Abstract
Many pathogenic viruses use programmed −1 ribosomal frameshifting to regulate translation of their structural and enzymatic proteins from polycistronic mRNAs. Frameshifting is commonly stimulated by a pseudoknot located downstream from a slippery sequence, the latter positioned at the ribosomal A and P sites. We report here the structures of two crystal forms of the frameshifting RNA pseudoknot from beet western yellow virus at resolutions of 1.25 and 2.85 Å. Because of the very high resolution of 1.25 Å, ten mono- and divalent metal ions per asymmetric unit could be identified, giving insight into potential roles of metal ions in stabilizing the pseudoknot. A magnesium ion located at the junction of the two pseudoknot stems appears to play a crucial role in stabilizing the structure. Because the two crystal forms exhibit mostly unrelated packing interactions and local crystallographic disorder in the high-resolution form was resolvable, the two structures offer the most detailed view yet of the conformational preference and flexibility of an RNA pseudoknot.
Keywords: metal ion coordination|ribosomal frameshifting|RNA tertiary structure|x-ray crystallography
Programmed −1 ribosomal frameshifting is an essential mechanism for regulating the relative expression of proteins that are encoded in two overlapping translational reading frames. Frameshifting is used by viruses (including all retroviruses), DNA insertion sequences, bacteria, and yeast (1, 2). The shift occurs at a heptanucleotide of general sequence X XXX YYZ and can reach efficiencies of up to 30%. In the majority of −1 type frameshifts, a pseudoknot motif is found 5–9 nucleotides downstream from the above slippery sequence (reviewed in ref. 3). HIV-1 is a notable exception to this rule in that a simple hairpin motif acts as the downstream stimulator for the frameshift (4, 5). The efficiency of frameshifting depends on the nature of the slippery sequence, as well as the sequence and complexity of the downstream pseudoknot motif, but the precise mechanism that triggers slipping of the tRNAs bound at the A and P sites from the zero to the −1 frame remains unknown. However, it appears that a protein factor mediating contacts between pseudoknot and ribosome does not play a primary role in the control of frameshifting efficiency (3). Thermodynamic or kinetic control of pseudoknot unfolding may be more important in regulating the efficiency of ribosomal frameshifting (3). Recent elucidation of the three-dimensional structure of the ribosome and the path of the mRNA (6) may make it possible to reformulate frameshifting in terms of ribosomal interactions. This possibility reinforces the importance of understanding the details of pseudoknot structure.
Beet western yellow virus (BWYV) belongs to the class of plant luteoviruses and its mRNA contains a 26-nucleotide pseudoknot motif 6 nucleotides downstream from the slippery sequence where it regulates the expression of an RNA-dependent RNA polymerase (7). We reported the crystal structure of a trigonal crystal form of the RNA pseudoknot from BWYV at 1.6 Å resolution (8). In addition, an extensive mutational analysis identified regions of the pseudoknot important for efficient frameshifting (9). Among the main geometric features revealed by the structure of the 28-nucleotide construct encompassing the pseudoknot (Fig. 1A) were a 48° rotation, a 5-Å helical displacement, and a 25° kink at the junction between stem 1 (S1) and stem 2 (S2). Other remarkable properties of the structure include the minor groove triplex between S1 and loop 2 (L2) and a quadruple base interaction in the major groove of S2, which involves the C12-G26 base pair and the protonated C8 from loop 1 (L1). A25 does not pair with U13 as part of S2; rather, U13 is looped out and A25 is stacked on L2. Remarkably, the number of tertiary hydrogen bonding interactions in the BWYV pseudoknot exceeds the number of hydrogen bonds resulting from Watson–Crick base pairing.
Figure 1.
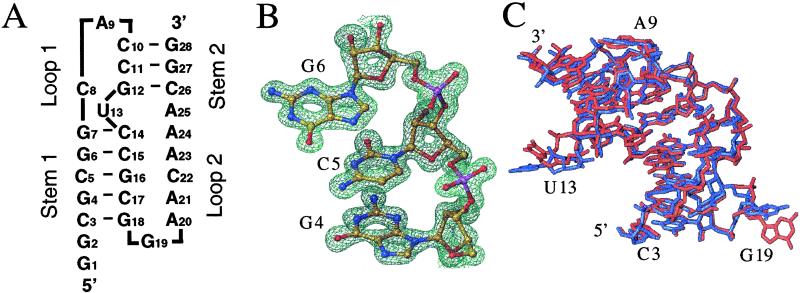
Sequence of the RNA construct and quality and conformational variation of the crystal structure as a function of the lattice. (A) Secondary structure diagram of the frameshifting pseudoknot from BWYV (residues C3–G28). (B) View of the final Fourier 2Fo − Fc sum electron density in the trigonal crystal form at 1.25 Å resolution and contoured at 2σ (= 1 e−/Å3) around residues G4, C5, and G6 from S1. RNA atoms are colored yellow, red, blue, and magenta for carbon, oxygen, nitrogen, and phosphorus, respectively. (C) Overall superposition of the pseudoknot structures in the trigonal (blue) and cubic (red) crystal forms.
In the original structure a magnesium ion was found to coordinate to the 5′-terminal triphosphate moiety and a sodium ion was tentatively assigned to the minor groove region of the S1–L2 triplex. To identify further mono- and divalent metal ions, the resolution of the trigonal crystal form was extended to 1.25 Å. Several of the metal ions newly observed in this structure may contribute to the stability of the pseudoknot. In addition, the structure of a cubic crystal form of the BWYV pseudoknot was refined to a resolution of 2.85 Å. Comparison of the trigonal and cubic crystal forms allows an assessment of lattice-independent conformational preferences of the frameshifting pseudoknot from BWYV.
Methods
Crystallization and Data Collection.
Crystallization conditions of the trigonal form have been described (8). A crystal was frozen without further cryoprotection and used for data collection. Separate low-, medium-, and high-resolution data sets were collected for a single crystal on the insertion device beam line of the DND-CAT at sector 5 of the Advanced Photon Source (Argonne, IL). Cubic crystals were grown using the sitting drop vapor diffusion technique. Droplets containing 2.7 mg/ml RNA, 1 M ammonium sulfate, and 33 mM sodium citrate (pH 5.0) were equilibrated over 2 M ammonium sulfate at 4°C for 1–2 weeks. Crystals were cryo-protected in 2.4 M lithium sulfate/100 mM sodium citrate/20% glycerol and frozen in liquid nitrogen. A complete data set from a single crystal was collected on the BL1–5 line at the Stanford Synchrotron Research Laboratory (Stanford, CA). All data were integrated and scaled with DENZO and SCALEPACK (10), respectively, and selected crystal data and data collection parameters are summarized in Table 1.
Table 1.
Selected crystal data and refinement parameters
| Parameter | Trigonal form | Cubic form |
|---|---|---|
| Space group | P3221 | I213 |
| Cell constants, Å | a, b = 30.16, c = 140.28 | 107.81 |
| Wavelength, Å | 1.00 | 1.08 |
| Unique reflections | 21,462 | 5,034 |
| Resolution, Å | 1.25 | 2.85 |
| Completeness, % | 99.1 | 100 |
| Rsym*, % | 7.9 | 5.4 |
| RNA atoms, waters | 709, 200 | 677, 14 |
| Metal ions | 6 Mg2+, 3 Na+, 1 K+ | — |
| R work†, R test‡ (%) | 14.2, 20.7 | 27.1, 27.5 |
| rms distance (Å), angles (°) | 0.005, 1.30 | 0.009, 1.00 |
Rsym = ∑hkl∑i|I(hkl)i − 〈I(hkl)〉|/∑hkl∑i〈I(hkl)i〉.
R = ∑hkl|F(hkl)o − F(hkl)c|/∑hklF(hkl)o.
For 10% of the data (30).
Structure Refinement.
The structure of the trigonal crystal form was isotropically refined with the program CNS (11). The 1.6-Å-resolution structure (8) served as a starting model. Regions displaying disorder were built into omit electron density maps, using the program TURBO FRODO (12). Anisotropic refinement was carried out with the program SHELX97 (13). The structure of the cubic crystal form was determined by the Molecular Replacement technique with the program AMORE (14), using the refined trigonal structure minus the looped out and capping residues as a starting model. Several regions were manually rebuilt and refinement was carried out subsequently with the program CNS (11).
Metal Ion Binding.
To potentially identify alkali metal ions (Na+ or K+) in the trigonal crystal form, a variety of native data sets were collected at low-energy wavelengths of >1.5 Å to maximize the anomalous contribution of potassium (15). In addition, trigonal crystals were soaked in solutions of potassium chloride to enhance the occupancy of metal ions putatively bound to the RNA. In an alternative approach, thallium acetate was added to droplets containing trigonal crystals such that the final concentration of Tl+ was between 25 and 50 mM. Diffraction data of soaked crystals were collected at a wavelength of 0.97626 Å (12.700 keV; the theoretical value for the L-III edge of Tl is 12.658 keV or 0.97950 Å). These data were then used to generate anomalous difference maps ([Fano − Fcalc] × φcalc) to determine whether thallium ions had potentially replaced potassium ions.
Coordinates.
Coordinates and structure factors for both crystal forms have been deposited in the Brookhaven Protein Databank [PDB ID codes 1L2X (trigonal) and 1L3D (cubic)].
Results and Discussion
Conformational Variations in the Two Crystal Forms.
Crystallization trials using the sparse matrix screening technique resulted in the isolation of two crystal forms of the BWYV RNA pseudoknot. Trigonal crystals were grown from either 2-methyl-2,4-pentanediol or sec-butanol as the precipitant and the cubic form was isolated from high gradients of ammonium sulfate (Table 1). The crystal structure of the pseudoknot in the trigonal form was initially determined by the multiple isomorphous replacement (MIR) method and refined to a resolution of 1.6 Å (8). To fully exploit the diffraction limit of trigonal crystals, data were subsequently collected on the DND-CAT 5-ID beamline at the Advanced Photon Source. The current data are virtually complete to 1.25 Å resolution and an anisotropic refinement was carried out for all RNA atoms and most of the 200 water molecules, as well as metal ions (Table 1). An example of the quality of the final electron density is depicted in Fig. 1B. The resolution of the trigonal crystal form is the highest reported to date for any RNA with significant tertiary structural interactions. By comparison, the cubic pseudoknot crystals diffracted only to relatively low resolution and data collected on the third-generation synchrotron 5-ID beamline were of similar quality as those initially obtained at the Stanford Synchrotron Research Laboratory (SSRL). The final model of the RNA pseudoknot in the cubic form is based on SSRL data with a resolution of 2.85 Å (Table 1).
The high resolution of the trigonal crystal form has allowed us to model alternative conformations observed for portions of the backbone and individual nucleotides. Comparison of the two forms provides insight on the preferred secondary and tertiary structure of the BWYV pseudoknot independent of lattice contacts (Fig. 2). Accordingly, the analysis of RNA fine structure and metal ion coordination described in the following paragraphs is based entirely on the high-resolution form. When the two structures are superimposed (omitting residues G1 and G2 that are not part of the wild-type BWYV pseudoknot sequence), an overall rms deviation of 0.97 Å results (Fig. 1C). Thus, the overall structures of the pseudoknot in the two crystal forms are fairly similar. In particular, the secondary structures are identical and, as previously reported for the trigonal form (8), U13 and A25 are not base paired, but are looped out and stacked on L2, respectively (Fig. 1A). As expected, the main differences between the conformations of the pseudoknot in the two forms are restricted to unpaired nucleotides residing on the surface. As illustrated in Fig. 1C, the positions of A9 that caps S2, the U13 bulge, and G19 at the S1-L2 junction are quite flexible. Additional slight deviations in conformation are observed for portions of L2 that are stacked in the minor groove of S1. The correspondence of secondary structure and overall similarities in tertiary and quaternary structure argue for the relevance of our structural results with regard to the preferred three-dimensional arrangement of the free (in the absence of a ribosome) RNA pseudoknot from BWYV.
Figure 2.
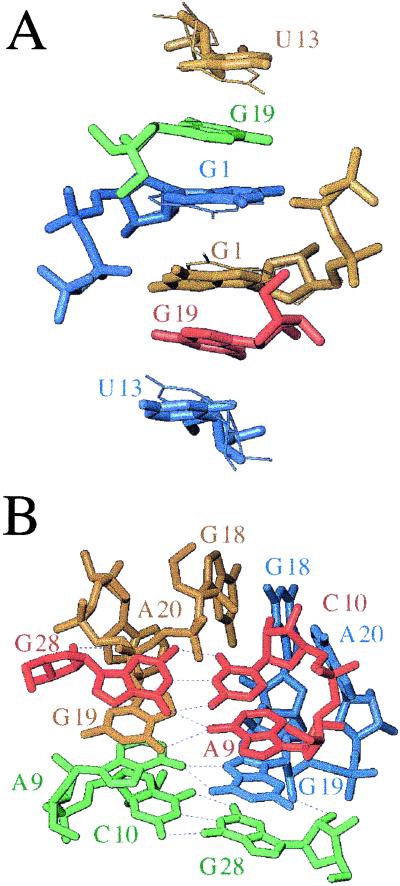
Packing modes in the two crystal forms of the pseudoknot from BWYV. Views of selected lattice interactions in the (A) trigonal and (B) cubic crystal forms. Nucleotides belonging to neighboring molecules are colored differently. In both lattices, a 2-fold rotation axis positioned roughly perpendicular to the plane of projection relates symmetry mates. Alternative conformations of nucleotides G1 and U13 observed in the trigonal crystal form are indicated with thick and thin bonds and hydrogen bonds are thin dashed lines.
Further insight regarding conformational flexibility in the pseudoknot structure is provided by regions exhibiting alternative geometries in the trigonal crystal form (Figs. 2A and 3A). Interestingly, the various conformational disorders were only partly apparent in the earlier structure at 1.6 Å and could not be resolved based on those data. For the model at 1.25 Å, three regions with two alternative conformations of equal occupancy were included in the final refinement. These regions include nucleotides G1, U13, and the sugar-phosphate backbone around A23. At the 5′ terminus, the base and sugar moieties of G1 exhibit alternative orientations (Figs. 2A and 3A), while there is just one major conformation for the triphosphate and the phosphate group of the adjacent residue G2. In the case of the alternative conformations adopted by U13, the disorder includes the nucleoside and the adjacent 5′- and 3′-phosphate groups (Figs. 2A and 3A). The glycosyl angles of the two nucleoside conformations differ by 3°, the sugar pucker of both is C2′-endo and the 5′- and 3′-phosphorus atoms display shifts of 1.4 Å and 3.9 Å, respectively. These geometric alterations result in an average shift of 0.9 Å for uracil base atoms. The third region of enhanced conformational flexibility concerns a portion of L2. There, the sugar moiety of A23 and the adjacent 5′- and 3′-phosphate groups display alternative conformations (Fig. 3A). In summary, the conformational variations observed between the two crystal forms and in the trigonal form alone are consistent with limited conformational freedom of unpaired nucleotides in the capping, junction, and triplex third-strand portions of the RNA pseudoknot, regions that may ultimately be important for recognition and function.
Figure 3.
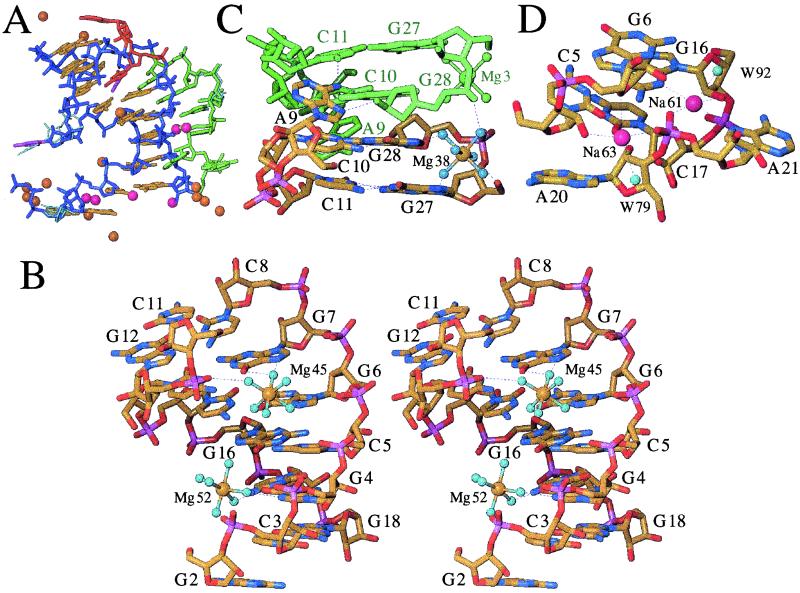
Coordination modes of metal ions in the trigonal crystal form. (A) View of individual magnesium (15) and monovalent ions (ref. 5; four Na+ with full occupancy and two K+ with occupancy 0.5) colored gold and magenta, respectively, surrounding each pseudoknot molecule. All metal ions are engaged in either direct or water mediated contacts to the RNA. The backbone and base pair portions of stem regions are colored blue and yellow, respectively. L1 and L2 residues are colored red and green, respectively, and the base portions of U13 and A25 are highlighted in purple. Alternative conformations of G1, U13, and A23 and surrounding regions are indicated with thin cyan lines. (B) Stereo view of magnesium hexahydrates Mg45 and Mg52 in the major groove of S1. (C) Magnesium hexahydrate Mg38 in the major groove of S2. (D) Sodium ions Na61 and Na63 in the minor groove of S1. RNA atoms are colored yellow, red, blue, and magenta for carbon, oxygen, nitrogen, and phosphorus, respectively, a symmetry related molecule and magnesium ion are colored green (Fig. 3C), hydrogen bonds and coordination spheres of metal ions are indicated with thin dashed lines and selected residues are labeled.
Lattice Contacts.
Although base stacking dominates the interactions between adjacent RNA pseudoknots in both lattices, the intermolecular contacts in the trigonal and cubic forms are significantly different. This finding suggests that the pseudoknot conformation is more or less packing-independent. The asymmetric unit of both crystals consists of a single RNA pseudoknot. In the trigonal form, predominance of stacking is evident from a hexanucleotide stack, involving unpaired nucleotides G1, U13, and G19 from four symmetry mates (Fig. 2A). Its significance for lattice formation and stability is supported by the observation that crystals cannot be grown from RNA constructs lacking either G1 or G2 (see below) or both.
Lattice interactions involving stacking in the cubic form include a junction formed by four molecules (Fig. 2B). Two molecules related by a dyad are stacked tail-to-tail (and head-to-head at the opposite end, a feature also encountered in the trigonal form), such that their A9 caps are spaced by about 2.8 Å along the S2 helix axis. The adenosines are engaged in asymmetric pairs with the extruded G19 residues from two additional molecules (both purines present their Hoogsteen face), the latter related to each other by the above dyad. Thus, the base sandwich exhibits overall 2-fold symmetry and is made up of terminal base pairs from two symmetry related pseudoknot stems (S2) that clamp two noncanonical A9–G19 pairs, the guanine bases contributed by molecules 3 and 4 (Fig. 2B). With G19 hydrogen bonded to the capping adenosine, G1 and U13 are forming a separate stack by themselves in the cubic form.
The above interactions are complemented by head-to-head stacking between stems (S1), a lattice contact observed in the trigonal and in the cubic crystal. At the stacking interface, G2 residues (the only remaining unpaired nucleotide) are facing the major groove edge of symmetry-related C3-G18 base pairs with their Watson–Crick side. Thus, N2 and N1 of G2 are hydrogen bonded to N7 and O6 of G18, respectively, and O6 of G2 is hydrogen bonded to N4 of C3 (data not shown).
Coordination Modes of Magnesium Ions.
Six ordered magnesium ions were found in the asymmetric unit of trigonal BWYV pseudoknot crystals. Every pseudoknot molecule has potential contacts to 15 Mg2+ (Fig. 3A) and, with the exception of two ions (Mg45 and Mg52, Table 2), all of them are shared between neighboring molecules. Three of the Mg2+ per asymmetric unit are engaged in at least two inner-sphere coordinations to RNA atoms (Mg29, Mg34, and Mg59; Table 2). Further, at least one ligand of these ions is constituted by an oxygen atom of nucleotide G1 (GTP) and, in addition, Mg29 and Mg34 are bound to a phosphate oxygen of G2 and G19, respectively. Mg29 was also found in the original structure of the BWYV pseudoknot at 1.6 Å resolution (8). However, nucleotides G1 and G2 are not part of the natural BWYV sequence. Therefore the metal ions coordinated to them are of less interest in terms of stability and function of the frameshifting pseudoknot. It is noteworthy though that the clustered magnesium ions at the 5′-terminal pppG1pG2 portion of the pseudoknot form an integral part of the lattice. This is evident from the observation that crystals cannot be grown from constructs featuring a 5′-terminal guanosine monophosphate (5′-pG1pG2).
Table 2.
Metal ion coordination
| Cation | Base | RNA ligand | Distance, Å | Water molecules | Average distance, Å |
|---|---|---|---|---|---|
| Mg29 | G1 | O1B* | 2.0 | W30, W31, W32, W33 | 2.1 |
| G2 | O1P | 2.0 | |||
| Mg34 | G1b#†‡ | O6 | 2.1 | W35a, W36, W37a, W37b | 2.1 |
| G19 | O2P | 2.1 | |||
| Mg38§ | W39, W40, W41, W42, W43, W44 | 2.1 | |||
| Mg45§ | W46, W47b, W48b, W49, W50, W51 | 2.1 | |||
| Mg52§ | W53, W54, W55, W56b, W57, W58 | 2.1 | |||
| Mg59 | G1 | O3G | 2.3 | W35b, W36#, W60 | 2.0 |
| G1 | O2B | 1.8 | |||
| G1a# | O6 | 2.2 | |||
| Na61 | G16 | O2′ | 2.9 | W92 | 2.7 |
| G16 | N3 | 2.8 | |||
| A21 | O2P | 2.7 | |||
| A21 | N7 | 3.1 | |||
| Na62 | C3 | O2′ | 2.7 | W104 | 2.8 |
| C3 | O2 | 2.7 | |||
| G18# | O2P | 2.7 | |||
| Na63 | C5 | O2′ | 2.6 | W79 | 2.9 |
| G6 | O4′ | 3.1 | |||
| A21 | O1P | 2.7 | |||
| K64¶ | G1 | O2′ | 3.0 | W65, W66, W102, W102# | 2.9 |
| G2 | N7 | 3.1 | |||
| G2 | O2P | 2.7 |
The letters A, B, and G refer to the α-, β-, and γ-phosphates, respectively, of G1.
The letters a and b refer to alternative positions of nucleotides and water molecules exhibiting disorder.
Symmetry related nucleotides or water molecules are marked by #.
Coordination shown in Fig. 3.
Occupancy = 0.5; a dyad dissects the K+—W65 bond and W66 is located on the dyad.
The three other magnesium ions are hexahydrates and reside in the major groove of either S1 (Mg45 and Mg52) or S2 (Mg38; Table 2). Water molecules coordinated to Mg45 form hydrogen bonds to the O6 and N7 atoms of both G6 and G7 (Fig. 3B). The coordination site is located at the upper end of S1, adjacent to the bulged U13 and thus right at the S1–S2 interface. Therefore, Mg45 takes on a crucial role in the stabilization of the relative orientation of the two stems. This is further illustrated by a hydrogen bond between one of its water ligands and a phosphate oxygen of G12. The particular twist angle between S1 and S2 leads to rotation of P12 into the major groove, where it enhances an already strongly negative electrostatic potential. By bridging electronegative moieties from opposite strands of S1 (G6, G7) and S2 (G12), Mg45 likely provides coulombic stabilization of the pseudoknot, thereby locking it in its particular tertiary structure.
The major groove edges of guanine offer preferred binding sites for Mg2+, either through inner- or outer-sphere types of coordination (16–18). In addition to the G6pG7 step occupied by Mg45, four more guanines are nearby: G4, G16, and G18 in S1, and G12. Mg52 lies more or less within the best plane defined by G4 base atoms and uses two of its water ligands to contact guanine O6 and N7 (Fig. 3B). The G4 nucleotide appears to offer ideal steric and electrostatic conditions for Mg2+ binding. The electrostatic conditions are the result of the strongly negative potential at the floor of the groove, as well as closely spaced phosphate groups at the periphery. Magnesium binding to G12 or G16 is most likely prohibited because of steric limitations; G12 is buried behind phosphate groups at the S1–S2 interface and is blocked by C8, and G16 is positioned too close to G4 and G6 to allow coordination of a further ion. Finally, G18 is engaged in lattice interactions (see section above) and therefore unavailable for metal ion coordination.
In the major groove of S2, the G27pG28 step harbors the third magnesium hexahydrate, Mg38 (Fig. 3C). Four of its coordinated water molecules are engaged in hydrogen bonds to RNA atoms: N7 of G27 and G28 and phosphate oxygens of G27 and G28, the latter nucleotide contributed by a symmetry-related molecule. It is likely that Mg38 can also coordinate to the S2 major groove when the interaction to a neighboring molecule observed in the crystal lattice is not present. However, compared with Mg45 that occupies a key site in the pseudoknot structure, it is more difficult to assess a potential role of Mg38 in the stability and function of the BWYV RNA pseudoknot.
Coordination Modes of Monovalent Metal Ions.
Four putative alkali metal ion coordination sites were identified in the asymmetric unit of trigonal crystals. Each pseudoknot molecule may interact with six monovalent ions, two of which feature occupancy 0.5 (Fig. 3A). Initially, all solvent peaks in the electron density maps were refined as water molecules. Four of these exhibited unusually low temperature factors following the preliminary isotropic least square refinement. All four displayed contacts to at least four RNA atoms or water molecules with an upper distance limit of 3 Å, and one of the RNA atoms was a phosphate oxygen. The remainder of the coordination spheres was made up of RNA atoms such as cytosine O2, guanine N7, or 2′-hydroxyl groups, as well as waters (Table 2).
To further investigate the identity of these peaks, we performed single-wavelength anomalous diffraction (SAD) data collections both at low and high energy. The latter was conducted with crystals that were soaked in thallium acetate, a strategy that had previously been shown to be of use for pinpointing K+ binding sites in group-I intron crystals (19). At relatively low energy, equivalent to wavelengths >1.5 Å, the anomalous contribution of K+ can be exploited for locating it in crystals (15). Detecting Tl+ is more straightforward in principle because this ion exhibits significant anomalous scattering both in the long and short wavelength ranges, the latter at the L-III edge. Moreover, Tl+ is a strong scatterer and can be identified simply by its peak height in standard electron density maps. The only one among the above peaks that showed significant anomalous density in maps based both on data collected at short (Tl+) and long wavelength (Tl+, K+) was peak no. 64, which was consequently interpreted as a potassium ion (Table 2).
The coordination sphere of K64 consists of atoms from G1 and G2 and three water molecules. As in the case of three of the Mg2+ ions above, K64 is therefore not relevant with regard to the stability or function of the wild-type pseudoknot. Peaks 61, 62, and 63 that did not reveal Tl+ occupancy in the anomalous maps were refined as Na+ (Table 2). Trigonal crystals were grown from solutions that are about equimolar in potassium Mops buffer and sodium chloride. It is possible that Na+ ions initially occupying certain sites were not displaced by Tl+ under the conditions used. In terms of a potential structure-stabilizing effect, the locations of Na61 and Na63 are the most interesting among the four putative alkali metal ion binding sites. They are bound adjacent to the S1–L2 junction, where the minor groove triplex originates (Fig. 3D). There the phosphate group of A21 sits in the center of the S1 minor groove, with the sodium ions bridging its oxygen atoms to 2′-hydroxyl groups from both strands, as well as to base atoms at the floor of the S1 groove and from the third strand. Therefore, the ions could help relieve a potentially unfavorable electrostatic interaction between the phosphate group and the negatively polarized edge of the C5–G16 base pair. The remaining ion, Na62, occupies a site adjacent to the base of S1 and coordinates to atoms of both C3 and G18.
Conclusions
The crystal structures of the BWYV pseudoknot provide a detailed picture of the dynamics of the RNA molecule and ordered metal ions coordinated to it. When interpreting the results of a crystallographic analysis of metal ion binding, it is important to bear in mind that crystal structures even at high resolution cannot accurately represent delocalized or mobile ions that may nevertheless contribute to the overall stability of a fold (20–22). In the trigonal crystal form, the combined positive charges of the mono- and divalent metal ions visible in electron density maps amount to neutralization of half the phosphate groups. In addition to the two general classes of ions, represented by those with highly specific and ordered structure on the one hand and delocalized ones on the other, crystal structures can contain metal ions sites that are solely the result of particular lattice interactions. Magnesium ions are known to stabilize pseudoknots (see, for example, ref. 23) and the Mg45 hexahydrate complex bound at the S1–S2 helical interface is a prime candidate for a stabilizing and conformation-specific ion coordination in an RNA pseudoknot. It has previously been pointed out that modification of bases near the junction destabilizes frameshifting (9). This finding suggests that Mg45 may play a special role in stabilizing the pseudoknot, and that unwinding is preceded by ejection of that ion. The observation here of divalent metal ion coordination to the pseudoknot major groove is in line with the results of other structural studies of pseudoknots in solution and in crystals (24, 25). Thus, the helix–helix junction emerges as one of the regions where metal ions may be capable of providing specific contributions to the thermodynamic stability of a pseudoknot.
Specific binding by alkali metal ions and in particular by K+ has been demonstrated to be important for RNA folding and stability (26, 27). Studies of the thermodynamics of the tertiary unfolding of the BWYV pseudoknot as a function of the nature of the monovalent ion render it unlikely that a particular type of alkali metal ion contributes preferentially to stability (28). The number of counter ions released as a result of the unfolding transition amounted to 0.7. The BWYV pseudoknot is unique in that significant tertiary structural stabilization is derived from stem–loop interactions (3, 8). Therefore, the putative Na61 ion in the minor groove of the S1–L2 triplex deserves particular attention in terms of a specific ion coordination and potential stabilization.
Even at high resolution, crystal structures cannot possibly answer all issues regarding ion coordination. For example, it is unclear to what extent monovalent metal ions can replace divalent metal ions at individual coordination sites. The recent finding that a specific site in the major groove of an A-form duplex was occupied by alkali, as well as earth alkali metal ions is worth mentioning in this respect (15). In particular, the identification of binding sites in RNA pseudoknots that are fully or partially occupied by monovalent cations may require alternative approaches beyond the structural approaches described here. Nucleotide analog interference screening (NAIS; ref. 29) may provide a complementary strategy for further investigation of this issue.
Finally, we note that the mechanism by which pseudoknots stimulate −1 translational frameshifting cannot be revealed by studies of the structure and function of the pseudoknot RNA alone. Rather, shedding light on mechanistic aspects will require a detailed structure of the complex between the ribosome and mRNA containing a pseudoknot. The recent tracing of the path of mRNA through the E. coli ribosome at 7 Å resolution (6) suggests that we may soon be able to visualize the detailed interactions between a pseudoknot and the S3, S4, and S5 proteins at the downstream tunnel of the ribosome, where the mRNA enters.
Acknowledgments
Assistance by Mike Soltis at the Stanford Synchrotron Research Laboratory is gratefully acknowledged, and we thank Dr. Joseph Brunzelle for help with various programs. This work was supported by National Institutes of Health Grant AI-47299 (to M.E. and A.R.). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. The DuPont–Northwestern–Dow Collaborative Access Team Synchrotron Research Center at the Advanced Photon Source (Sector 5-ID) is supported by E. I. DuPont de Nemours & Co., The Dow Chemical Company, the National Science Foundation, and the State of Illinois.
Abbreviation
- BWYV
beet western yellow virus
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes 1L2X (trigonal) and 1L3D (cubic)].
References
- 1.Farabaugh P J. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gesteland R F, Atkins J F. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 3.Giedroc D P, Theimer C A, Nixon P L. J Mol Biol. 2000;298:167–185. doi: 10.1006/jmbi.2000.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacks T, Power M D, Masiarz F R, Lucius P A, Barr P J, Varmus H E. Nature (London) 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 5.Parkin N T, Chamorro M, Varmus H E. J Virol. 1992;66:5147–5151. doi: 10.1128/jvi.66.8.5147-5151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yusupova G Z, Yusupov M M, Cate J H D, Noller H F. Proc Natl Acad Sci USA. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 7.Garcia A, van Duin J, Pleij C W A. Nucleic Acids Res. 1993;21:401–406. doi: 10.1093/nar/21.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su L, Chen L, Egli M, Berger J M, Rich A. Nat Struct Biol. 1999;6:285–292. doi: 10.1038/6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y-G, Su L, Maas S, O'Neill A, Rich A. Proc Natl Acad Sci USA. 1999;96:14232–14239. doi: 10.1073/pnas.96.25.14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 11.Brünger A T. CNS: Crystallography & NMR System. New Haven, CT: Yale Univ.; 1998. , Version 0.9. [Google Scholar]
- 12.Cambillau C, Roussel A. TURBO FRODO. Marseille, France: Université Aix-Marseille II; 1997. , Version OpenGL.1. [Google Scholar]
- 13.Sheldrick G M, Schneider T R. Methods Enzymol. 1997;277:319–343. [PubMed] [Google Scholar]
- 14.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 15.Tereshko V, Wilds C J, Minasov G, Maier M A, Prakash T, Howard A, Wawrzak Z, Manoharan M, Egli M. Nucleic Acids Res. 2001;29:1208–1215. doi: 10.1093/nar/29.5.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egli M, Gessner R V, Williams L D, Quigley G J, van der Marel G A, van Boom J H, Rich A, Frederick C A. Proc Natl Acad Sci USA. 1990;87:3235–3239. doi: 10.1073/pnas.87.8.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minasov G, Tereshko V, Egli M. J Mol Biol. 1999;291:83–99. doi: 10.1006/jmbi.1999.2934. [DOI] [PubMed] [Google Scholar]
- 18.Chiu T K, Dickerson R E. J Mol Biol. 2000;301:915–945. doi: 10.1006/jmbi.2000.4012. [DOI] [PubMed] [Google Scholar]
- 19.Basu S, Rambo R P, Strauss-Soukup J, Cate J H, Ferré-D'Amaré A, Strobel S A, Doudna J A. Nat Struct Biol. 1998;5:986–992. doi: 10.1038/2960. [DOI] [PubMed] [Google Scholar]
- 20.Laing L G, Gluick T C, Draper D E. J Mol Biol. 1994;237:577–587. doi: 10.1006/jmbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- 21.Murthy V L, Rose G D. Biochemistry. 2001;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- 22.Misra V K, Draper D E. Proc Natl Acad Sci USA. 2001;98:12456–12461. doi: 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nixon P L, Giedroc D P. Biochemistry. 1998;37:16116–16129. doi: 10.1021/bi981726z. [DOI] [PubMed] [Google Scholar]
- 24.Gonzales R L, Jr, Tinoco I., Jr J Mol Biol. 1999;289:1267–1282. doi: 10.1006/jmbi.1999.2841. [DOI] [PubMed] [Google Scholar]
- 25.Nix J, Sussman D, Wilson C. J Mol Biol. 2000;296:1235–1244. doi: 10.1006/jmbi.2000.3539. [DOI] [PubMed] [Google Scholar]
- 26.Shiman R, Draper D E. J Mol Biol. 2000;302:79–91. doi: 10.1006/jmbi.2000.4031. [DOI] [PubMed] [Google Scholar]
- 27. Conn, G. L., Gittis, A. G., Lattman, E. E., Misra, V. K. & Draper, D. E. (2001) J. Mol. Biol., in press. [DOI] [PubMed]
- 28.Nixon P L, Giedroc D P. J Mol Biol. 2000;296:659–671. doi: 10.1006/jmbi.1999.3464. [DOI] [PubMed] [Google Scholar]
- 29.Basu S, Strobel S A. Methods. 2001;23:264–275. doi: 10.1006/meth.2000.1137. [DOI] [PubMed] [Google Scholar]
- 30.Brünger A T. Nature (London) 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]