | T258 | R259 | E262 | E266 | Y283 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 library | X* | X | X | X | X | I | T | R | E | E | X |
| C1(+) #1 | G | V | T | S | A | I | T | R | E | E | Y |
| C2 library | M | V | T | Q | H | X | X | X | X | X | Y |
| C2(+) #1 | M | V | T | Q | H | L | N | S | G | G | Y |
| C2(±) #4 | M | V | T | Q | H | A | L | S | H | G | Y |
X indicates a random amino acid residue; mutations relative to wild-type Cre are shown in bold.
Figure 5.
In vivo activity of recombinase variants. Recombination activity is assessed as a ratio of EYFP (yellow) and GFPuv (green) expression, reflecting the relative concentrations of the two reporter plasmid arrangements in E. coli. Cells containing an active recombinase should contain approximately equal concentrations of the two reporter arrangements and express EYFP and GFPuv in roughly equal abundance.
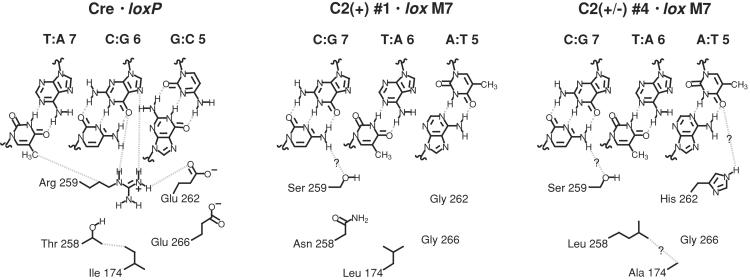
Two residues within wild-type Cre, R259 and E262, make contacts with base pairs 5–7 of wild-type loxP (Fig. 6). Three additional residues, I174, T258, and E266, may contribute to Cre/loxP stability through contacts with R259 and E262 and water molecules. Large pockets resulting from mutations within the C2(+) #1 and C2(±) #4 variants may be accommodated by structural shifts and/or bridging water molecules, which might play important roles in DNA recognition. The C2(±) #4 recombinase contains mutations at all five randomized positions (Table 1; Fig. 6); these changes may facilitate recognition of lox M7. A Ser residue that substitutes for Arg-259 may accept a hydrogen bond from the exocyclic amine of the cytosine of base pair 7 within lox M7 and a His residue that substitutes for Glu-262 may interact with base pair 5 of lox M7, although some structural rearrangement would be necessary to allow these interactions. The remaining substitutions, I174 → A, T258 → L, and E266 → G, may indirectly facilitate recognition of lox M7 or relieve unfavorable interactions of the wild-type residues at these positions. Despite the functional differences between C2(+) #1 and C2(±) #4, the two Cre variants share some sequence similarities (Table 1). Both variants have R259 → S and E266 → G mutations and a Leu substitution at either position 174 or 258 (Fig. 6), suggesting that these mutations within C2(±) #4 are not responsible for its selectivity in recognizing the lox M7 site. The C2(+) #1 variant has an additional Gly residue at position 262 that may create a pocket near the DNA–protein interface, which may allow this recombinase to tolerate multiple base pairs at positions 5 and 6. A comparison of substitutions within the two Cre variants suggests that the His-262 substitution of C2(±) #4 may be the key to its ability to achieve selectivity in recognizing the lox M7 site, perhaps by making contact with base pair 5 of lox M7. Structural analyses and mutagenesis experiments will be necessary to test these hypotheses.
Figure 6.
Models of interactions of wild-type and C2 variant recombinases with loxP and lox M7. Interactions between Cre residues varied within the C2 library and base pairs 5–7 of loxP (Left) are based on crystal structural analysis (4–6). Possible interactions between C2(+) #1 and lox M7 (Center) and C2(±) #4 and lox M7 (Right) are indicated by dashed lines.
In Vitro Activity of a Recombinase with Altered Site Recognition.
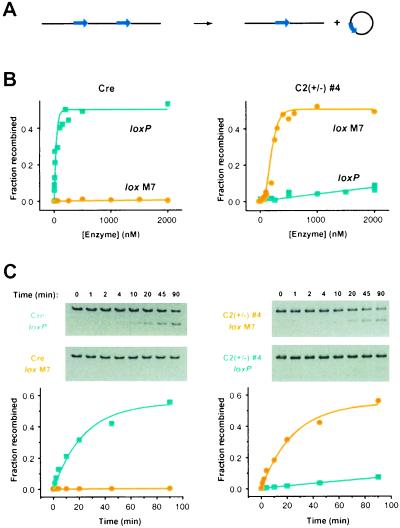
To further compare the activity differences and substrate preferences of C2(±) #4 and Cre, the proteins were overexpressed in E. coli and purified to homogeneity. An in vitro assay for recombination of linear double-stranded DNA substrates containing a pair of either wild-type loxP or lox M7 sites in parallel orientation (Fig. 7A) was first used to compare the two enzymes with respect to the concentration dependence of their recombination of the DNA substrates (Fig. 7B). Because four recombinase subunits must bind to each DNA substrate to catalyze recombination, single-turnover reaction conditions were used with a range of protein concentrations in large excess over a fixed concentration of DNA substrate. Reactions were allowed to proceed for a fixed period and the products were analyzed by gel electrophoresis. Data were fit to the Hill equation: F = Feq [E]h/([E50]h + [E]h), where F is the fraction of substrate recombined within a reaction containing an enzyme concentration [E], Feq is the fraction of substrate recombined at equilibrium, [E50] is the concentration of enzyme needed to recombine the substrate to an extent of 50% of Feq, and h is the Hill coefficient. This analysis revealed that the apparent affinity of C2(±) #4 for its preferred substrate, lox M7 (220 nM), is about 8-fold lower than that of Cre for its preferred substrate, loxP (28 nM). From these analyses, the binding of C2(±) #4 to lox M7 (h = 3.1) appears to be more cooperative than the binding of Cre to loxP (h = ≈1.0), although this difference may reflect, in part, the larger error associated with determining the value of h when [E50] is low.
Figure 7.
In vitro activity of C2(±) #4 and wild-type Cre recombinases. (A) Schematic representation of the in vitro recombination reaction. Double-stranded DNA is shown in black; lox sites are shown as blue arrows. Recombination results in excision. (B) Enzyme concentration dependence of recombination. Reactions were carried out with in the presence of 0.1 nM substrate for 2 h. (C) Time courses of recombination. Agarose gels showing the substrate and one of the reaction products were ethidium bromide stained and analyzed by densitometry. Reactions were carried out in the presence of 0.5 nM substrate and 1000 nM enzyme.
The in vitro assay also was used to compare C2(±) #4 and Cre with respect to the rate at which they approach equilibrium in recombining the lox M7 and wild-type loxP-containing DNA substrates (Fig. 7C). For each reaction, a high concentration of each recombinase was incubated with a DNA substrate and aliquots were removed and analyzed following varying incubation periods. Data were fit to the equation: F = Feq (1 − e−kt), where F is the fraction of substrate recombined at time t, Feq is the fraction of substrate recombined at t = ∞, and k is the rate of approach to equilibrium for recombination. The rates of approach to equilibrium for C2(±) #4 and Cre in recombining their preferred substrates are nearly identical (0.040 and 0.042 min−1, respectively), indicating that the binding preference of C2(±) #4 does not perturb its catalytic rate. The lox M7 site is recombined very slowly by Cre (0.00003 min−1) and wild-type loxP is recombined very slowly by C2(±) #4 (0.001 min−1), even in the presence of a high concentration of protein. It was not possible from these experiments to determine whether the low activity was a result of a reduction in catalytic rate, binding affinity, or some combination thereof. The apparent affinity and rate of approach to equilibrium observed for Cre/loxP are each about an order of magnitude lower than reported previously (10), which may reflect the use of a his-tagged version of Cre in these experiments or to differences in substrates used in the two measurements.
Relaxing Versus Switching Site Specificity.
In principle, selection for recombinase variants that recognize new recombination sites can result in either a relaxation or a switching of specificity. The objective of these experiments was to develop a system that would allow a switching of Cre site recognition specificity. Previous efforts to use positive selective pressure to identify, from a globally randomized library, bacteriophage λ integrase variants that could recognize bacteriophage HK022 sites resulted in integrases with relaxed specificity (13, 14). Similar results were obtained with the application of positive selection pressure alone to the evolution of the site recognition of Cre from a nontargeted library (9).
The FACS-based screening system described here was designed to allow the application of negative selection pressure, if necessary, as well as positive pressure. Screening experiments were carried out using either positive selection pressure alone or a combination of both positive and negative selection pressure. Surprisingly, the two processes yielded quite different results. Positive screening for recognition of the lox M5 variant site resulted in the identification of a recombinase with relaxed site recognition specificity (Fig. 5), while a combination of positive and negative screening failed to identify active recombinases. The latter result suggests that no individuals within the starting C1 library were able to satisfy both the positive and negative selection requirements. Positive screening for recognition of the lox M7 variant site resulted in several different recombinases, most of which retained high levels of activity with the wild-type loxP site. In contrast, a combination of positive and negative screening resulted in recombinases that selectively recognized the lox M7 site. These results indicate that recombinase variants with relaxed specificity are more abundant than those with switched specificity in random libraries, even those constructed using targeted randomization. The relative abundance of recombinases with relaxed versus switched specificity may depend on the degree to which the variant and wild-type loxP sites are related. Recognition of both loxP and a significantly less similar variant site by the same enzyme may be more difficult, assuming that the number of nonspecific contacts remains constant. Thus, targeting loxP variant sites with greater differences from loxP might obviate the need for applying negative selection pressure.
Applying Evolved Recombinases.
The sequence-specific manipulation of DNA by Cre recombinase represents a powerful approach to studying gene function. Cre has been used to conditionally express, mutagenize, and replace genes (15–20), as well as rearrange chromosomes (21–24) in mice, resulting in major advances in the understanding of gene function. The requirement for preexisting genomic loxP sites complicates the use of Cre in studies of gene function and is an impediment to the use of Cre in applications such as gene therapy. The results described here and elsewhere (9) represent steps toward the custom-evolution of recombinases that recognize unique sites within mammalian genomes. Such systems may eventually allow simultaneous control over multiple genes within whole animals, excision of genes involved in pathological processes, replacement of nonfunctional genes, or insertion of genes of therapeutic value. To realize these possibilities, more work will be needed to demonstrate the practicality of directing the evolution of Cre to recognize sites that are significantly dissimilar to loxP. In addition, future experiments will likely address the question of whether Cre variants can be made to function as part of a heterotetrameric system to allow the recombination of asymmetric sites.
Acknowledgments
We thank Alan Saluk, Cheryl Silao, and Eric O'Connor of The Scripps Research Institute Flow Cytometry Core Facility. This work was supported by the Skaggs Institute for Chemical Biology. S.W.S. is a fellow of the Jane Coffin Childs Memorial Fund for Medical Research. This is manuscript number 14708-CH of The Scripps Research Institute.
Abbreviation
- FACS
fluorescence-activated cell sorting
References
- 1.Abremski K, Hoess R H, Sternberg N. Cell. 1983;32:1301–1311. doi: 10.1016/0092-8674(83)90311-2. [DOI] [PubMed] [Google Scholar]
- 2.Abremski K, Hoess R. J Biol Chem. 1984;259:1509–1514. [PubMed] [Google Scholar]
- 3.Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Cold Spring Harbor Symp Quant Biol. 1981;1:297–309. doi: 10.1101/sqb.1981.045.01.042. [DOI] [PubMed] [Google Scholar]
- 4.Guo F, Gopaul D N, Van Duyne G D. Nature (London) 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 5.Gopaul D N, Guo F, Van Duyne G D. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo F, Gopaul D N, Van Duyne G D. Proc Natl Acad Sci USA. 1999;96:7143–7148. doi: 10.1073/pnas.96.13.7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagy A. Genetics. 2000;26:99–109. [Google Scholar]
- 8.Sauer B. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- 9.Buchholz F, Stewart A F. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 10.Ringrose L, Lounnas V, Ehrlich L, Buchholz F, Wade R, Stewart A F. J Mol Biol. 1998;284:363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 11.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartung M, Kisters-Woike B. J Biol Chem. 1998;273:22884–22891. doi: 10.1074/jbc.273.36.22884. [DOI] [PubMed] [Google Scholar]
- 13.Yagil E, Dorgai L, Weisberg A. J Mol Biol. 1995;252:163–177. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 14.Dorgai L, Yagil E, Weisberg A. J Mol Biol. 1995;252:178–188. doi: 10.1006/jmbi.1995.0486. [DOI] [PubMed] [Google Scholar]
- 15.Lakso M, Sauer B, Mosinger B, Jr, Lee E J, Manning R W, Yu S H, Mulder K L, Westphal H. Proc Natl Acad Sci USA. 1992;89:6232–6236. doi: 10.1073/pnas.89.14.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiering S, Kim C G, Epner E M, Groudine M. Proc Natl Acad Sci USA. 1993;90:8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Russ A P, Friedel C, Ballas K, Kalina U, Zahn D, Strebhardt K, von Melchner H. Proc Natl Acad Sci USA. 1996;93:15279–8473. doi: 10.1073/pnas.93.26.15279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gu H, Marth J D, Orban P C, Mossman H, Rajewsky K. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 20.Zou Y R, Muller W, Gu H, Rajewsky K. Curr Biol. 1994;4:1099–1103. doi: 10.1016/s0960-9822(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 21.Qin M, Bayley C, Stockton T, Ow D W. Proc Natl Acad Sci USA. 1994;91:1706–1710. doi: 10.1073/pnas.91.5.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Deursen J, Fornerod M, Van Rees B, Grosveld G. Proc Natl Acad Sci USA. 1995;92:7376–7380. doi: 10.1073/pnas.92.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Justice M J, Zheng B, Woychik R P, Bradley A. Methods. 1997;13:423–436. doi: 10.1006/meth.1997.0548. [DOI] [PubMed] [Google Scholar]
- 24.Su H, Wang X, Bradley A. Nat Genet. 2000;24:92–95. doi: 10.1038/71756. [DOI] [PubMed] [Google Scholar]