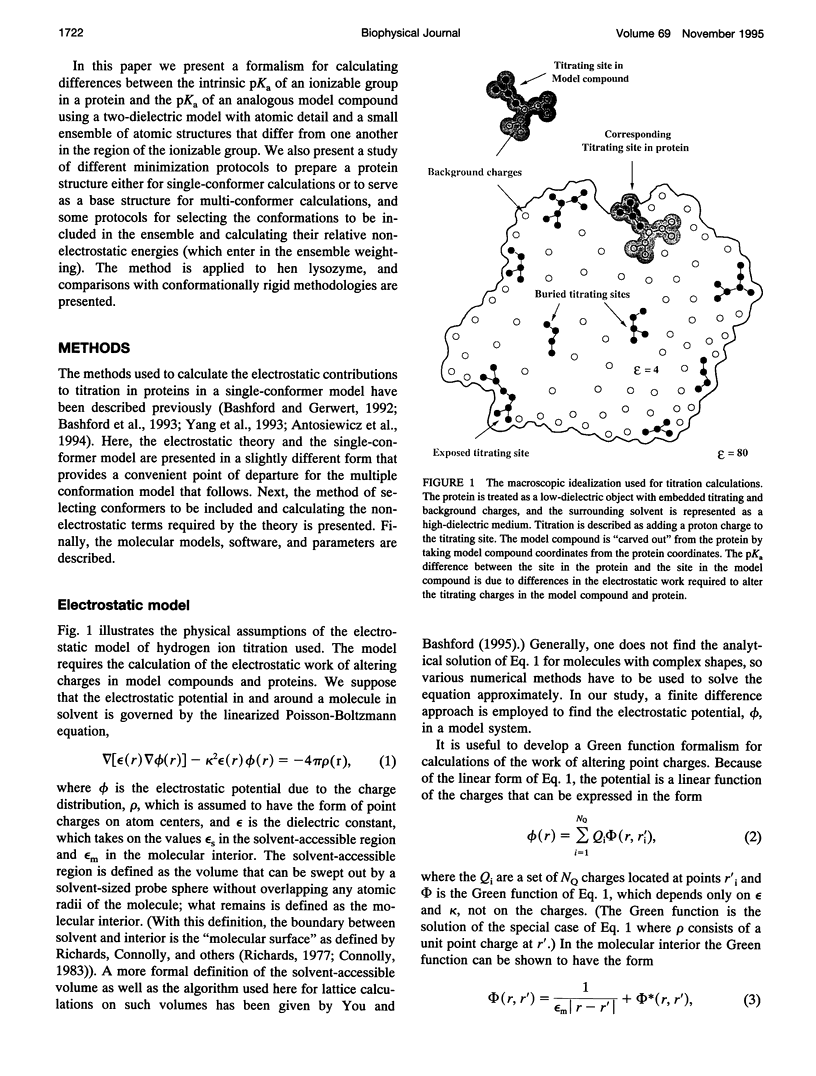
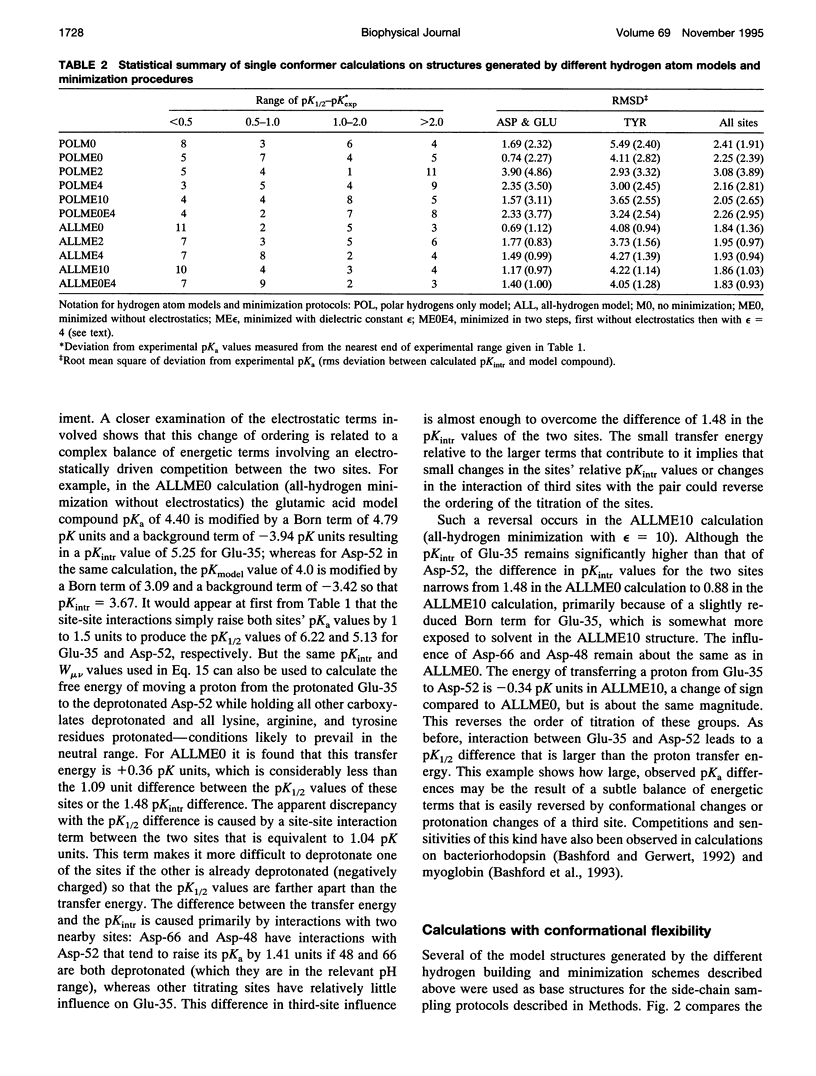
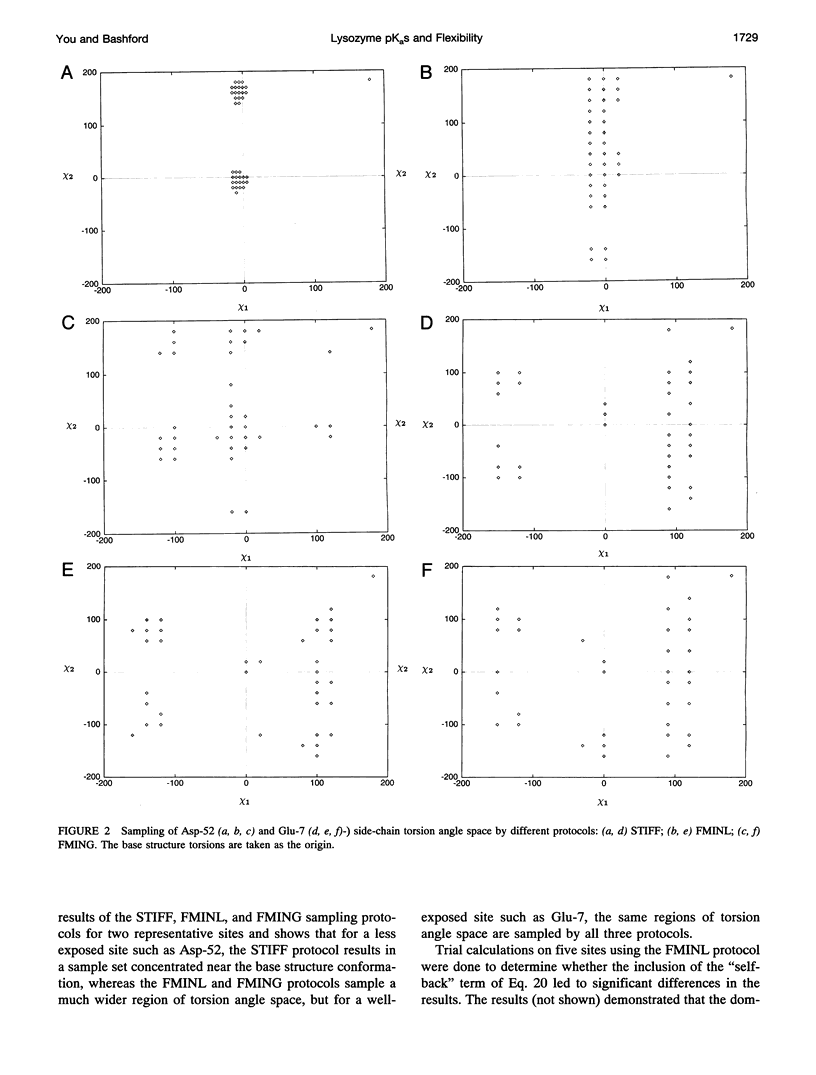
Abstract
A new method for including local conformational flexibility in calculations of the hydrogen ion titration of proteins using macroscopic electrostatic models is presented. Intrinsic pKa values and electrostatic interactions between titrating sites are calculated from an ensemble of conformers in which the positions of titrating side chains are systematically varied. The method is applied to the Asp, Glu, and Tyr residues of hen lysozyme. The effects of different minimization and/or sampling protocols for both single-conformer and multi-conformer calculations are studied. For single-conformer calculations it is found that the results are sensitive to the choice of all-hydrogen versus polar-hydrogen-only atomic models and to the minimization protocol chosen. The best overall agreement of single-conformer calculations with experiment is obtained with an all-hydrogen model and either a two-step minimization process or minimization using a high dielectric constant. Multi-conformational calculations give significantly improved agreement with experiment, slightly smaller shifts between model compound pKa values and calculated intrinsic pKa values, and reduced sensitivity of the intrinsic pKa calculations to the initial details of the structure compared to single-conformer calculations. The extent of these improvements depends on the type of minimization used during the generation of conformers, with more extensive minimization giving greater improvements. The ordering of the titrations of the active-site residues, Glu-35 and Asp-52, is particularly sensitive to the minimization and sampling protocols used. The balance of strong site-site interactions in the active site suggests a need for including site-site conformational correlations.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antosiewicz J., McCammon J. A., Gilson M. K. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994 May 6;238(3):415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- Bartik K., Redfield C., Dobson C. M. Measurement of the individual pKa values of acidic residues of hen and turkey lysozymes by two-dimensional 1H NMR. Biophys J. 1994 Apr;66(4):1180–1184. doi: 10.1016/S0006-3495(94)80900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford D., Case D. A., Dalvit C., Tennant L., Wright P. E. Electrostatic calculations of side-chain pK(a) values in myoglobin and comparison with NMR data for histidines. Biochemistry. 1993 Aug 10;32(31):8045–8056. doi: 10.1021/bi00082a027. [DOI] [PubMed] [Google Scholar]
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Bashford D., Karplus M. pKa's of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990 Nov 6;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Karplus M. Polar hydrogen positions in proteins: empirical energy placement and neutron diffraction comparison. Proteins. 1988;4(2):148–156. doi: 10.1002/prot.340040208. [DOI] [PubMed] [Google Scholar]
- Connolly M. L. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983 Aug 19;221(4612):709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. Calculation of the total electrostatic energy of a macromolecular system: solvation energies, binding energies, and conformational analysis. Proteins. 1988;4(1):7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- Hodsdon J. M., Brown G. M., Sieker L. C., Jensen L. H. Refinement of triclinic lysozyme: I. Fourier and least-squares methods. Acta Crystallogr B. 1990 Feb 1;46(Pt 1):54–62. doi: 10.1107/s0108768189009183. [DOI] [PubMed] [Google Scholar]
- Kuramitsu S., Hamaguchi K. Analysis of the acid-base titration curve of hen lysozyme. J Biochem. 1980 Apr;87(4):1215–1219. [PubMed] [Google Scholar]
- Matthew J. B., Gurd F. R. Calculation of electrostatic interactions in proteins. Methods Enzymol. 1986;130:413–436. doi: 10.1016/0076-6879(86)30019-3. [DOI] [PubMed] [Google Scholar]
- McGrath M. E., Vásquez J. R., Craik C. S., Yang A. S., Honig B., Fletterick R. J. Perturbing the polar environment of Asp102 in trypsin: consequences of replacing conserved Ser214. Biochemistry. 1992 Mar 31;31(12):3059–3064. doi: 10.1021/bi00127a005. [DOI] [PubMed] [Google Scholar]
- Perutz M. F. Electrostatic effects in proteins. Science. 1978 Sep 29;201(4362):1187–1191. doi: 10.1126/science.694508. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Nakamura H., Wada A. Electrostatic forces in two lysozymes: calculations and measurements of histidine pKa values. Biopolymers. 1992 Aug;32(8):897–909. doi: 10.1002/bip.360320802. [DOI] [PubMed] [Google Scholar]
- Tanford C., Roxby R. Interpretation of protein titration curves. Application to lysozyme. Biochemistry. 1972 May 23;11(11):2192–2198. doi: 10.1021/bi00761a029. [DOI] [PubMed] [Google Scholar]
- Warshel A., Russell S. T. Calculations of electrostatic interactions in biological systems and in solutions. Q Rev Biophys. 1984 Aug;17(3):283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- Warwicker J., Watson H. C. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. J Mol Biol. 1982 Jun 5;157(4):671–679. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- Yang A. S., Gunner M. R., Sampogna R., Sharp K., Honig B. On the calculation of pKas in proteins. Proteins. 1993 Mar;15(3):252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]
- Yang A. S., Honig B. Structural origins of pH and ionic strength effects on protein stability. Acid denaturation of sperm whale apomyoglobin. J Mol Biol. 1994 Apr 15;237(5):602–614. doi: 10.1006/jmbi.1994.1258. [DOI] [PubMed] [Google Scholar]