Abstract
We have used small angle x-ray scattering and computer simulations with a coarse-grained model to provide a time-resolved picture of the global folding process of the Tetrahymena group I RNA over a time window of more than five orders of magnitude. A substantial phase of compaction is observed on the low millisecond timescale, and the overall compaction and global shape changes are largely complete within one second, earlier than any known tertiary contacts are formed. This finding indicates that the RNA forms a nonspecifically collapsed intermediate and then searches for its tertiary contacts within a highly restricted subset of conformational space. The collapsed intermediate early in folding of this RNA is grossly akin to molten globule intermediates in protein folding.
In the process of adopting a functional structure, biological macromolecules must fold from a highly disordered polymer to a discrete structure. In this process, the number of accessible conformational states starts out extraordinarily large in the unfolded state but is radically reduced in the folded, functional structure. As originally pointed out by Levinthal (1), there is insufficient time to explore all conformations in the search for the correct folded structure, indicating that macromolecular folding must proceed through intermediate states with increased bias to continue the folding process.
A fundamental property of folding is compaction from a highly flexible and dynamic set of unfolded conformations to a tightly packed functional structure. This process of compaction severely limits the number of accessible conformations, so formation of a compact intermediate may aid folding by restricting the conformations of the macromolecule to a subset of states that includes the functional state (2). Alternatively, or in addition, a compact state can slow folding by fostering formation of nonnative interactions and by providing steric barriers to formation of the native state, generating long-lived, off-pathway folding intermediates (3, 4).
Many early discussions in the protein folding field centered on the “molten globule” (5). Under nonnative conditions, species with considerable hydrophobic collapse and secondary structure, but little or no set tertiary structure, are often observed and have been generically referred to as molten globules. Kinetic experiments have established that molten globule intermediates are formed early in folding for several proteins (6, 7), and, in a limited number of cases, early compaction has been observed directly by small angle x-ray scattering (SAXS; refs. 8 and 9). Early compaction is not obligatory for proteins, however, as folding in the absence of early collapse also has been observed (10). Further, numerous proteins have been observed to fold with two-state kinetics, suggesting that accumulation of a collapsed intermediate is not a general feature of protein folding (11).
RNA macromolecules also fold to biologically functional structures. Revealing the properties of RNA as it folds in vitro is important for understanding the constraints on the in vivo process conferred by the nature of the polymer. In addition, RNA folding studies provide a distinct and potentially valuable perspective on the general problem of the conformational search of a polymer for its functional state. There are major differences between RNA and proteins, including a lower information content of RNA (with 4 side chains instead of the 20 protein side chains), which may render folding to a discrete state more difficult (12, 13), and the polyelectrolyte nature of the RNA backbone (which presents an electrostatic barrier to formation of a compact, functional structure). Further, unlike proteins, RNA readily forms stable secondary structure in the absence of enforcing tertiary structure, suggesting that RNA folding may be largely hierarchical and thus easier to understand than that of proteins (12, 14).
For RNA, the question of whether overall compaction precedes or is concomitant with tertiary structure formation has not been addressed. Several studies have raised the possibility of a rapid collapse (15–18), prompting suggestions of an early collapse for RNA folding in general, with accumulation of nonspecifically collapsed species (19, 20). However, none of these previous studies directly probed the global structure of the RNA at times earlier than required for tertiary structure formation.
SAXS provides a unique vantage point from which to view the folding process. This solution scattering method lacks the atomic resolution possible with x-ray scattering from a crystal lattice, but it can describe the overall size and shape of a macromolecule in solution. Unlike scattering from a crystal, SAXS can follow the evolution of macromolecular size and shape during folding (21, 22).
Therefore, we have used SAXS to follow the folding of the Tetrahymena group I ribozyme (Fig. 1A). This RNA was previously shown to exhibit a large difference in SAXS between the unfolded and folded states, and a late folding intermediate was shown to be compact (15). Here, we directly follow compaction of the ribozyme during folding by SAXS. Compaction is found to be substantially faster than stable formation of any known tertiary contacts, which are detectable by protection from solution radicals (23) and, in some cases, by protection from complementary oligonucleotides (24). The results suggest that there is global collapse of the RNA before there is significant formation of specific tertiary structure.
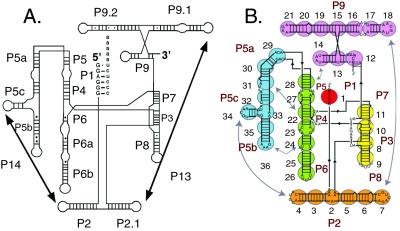
Figure 1.
The Tetrahymena ribozyme. (A) Secondary structure of the ribozyme. (B) The course-grained model used in simulations. In this model, each group of approximately five base pairs or five residues of a single-stranded region is represented as a sphere (see Methods).
Methods
SAXS Measurements.
All experiments herein used the standard L-21 ScaI form of the Tetrahymena ribozyme (25), which was prepared by in vitro transcription and column purification as described (26). SAXS experiments at Advanced Photon Source (APS; 5–50 ms folding time) were performed at the 8-ID beamline of IBM/Massachusetts Institute of Technology/McGill University/Yale University/University of Toronto-Whitehead Institute-Collaborative Access Team (IMMYT-Whitehead-CAT). Folding experiments were performed at 25°C by flowing 1.9 mg/ml (15 μM) ribozyme in 50 mM Na-Mops [3-(N-morpholino)propanesulfonic acid], pH 7.0, through the inlet channel of the microfabricated flow cell to mix with equivalent buffer solution containing 10 mM MgCl2.
Data were collected (1 min per time point) by using pink beam (27, 28), with defining slits set to 10 μm in the vertical dimension and 40 μm along the direction of flow. These conditions give a beam flux of ≈3 × 1010 photons per s; with a flow rate of 86 mm/s along the outlet channel, these slits give an exposure time of 400 μs. In addition to providing high temporal resolution, the short exposure time of each RNA molecule to the x-ray beam minimizes the potential for radiation damage to the RNA. Consideration of the beam intensity used in synchrotron hydroxyl radical cleavage experiments (23) suggests that only ≈1% of the RNA is cleaved during the 400-μs SAXS exposure.
SAXS experiments at Stanford Synchrotron Radiation Laboratory (SSRL, beamline 4–2) were performed by stopped-flow and manual mix methods as described (8, 15, 29). Equal volumes of buffer solutions containing 3.2 mg/ml (25 μM) ribozyme and 30 mM MgCl2 were mixed to initiate folding of the ribozyme. Previous experiments showed that the scattering was essentially independent of ribozyme concentration from 1–4 mg/ml, suggesting that the ribozyme is predominantly monomeric in this concentration range (15). A significant amount of the total Mg2+ after mixing (15 mM) is expected to be bound by the ribozyme (30), such that the free Mg2+ concentration is expected to be similar to that in the continuous-flow experiments. (In the continuous flow experiments there is a large excess of the 10 mM Mg2+ buffer, so that the Mg2+ concentration can equilibrate to 10 mM in the sample stream.) X-ray energy was selected by using a pair of Mo/B4C multilayer monochromating crystals (29). For stopped-flow measurements, 10 identical experiments were performed sequentially, and for each time point scattering profiles from the 10 experiments were summed.
Quantitative Analysis of SAXS Data.
SAXS data for folding times from 5 ms to 1,000 s were fit by using singular value decomposition (SVD) analysis (22, 31). SVD analysis allows determination of the smallest number of independent curves required to reconstruct a series of related experimental curves. In this analysis, the series of scattering profiles acquired at different times is represented as a two-dimensional matrix, with each column corresponding to a scattering profile and each row corresponding to scattering intensities for a small range of S values. An SVD algorithm (MATLAB) transforms this data matrix into the product of three matrices UWVT. The matrix U consists of the independent basis curves that can reconstruct the data, and W contains the so-called singular values, one for each basis curve in U, which provides a measure of the weights of each basis curve required to reconstruct the data. The number of significant singular values (i.e., with a magnitude significantly larger than noise) indicates the number of independent basis curves required to completely reconstruct the data.
SVD analysis of the data herein yielded only two singular values that were clearly significant. The presence of two major singular values indicated that two independent curves were sufficient to reconstruct the data. Therefore, we analyzed the data by determining the projection of each scattering profile onto two physically meaningful states, unfolded and folded, by using a least squares fit to minimize the difference between the linear combination and the scattering profile at each time point. For most of the data, the residuals of these fits are indistinguishable from random noise (see text in supporting information, which is published on the PNAS web site, www.pnas.org). However, for some data sets, small amounts of systematic deviation were observed when using only two SVD components. The magnitude of this deviation varied between data sets, and inclusion of a third SVD component to account for the deviation did not significantly affect the relative weights of the primary two components (data not shown), so the simpler two-component analysis is presented here.
There were small differences in scattering of the unfolded state at the lowest values of S between data collected at APS and at SSRL. Because of the differences in scattering of the unfolded states, two-state fits to each time point were performed by using the scattering profile of the unfolded state that was collected at the same facility as the time point. Fitting each point by using the unfolded state data collected at the other facility instead gave small changes in the weights of the unfolded and folded states in the fit (<10% of the values) but had no significant effects on the rate constants for the two phases of compaction (data not shown). The SAXS profile of the long-lived misfolded state was used in all two-state fits as an approximate description of the folded form, because under all conditions herein, most of the ribozyme population folds to the misfolded form (26, 32); the SAXS profiles of this misfolded form and the native state are similar (ref. 15, and R.R., I.M., S.D., and D.H., unpublished results).
Simulations.
For computational tractability and simplicity of interpretation, we have used a coarse-grained model in which five- to six-bp segments and five- to six-residue single-stranded segments of the ribozyme are represented by single spheres. The folded structure of the ribozyme was taken from the model developed by Michel, Westhof, and colleagues (33). More information about the simulations is provided in supporting information.
To select the frame of a simulation that gave the scattering profile most similar to a given experimental time point, each frame of a simulation was compared with the experimental scattering data by linear regression [I(S)simulation vs. I(S)experimental]. Least-squares analysis of the plots were performed with the residuals weighted by S2 to emphasize low S values in Kratky plots, rendering the weighting approximately inversely proportional to the SD of the experimental data (data not shown).
Results
Previous studies have established that stable tertiary structure is formed in the Tetrahymena ribozyme under standard in vitro conditions (pH 7–8, 10 mM Mg2+, ≈10 mM Na+, 37–42°C), with rate constants for different parts of the structure ranging from ≈1 s−1 to 0.02 s−1 (23, 24). To determine whether collapse precedes tertiary structure formation, we would need to collect SAXS data at folding times sufficiently short (<100 ms) so that even the tertiary structure that forms first, the P4-P6 domain, would not have formed to a significant extent. To allow direct comparison with the earlier results, the SAXS experiments herein were performed under solution conditions similar to these previous studies. (As described in Discussion, it has recently been shown that increased ionic strength increases the rate of tertiary structure formation in P4-P6; ref. 34).
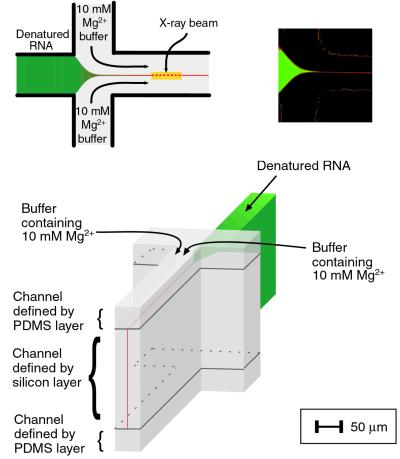
To achieve the high temporal resolution necessary in an apparatus sufficiently resistant to radiation damage from the intense x-ray source needed, we improved a previous mixing device (ref. 9; Fig. 2). As the RNA solution passes through the microfabricated flow cell, incoming Mg2+-containing buffer from the side channels focuses the RNA solution to a narrow stream that flows rapidly through the outlet channel of the cell. The narrow width of the RNA stream allows Mg2+ to mix diffusely on the submillisecond time scale, initiating folding of the RNA. Time-resolved SAXS data are collected by acquiring a series of scattering profiles at different locations along the outlet channel, corresponding to well defined times after mixing with Mg2+. The rapid flow of the RNA stream allows experimental access to very short times after mixing and gives high temporal resolution by illuminating only a short length of the jet, corresponding to a narrow time window (<0.5 ms under these conditions).
Figure 2.
Schematics of the flow cell used to collect SAXS data on the low millisecond timescale. (Upper) Principle of operation of the flow cell. (Upper Left) Cross section through the center of the flow cell indicating one possible location of the x-ray beam. Details of operation are given in the text. (Upper Right) Calibration of the mixing time for the device. The mixing time for ions from the side channels was determined by monitoring Ca2+ concentration with multiphoton microscopy.†† Changes in free Ca2+ concentration, representing diffusion of Ca2+ across the focused stream, were determined from the emission characteristics of the Ca2+-sensitive dye Indo-1 (Molecular Probes) under the flow conditions of the folding experiments. Binding of Ca+ to the dye results in a shift of the emission peak from ≈475 nm in the absence of bound Ca+ (shown in false color as green) to ≈400 nm with bound Ca+ (shown as red). Determination of the distance from the junction of the side channels with the inlet channel to the point at which the complete color change was observed, combined with the flow speed (86 mm/s), gave a value for the mixing time of 400 μs. Flow speeds were determined by using fluorescence correlation spectroscopy (J. Korlach, L.W.K., and L.P., unpublished work). (Lower) A three-dimensional schematic of the flow cell. For clarity, only the channels of the microfluidic mixer are shown in the drawing. The cell is constructed in three layers. At its center is a silicon wafer with two through-etched crossed channels (9). The addition of top and bottom layers, fabricated from polydimethyl siloxane (PDMS; ref. 46), prevents the macromolecules from sticking to the surfaces and produces the uniform flow profile for the RNA stream shown.§§
SAXS data for folding times from 5 ms to 45 ms were collected by using this mixing device at the APS. For longer folding times, data were collected at the SSRL by using stopped-flow (0.1–30 s; refs. 8 and 35) and manual mix (80–1,000 s) methods. Multiple independent data sets for the stopped-flow and manual mix data were in reasonable agreement with each other and were only weakly dependant on temperature between 15 and 37°C. Robustness of the methodology and the global folding properties was established further by the continuity that was observed between the continuous-flow (<50 ms) and stopped-flow (>50 ms) data, despite differences in mixing devices and beam conditions.
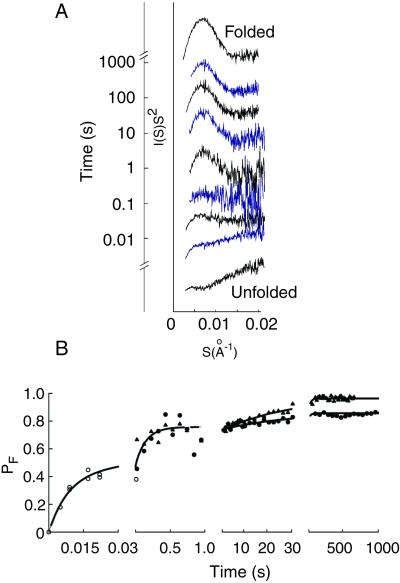
Representative scattering profiles for RNA folding times ranging over five orders of magnitude (5 ms to 1,000 s) are shown in Fig. 3 A as Kratky plots [I(S)S2 vs. S (21); see legend to Fig. 3], which allow features of the scattering profiles indicative of global structural changes to be readily discerned. The Kratky plot of a random coil gives a continuously increasing function over a wide range of S, whereas the Kratky plot of a compact molecule gives a decrease at large S, producing a maximum value and the appearance of a peak. In the unfolded state there is a rising tail (S > 0.008 Å−1), indicating a highly extended structure. As folding begins in the low millisecond time regime, this rising tail disappears, and a small bulge appears at low S (0.005–0.01 Å−1), consistent with a more compact species. At times longer than 50 ms, the bulge turns into a distinct peak, indicative of substantial globular nature. Between 2 s and 30 s, there is a gradual decrease in the high S tail, leading to stronger definition of the peak. The intensification of this peak indicates that the ribozyme has increased further in globularity.
Figure 3.
Time course of shape changes in ribozyme folding. (A) The progression of Kratky plots (IS2 vs. S; S = 2sinθ/λ, where 2θ is the scattering angle and λ is the x-ray wavelength) over the entire time course of these experiments. Each Kratky plot is positioned by its folding time, which increases logarithmically from bottom to top. (B) Quantitative analysis of folding. To fit the data, we projected each experimental scattering curve onto two static states of the RNA representing the beginning and ending points of folding: the unfolded (U) and folded (F) states. The fractional weight of F in the projection (PF) is shown as a function of time for data from the APS at 25°C (○) and data collected at SSRL at 15°C (●) and 37°C (▴). The time course was described well by three successive first-order processes. The fastest process occurs with a time constant of 7 ± 1 ms, and a second phase gives a time constant of 140 ± 30 ms.¶¶ Slower third transitions, which do not give large changes in overall shape (Fig. 3a) and, therefore, have smaller amplitudes than the initial two phases, give time constants of 26 ± 9 s (15°C) and 26 ± 3 s (37°C). Note that although the time constants for the slow transitions at 15°C and 37°C are identical within error, the amplitudes are different, giving the observed deviation of the two curves from each other.
A quantitative analysis of the progress of folding was obtained by performing SVD analysis of the time-resolved data (8). This analysis showed that the data across the entire time course (5 ms–1,000 s) are well represented by a linear combination of two independent states (see Methods). To provide a conceptual measure of the progress of folding, the experimental scattering profile at each time is represented as a linear combination of the starting (unfolded) state and the folded state (Fig. 3B). The compactness at each time point is represented by the relative weight of the folded conformation in the fit. At the earliest times after Mg2+ addition, substantial progress in folding is apparent, followed by a plateau region in the range of 20–40 ms. These data indicate that a discrete transition occurs with a time constant of 7 ± 1 ms.
At folding times between 40 ms and 1 s, a second transition is observed, giving a time constant of 140 ± 30 ms.§§ Fluorescence resonance energy transfer experiments have detected a folding transition with a similar time constant of 130 ± 50 ms (X. Zhuang, H. Babcock, R.R., D.H., and S. Chu, unpublished results), suggesting that both approaches can follow this second phase of compaction.
Upon completion of these two early folding transitions (t > 1 s), the ribozyme gives a scattering profile similar to that of the fully folded form, indicating that most of the global shape change is complete (see Fig. 3A). The earliest stable tertiary structure detected by protection from solution radicals, the P4-P6 domain (Fig. 1A), is formed at least 5-fold slower than this compaction under similar conditions (23), providing direct evidence that a collapsed intermediate lacking the known tertiary contacts is formed early in folding. Additional small but significant changes were observed in the scattering profile between 1 and 1,000 s, consistent with rearrangements that give the slower formation of tertiary contacts, as observed by other approaches (23, 24).
As noted above, the changes in shape of the scattering profiles indicate substantial compaction over the millisecond time scale (Fig. 3). Nevertheless, the scattering profiles do not directly provide a physical description of the intermediate species. Therefore, we used folding simulations with a simplified ribozyme model to obtain molecular shapes representative of the observed scattering profiles (Fig. 4).
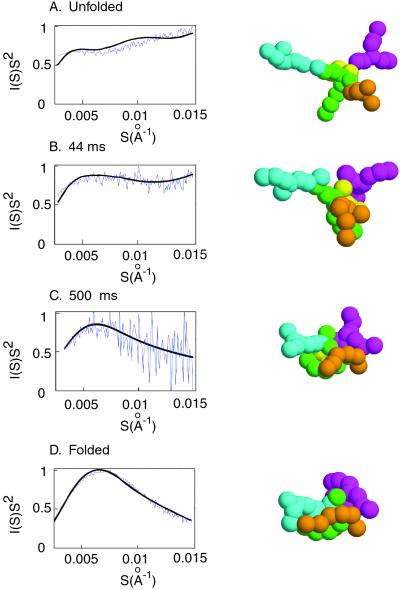
Figure 4.
Comparison of experimental and simulated SAXS profiles. Each panel shows the experimental SAXS profile from a given folding time or condition (blue) superimposed on the simulated SAXS profile that gave the best fit to the experimental data (black). The corresponding simulated structures are shown adjacent to the plots.
This coarse-grained model represents groups of residues as spheres, as depicted in Fig. 1B. Folding simulations were begun from a model for the unfolded ribozyme, which was generated by applying a universal repulsive Coulomb potential intended to mimic the low ionic strength conditions in the absence of Mg2+. SAXS profiles generated from the folding simulations were systematically compared with the experimental SAXS profiles to identify the simulated intermediates that give scattering profiles most similar to the experimental profiles.
Intermediates from the folding simulations that best fit the observed scattering profiles for the unfolded ribozyme, and for two early time points, are shown in Fig. 4 A–C. SAXS data for the unfolded ribozyme are best approximated by conformations of the model in which the ribozyme domains are extended away from each other, suggesting that the structure is dominated by electrostatic repulsion. Some structural elements are in close proximity with each other even at 40 ms, when the first kinetic phase is essentially complete, and there is considerable globular character to the overall structure by 500 ms. Further, the scattering profile of the fully folded ribozyme is best fit by simulated structures that are only slightly more compact and ordered than the 500-ms structure (Fig. 4D). Additional simulations and experimental time points support this general picture of these folding intermediates (data not shown). The simulations provide a useful tool for visualization of the global folding process, and we anticipate that coordination of simulation with additional physical data will prove valuable in interpreting experiments, deriving folding models, and designing decisive experimental tests.
Discussion
As a macromolecule folds to its functional form, it must undergo compaction from a disordered chain to a specific structure. Here, we have used SAXS to directly monitor the compaction during folding of a structured RNA, the Tetrahymena ribozyme. We wanted to determine whether compaction occurs as an early step in folding, before specific tertiary structure formation, or whether it occurs as long-range contacts form, which by their nature necessitate collapse. The results show that compaction is largely complete in less than one second, whereas under similar conditions, the earliest detected tertiary structure is formed 5-fold slower (23). Thus, compaction largely precedes specific tertiary structure formation, indicating that a nonspecifically collapsed intermediate is formed and transiently accumulates during folding. The direct observation of an early collapse for RNA provides support for interpretations of earlier experiments that have suggested the possibility of rapid collapse for this and related RNAs (15, 16, 18).
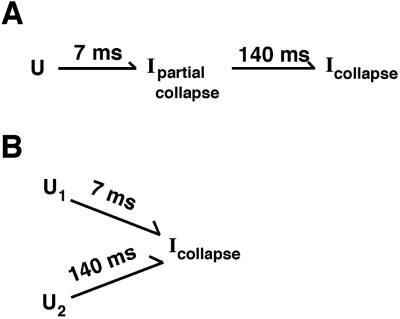
The early collapse was found to occur in two distinct kinetic phases, giving time constants of 7 ms and 140 ms. Two general models can account for the presence of two kinetic phases: in Model 1, compaction proceeds in two steps, with the transient accumulation of an intermediate that has undergone partial compaction (see Scheme S1A), whereas in Model 2, the two phases arise because there are two populations of unfolded ribozyme that undergo compaction with distinct rates (see Scheme S1B). Although the data describing the early collapse were adequately represented by combinations of two states in SVD analysis (Fig. 3B), the presence of two kinetic phases necessitates at least three states. There is no inherent contradiction; the presence of two states in SVD analysis indicates the existence of at least two species. The scattering profile of the third state identified by the kinetic fitting can be reasonably described by a combination of the scattering profiles of the other two states.
Scheme 1.
Model 1 requires that the SAXS profile of the partially collapsed intermediate be approximated by a combination of the SAXS profiles of the folded and unfolded states, whereas Model 2 requires that there be two populations of starting states that do not interconvert with each other and collapse with different rates. Model 1 is favored because of its simplicity—there is no evidence for multiple pathways during compaction under these conditions, and evidence does exist for a single pathway later in folding (32). However, as there is ample evidence for multiple pathways under different conditions and for different versions of this RNA (26, 36–38), Model 2 is plausible. The dependence of the kinetics of collapse on solution conditions before and during Mg2+-induced folding (38) should be valuable in distinguishing the models above and in beginning to probe the physical origins of the collapse.
What structural features could be present in the partially collapsed intermediate postulated in Model 1 (Scheme S1A, Ipartial collapse)? Comparison with simulation suggests that an intermediate in which a subdomain is collapsed but other parts of the structure remain largely extended could give the observed SAXS profile (Fig. 4B). A recent study has shown that under higher ionic strength conditions, upon addition of Mg2+, the P4-P6 domain forms its native tertiary structure on the timescale of milliseconds, indicating that any compaction of P4-P6 must also occur at least this fast, at least under the high ionic strength conditions (34). As formation of the native structure requires the P5abc element within P4-P6 to bend over and onto the rest of the subdomain (Fig. 1A), substantial compaction from an extended structure is necessary. This result raises the possibility that compaction of P4-P6 is also comparably fast under the low ionic strength conditions used here and in previous studies (e.g., ref. 23), but formation of an ensemble of misfolded species slows the onset of specific native structure. Early compaction of P4-P6 is supported by the finding that native structure within P5abc is formed within 30 ms at low ionic strength when P4-P6 is prevented from bending over (by mutations that rigidify the hinge region between P5 and P5a) but not for the wild-type P4-P6. This difference suggests that the wild-type P4-P6 bends at the P5-P5a hinge in less than 30 ms to give a more compact species (16).
Thus, it is possible that the partial collapse seen by SAXS represents formation of a family of intermediates in which P4-P6 has collapsed by bending over upon itself, but other subdomains of the ribozyme are not yet fully collapsed. Partial compaction of other domains also could contribute to the rapid phase of compaction, as could formation of small amounts of local secondary structure not formed in the starting population (39). Further compaction of this partially collapsed intermediate may require disrupting fortuitous nonnative contacts and/or rearrangements that are hindered by topological barriers, giving rise to the slower phase of compaction.
The observation that both phases of compaction are completed before any stable tertiary structure is detected indicates that a nonspecifically collapsed folding intermediate is populated. It is possible that subsets of the known contacts are formed transiently in this state, with each contact formed in a sufficiently low fraction of the population so as not to give detectable protection from solution radicals. It is also possible that contacts are formed in the collapsed intermediate that do not give burial of the ribose moiety, and so do not give protection from cleavage by solution radicals. However, the solution radical protection pattern of the folded ribozyme identifies all known tertiary contacts and is globally consistent with solvent accessibility in a model for the overall structure (23, 33, 40), indicating that the protection approach is a sensitive probe for tertiary structure. Thus, the absence of any protections for the early collapsed intermediate strongly suggests that there is little or no stable tertiary structure in the collapsed state.
On the one hand, the ability of RNA to rapidly compact opens up the possibility of rapid folding—this can be imagined if the preformed secondary structural elements are correctly formed and if they collapse in the vicinity of their positions in the final folded structure. Indeed, under certain conditions, the overall folding of a fraction of the ribozyme population gives a rate constant of ≥1 s−1 and may be as fast as the compaction observed here (41, 42). On the other hand, the compact state could hinder the conformational search for the native state for the fraction of molecules that collapses incorrectly by introducing topological barriers or by favoring the formation of fortuitous nonnative interactions.
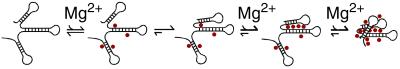
The collapsed intermediate in RNA folding is grossly akin to the molten globule intermediate in protein folding (5), but the physical origin is presumably distinct. A general model to describe rapid and highly cooperative collapse of an RNA molecule upon addition of divalent cations is shown in Fig. 5. Divalent cations localize near one or more helices, increasing the probability that the helices come together; when they do come together, the local negative charge is increased, facilitating the localization of still more cations, thereby giving progressively increased cation localization and compaction. Collapse of polyanions upon the addition of multivalent cations has been suggested on theoretical grounds from ion–ion correlation effects (43) and entropic effects arising from delocalization of counterions in a collapsed state (44). As the collapse for the Tetrahymena ribozyme occurs before tertiary structure formation, it may result from basic features of RNA and, therefore, may be general for RNA. Understanding the physical origins of the collapse provides exciting new challenges and an opportunity to enhance our understanding of the fundamental behavior of RNA molecules and how these molecules traverse complex energetic landscapes to find their native functional structures.
Figure 5.
A model for rapid, Mg2+-induced collapse of RNA. A simple RNA is shown, consisting of duplex regions connected by short single-stranded segments. Mg2+ ions, shown as red circles, bind rapidly to RNA duplexes, neutralizing negative charge and, therefore, favoring convergence of structural elements to give a global collapse (see text). This process could be aided or hindered by fortuitous interactions within the RNA (e.g., refs. 13 and 47).
Supplementary Material
Acknowledgments
We thank Rhiju Das for valuable discussions, Xiaowei Zhuang, Hazen Babcock, and Steve Chu for allowing us to cite unpublished fluorescence experiments, and G. Toombes, J. Korlach, L. Lurio, A. Rühm, V. Genova, and A. Goldman for experimental assistance. We also thank H. Tsuruta for help on Stanford Synchrotron Radiation Laboratory (SSRL) beamline 4–2 and K. Hodgson for his support. L.W.K. was supported by a National Institutes of Health training grant. R.R. was supported by a National Institutes of Health postdoctoral fellowship. This work was supported by a National Institutes of Health grant (to D.H.), a U.S. Department of Energy-Office of Biological and Environmental Research grant (to S.M.G.), and the Nanobiotechnology Center at Cornell (to L.P.). SSRL is supported by the Department of Energy and the National Institutes of Health. Use of the Advanced Photon Source (experiments were performed at IMMYT-Whitehead-CAT on beamline ID-8) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science. These experiments made use of the Cornell Nanofabrication Facility, supported by the National Science Foundation, Cornell University, industrial affiliates, and the Developmental Resource for Biophysical Imaging Opto-Electronics at Cornell, a National Institutes of Health-National Center for Research Resources facility.
Abbreviations
- SAXS
small angle x-ray scattering
- SVD
singular value decomposition
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Ca2+ was used for calibration instead of Mg2+ because of the availability of well characterized calcium-sensitive dyes. As the hydrated radii of Ca2+ and Mg2+ are nearly equal (45), their diffusive mixing times are expected to be similar.
The PDMS layers contain channels that are directly above and below the side channels and the outlet channel of the silicon device, but not above or below the inlet channel. Buffer solutions that do not contain RNA flow through these channels against the top and bottom sealed surfaces of the chip. Thus, the RNA solution in the outlet channel is surrounded by buffer on all sides, minimizing surface effects on the flow profile of the RNA solution.
Additional uncertainty, beyond the reported value obtained from the fit, exists in the time constant for this 140-ms phase. This additional uncertainty arises because of uncertainty in the amplitude of the fast phase under the experimental conditions used to measure the slow phase (25°C for the fast phase vs. 15°C and 37°C for the slower phase). Changes in the amplitude of the fast phase would give changes in the time constant of the slower phase by changing the “starting” point (the y axis value in Fig. 3B) of the slower transition. This uncertainty does not affect the conclusion that both phases of compaction are completed more rapidly than stable tertiary structure has been shown to form.
References
- 1.Levinthal C. In: Proceedings of a Meeting held at Allerton House, Monticello, IL. Debrunner P, Tsibris J C M, Münck E, editors. Urbana: Univ. of Illinois Press; 1969. pp. 22–24. [Google Scholar]
- 2.Dill K A. Biochemistry. 1985;24:1501–1509. doi: 10.1021/bi00327a032. [DOI] [PubMed] [Google Scholar]
- 3.Sosnick T R, Mayne L, Hiller R, Englander S W. Nat Struct Biol. 1994;1:149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- 4.Pandya M J, Williams P B, Dempsey C E, Shewry P R, Clarke A R. J Biol Chem. 1999;274:26828–26837. doi: 10.1074/jbc.274.38.26828. [DOI] [PubMed] [Google Scholar]
- 5.Ptitsyn O B. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin R L. Curr Opin Struct Biol. 1993;3:84–91. [Google Scholar]
- 7.Chamberlain A K, Marqusee S. Adv Protein Chem. 2000;53:283–328. doi: 10.1016/s0065-3233(00)53006-x. [DOI] [PubMed] [Google Scholar]
- 8.Chen L, Wildegger G, Kiefhaber T, Hodgson K O, Doniach S. J Mol Biol. 1998;276:225–237. doi: 10.1006/jmbi.1997.1514. [DOI] [PubMed] [Google Scholar]
- 9.Pollack L, Tate M W, Darnton N C, Knight J B, Gruner S M, Eaton W A, Austin R H. Proc Natl Acad Sci USA. 1999;96:10115–10117. doi: 10.1073/pnas.96.18.10115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plaxco K W, Millett I S, Segel D J, Doniach S, Baker D. Nat Struct Biol. 1999;6:554–556. doi: 10.1038/9329. [DOI] [PubMed] [Google Scholar]
- 11.Jackson S E. Folding Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 12.Sigler P B. Annu Rev Biophys Bioeng. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 13.Herschlag D. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 14.Tinoco I, Jr, Bustamante C. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 15.Russell R, Millett I S, Doniach S, Herschlag D. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 16.Deras M L, Brenowitz M, Ralston C Y, Chance M R, Woodson S A. Biochemistry. 2000;39:10975–10985. doi: 10.1021/bi0010118. [DOI] [PubMed] [Google Scholar]
- 17.Buchmueller K L, Webb A E, Richardson D A, Weeks K M. Nat Struct Biol. 2000;7:362–366. doi: 10.1038/75125. [DOI] [PubMed] [Google Scholar]
- 18.Webb A E, Weeks K M. Nat Struct Biol. 2001;8:135–140. doi: 10.1038/84124. [DOI] [PubMed] [Google Scholar]
- 19.Thirumalai D, Lee N, Woodson S A, Klimov D. Annu Rev Phys Chem. 2001;52:751–762. doi: 10.1146/annurev.physchem.52.1.751. [DOI] [PubMed] [Google Scholar]
- 20.Treiber D K, Williamson J R. Curr Opin Struct Biol. 2001;11:309–314. doi: 10.1016/s0959-440x(00)00206-2. [DOI] [PubMed] [Google Scholar]
- 21.Glatter O, Kratky O. Small Angle X-Ray Scattering. London: Academic; 1982. [DOI] [PubMed] [Google Scholar]
- 22.Doniach S. Chem Rev. 2001;101:1763–1778. doi: 10.1021/cr990071k. [DOI] [PubMed] [Google Scholar]
- 23.Sclavi B, Sullivan M, Chance M R, Brenowitz M, Woodson S A. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- 24.Zarrinkar P P, Williamson J R. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 25.Zaug A J, Grosshans C A, Cech T R. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 26.Russell R, Herschlag D. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 27.Sandy A R, Lurio L B, Mochrie S G J, Malik A, Stephenson G B, Pelletier J F, Sutton M. J Synchrotron Radiat. 1999;6:1174–1184. [Google Scholar]
- 28.Pollack L, Tate M W, Finnefrock A C, Kalidas C, Trotter S, Darnton N C, Lurio L, Austin R H, Batt C A, Gruner S M, Mochrie S G J. Phys Rev Lett. 2001;86:4962–4965. doi: 10.1103/PhysRevLett.86.4962. [DOI] [PubMed] [Google Scholar]
- 29.Tsuruta H, Brennan S, Rek Z U, Irving T C, Tompkins W H, Hodgson K O. J Appl Crystallogr. 1998;31:672–682. [Google Scholar]
- 30.Beebe J A, Kurz J C, Fierke C A. Biochemistry. 1996;35:10493–10505. doi: 10.1021/bi960870m. [DOI] [PubMed] [Google Scholar]
- 31.Henry E R, Hofricter J. Methods Enzymol. 1992;210:129–192. [Google Scholar]
- 32.Russell R, Herschlag D. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 33.Lehnert V, Jaeger L, Michel F, Westhof E. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 34.Silverman S K, Deras M L, Woodson S A, Scaringe S A, Cech T R. Biochemistry. 2000;39:12465–12475. doi: 10.1021/bi000828y. [DOI] [PubMed] [Google Scholar]
- 35.Tsuruta H, Nagamura T, Kimura K, Igarashi Y, Kajita A, Wang Z X, Wakabayashi K, Amemiya Y, Kihara H. Rev Sci Instrum. 1989;60:2356–2358. [Google Scholar]
- 36.Pan J, Thirumalai D, Woodson S A. J Mol Biol. 1997;273:7–13. doi: 10.1006/jmbi.1997.1311. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Deras M L, Woodson S A. J Mol Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 38.Russell R, Zhuang X, Babcock H P, Millett I S, Doniach S, Chu S, Herschlag D. Proc Natl Acad Sci USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaeger J A, Zuker M, Turner D H. Biochemistry. 1990;29:10147–10158. doi: 10.1021/bi00496a002. [DOI] [PubMed] [Google Scholar]
- 40.Latham J A, Cech T R. Science. 1989;245:276–282. doi: 10.1126/science.2501870. [DOI] [PubMed] [Google Scholar]
- 41.Zhuang X, Bartley L E, Babcock H P, Russell R, Ha T, Herschlag D, Chu S. Science. 2000;288:2048–2051. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 42.Heilman-Miller S L, Thirumalai D, Woodson S A. J Mol Biol. 2001;306:1157–1166. doi: 10.1006/jmbi.2001.4437. [DOI] [PubMed] [Google Scholar]
- 43.Khan M O, Jönsson B. Biopolymers. 1999;49:121–125. [Google Scholar]
- 44.Murthy V L, Rose G D. Biochemistry. 2000;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- 45.Israelachvili J N. Intermolecular and Surface Forces. London: Academic; 1992. [Google Scholar]
- 46.Duffy D C, McDonald J C, Schueller O J A, Whitesides G M. Anal Chem. 1998;70:4974–4984. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 47.Cate J H, Gooding A R, Podell E, Zhou K, Golden B L, Kundrot C E, Cech T R, Doudna J A. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.