Abstract
In recent years, short coiled coils have been used for applications ranging from biomaterial to medical sciences. For many of these applications knowledge of the factors that control the topology of the engineered protein systems is essential. Here, we demonstrate that trimerization of short coiled coils is determined by a distinct structural motif that encompasses specific networks of surface salt bridges and optimal hydrophobic packing interactions. The motif is conserved among intracellular, extracellular, viral, and synthetic proteins and defines a universal molecular determinant for trimer formation of short coiled coils. In addition to being of particular interest for the biotechnological production of candidate therapeutic proteins, these findings may be of interest for viral drug development strategies.
Keywords: protein engineering, sequence-to-structure rules, protein–protein interaction, salt bridges, x-ray crystallography
The potential of short α-helical coiled coils for protein engineering, biotechnological, biomaterial, basic research, and medical applications has recently been recognized (1–12). The wide range of applications underscores the need for topological control of coiled-coil peptides. Although seemingly simple, coiled coils can form a variety of different assemblies ranging from dimers to pentamers (13, 14). Furthermore, coiled coils can form homomers or heteromers with their chains arranged in a parallel or antiparallel fashion. To understand how side-chain–side-chain interactions determine a particular structure, it is important to discriminate the factors responsible for specific interhelical interactions from those that are common to all coiled coils such as the heptad repeat.
It is generally acknowledged that the detailed packing geometry of hydrophobic core residues correlates with the oligomerization state (15, 16). In dimers the side chains of the residues at a and d positions pack in a manner termed parallel and perpendicular, respectively. In the four-stranded state the dimer mode is reversed. Trimeric coiled coils are characterized by an intermediate geometry of these residues, termed acute. Previous studies found that these oligomerization states are determined in particular by the distribution of isoleucine and leucine residues that prefer the parallel and acute, and the acute and perpendicular geometries, respectively (15, 16).
Buried polar residues in hydrophobic interfaces also play an important role in determining the number and orientation of strands in coiled coils. Many two-stranded leucine zippers of transcription regulators contain at least one conserved polar residue. Frequently an asparagine or a lysine residue occupies heptad position a toward the center of the sequence, suggesting a common mechanism for determining dimer specificity in these molecules (17). Polar residues like glutamine and threonine at positions a and d, respectively, can favor trimer formation (18, 19). Interhelical interactions between side chains of residues at the e and g positions as well as the packing of these amino acids against the hydrophobic a and d core residues have also been shown to contribute determining the number, orientation, and selection of strands in a coiled coil (1).
The characteristics of coiled coils have allowed the development of a variety of statistics- and pattern-based methods that predict the occurrence of the motif with a high degree of confidence (17, 20–28). However, the reliable prediction of the specific oligomerization state of many native coiled coils remains difficult. Although predicted to be dimeric, many short coiled coils form three-stranded parallel coiled-coil structures. This observation is strikingly exemplified by the three-stranded, 32-residue coiled-coil domain of the actin-associated protein coronin 1 (referred to as ccCor1; ref. 29). In contrast to experimental evidence, ccCor1 is predicted both by scorer (24) (Δ-score of 2.49) and multicoil (23) (probability of 0.98) to form a two-stranded structure. It exhibits even higher scores than the archetypical dimeric 33-residue coiled coil of the yeast transcriptional activator GCN4 (denoted GCN4-p1; Δ-score of 1.55 and probability of 0.67, respectively). To understand the structural basis of this discrepancy, we have carried out a detailed biophysical study on ccCor1.
Materials and Methods
Peptide Preparations. All ccCor1 peptides (the WT sequence corresponds to residues Val-430–Lys-461 of mouse coronin 1A) and the ccβ-pR8K peptide were assembled on an automated continuous-flow synthesizer by using standard methods. The purity of the peptides was verified by reversed-phase analytical HPLC, and the identities were assessed by mass spectral analysis. Construction, recombinant expression, and purification of the TN-C-p1C64S/R125K (chicken tenascin-C, residues 34–139) and ccMat1-R487A (human matrilin-1, residues 454–496) mutants were performed as described (30, 31). Peptide stock solutions were prepared in either 10 mM Tris·HCl, pH 7.0 (for crystallization) or 5 mM sodium phosphate, pH 7.4 supplemented with 150 mM NaCl (PBS; for solution studies). Disulfide bridge exchange and shuffling in redox buffer to assess the oligomerization states of the TN-C-p1C64S/R125K and ccMat1-R487A mutants were performed as described (30, 31). Concentrations of peptide samples were determined by the Advanced Protein Assay (Cytoskeleton, Denver).
Crystal Structure Determination. Crystals were grown by mixing equal volumes of protein with the reservoir solution containing 0.1 M Mes/NaOH at pH 6.5, 10 mM ZnSO4, and 20% polyethylene glycol monomethyl ether 550. The ccCor1 crystals grew within 1 week from a 10 mg/ml stock solution at 4°C by using the hanging drop method. In-house data sets were collected by using CuKα radiation produced by an Enraf-Nonius Service (Delft, The Netherlands) FR591 rotating anode generator. Synchrotron data sets were collected at the Swiss Light Source (Villigen PSI, Switzerland) protein beam line X06SA. The ccCor1 structure exhibited space group P1 and was solved by molecular replacement by using a search model based on the structure of the GCN4-p1 Asn-16–Gln trimer [Protein Data Bank (PDB) ID code 1zim]. Iterative rounds of model building and conjugate gradient refinement resulted in a complete high-resolution model for ccCor1. The refined high-quality model (R = 0.16/Rfree = 0.20) consists of one ccCor1 trimer with 798 nonhydrogen protein atoms, 94 water molecules, and 7 Zn2+ ions. Data sets, refinement statistics, and superhelical parameters of the refined ccCor1 coiled-coil structure are given in Table 3, which is published as supporting information on the PNAS web site).
Analytical Ultracentrifugation (AUC), Static Light Scattering (SLS), and CD Spectroscopy. AUC was performed on an Optima XLA analytical ultracentrifuge (Beckman) equipped with an adsorption optical system and an An-60Ti rotor. Data were collected at 26,000, 32,000, 38,000, and 48,000 rpm for ccCor1; 26,000, 29,000, and 32,000 rpm for ccCor1-R450A and ccCor1-R450Nle; 28,000 and 34,000 rpm for ccCor1-R450K; and 36,000 and 44,000 rpm for both ccβ-p and ccβ-pR8K. A single ideal species model was fitted to the data. The partial specific volumes of the synthetic peptides were calculated from their amino acid sequence. Solvent density for PBS was taken as 1.005 g/ml.
SLS experiments were performed by using a miniDAWN TriStar with Optilab rex refractometer (Wyatt, Santa Barbara, CA) coupled to a Superdex 200 10/30 gel filtration column on an Agilent Technologies (Palo Alto, CA) 1100 Series HPLC. One hundred microliters of 1 mg/ml peptide solutions was injected on the column equilibrated in 20 mM Tris, pH 7.4 supplemented with 75 mM NaCl. Molecular weights were calculated by using the Wyatt astra, version 4.90.08 software package.
Far-UV CD spectroscopy was carried out on a J-810 spectropolarimeter (Jasco, Tokyo) equipped with a temperature-controlled quartz cell of 0.1-cm pathlength. The spectra shown are the averages of five accumulations and were evaluated with the Jasco software packages. A ramping rate of 1°C per min was used to record the thermal unfolding profiles. The apparent midpoints of the transitions, Tms, were taken as the maximum of the derivative d[Θ]222/dT.
Database Search. High-resolution structures of short coiled coils comprising ≤50 amino acid residues and harboring the trimerization motif were identified by cross-referencing the SOCKET (14), version 2.18 coiled-coil database (supplemented with PDB ID codes 2akf, 1qbz, 1jek, 1s9z, 1kyc, 1hqj, 1env, 1eo8, 2bez, and 1ij3) against a PROSITE (www.expasy.org/prosite) PDB ID code hit list obtained with the sequence pattern R-[ILVM]-X-X-[ILV]-E. A total of 50 parallel and 141 antiparallel dimers, 16 parallel and 11 antiparallel trimers, 1 parallel and 6 antiparallel tetramers, and 1 parallel pentamer were used for the search. The data set comprises a total of 226 nonredundant coiled coils.
Of the 16 parallel three-stranded coiled coils (PDB ID codes 2akf, 1qbz, 1jek, 1s9z, 1kyc, 1hqj, 1env, 1eo8, 2bez, 1ij3, 1aa0, 1aq5, 1coi, 1fzc, 1mof, and 1rtm), 12 structures contain the motif. From the four structures that do not harbor the motif (PDB ID codes 1coi, 1fzc, 1mof, and 1rtm), only two structures can be classified as autonomous (i.e., their parallel three-stranded coiled-coil structures are not influenced by flanking domains) coiled coils (coil-VaLd, PDB ID code 1coi; Moloney murine leukemia virus transmembrane subunit, PDB ID code 1mof). Thus 14 sequences were used to calculate the frequency of occurrence (12/14 = 0.86) of the trimerization motif in parallel three-stranded coiled coils. Finally, one antiparallel trimer (PDB ID codes 1bg1; frequency of occurrence 1/11 = 0.09), two parallel dimers (PDB ID codes 1fos and 1zta; frequency of occurrence 2/50 = 0.04), one antiparallel dimer (PDB ID code 1grj; frequency of occurrence 1/141 = 0.007), and no tetramers and pentamers were found to contain the motif.
Results
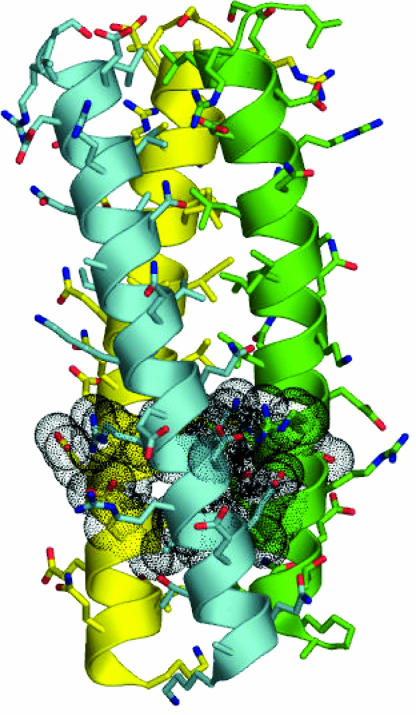
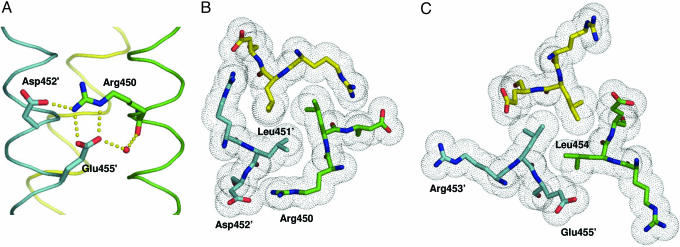
The ccCor1 Crystal Structure Reveals a Distinct Structural Motif. To understand the molecular basis of ccCor1 trimerization, we solved its crystal structure at 1.2-Å resolution. The overall structure of the trimer shows characteristics typical of left-handed parallel coiled coils (Fig. 1). The most prominent feature seen in the trimer is a distinct structural motif formed by the sequence segment Arg-450–Glu-455. As shown in Fig. 2, the motif encompasses both optimal networks of surface salt bridges and internal hydrophobic packing interactions. Interhelical salt bridges are formed by a bifurcated contact between Arg-450:Nε,Nη2 at position g of one chain and Glu-455′:Oε1,Oε2at position e′, and an interaction between Arg-450:Nη2 and Asp-452′:Oδ1 at position b′ of the neighboring chain (Fig. 2 A). A water-mediated hydrogen bond between Arg-450:O and Glu-455′:Oε2 completes the network. Tight interhelical hydrophobic contacts involve the side chain of Leu-451′ at position a′, which is optimally shielded from solvent exposure by the aliphatic side-chain portion of Arg-450 (Fig. 2B). Similarly, the Leu-454 residue at position d packs against the hydrophobic side chain moiety of Glu-455′ at position e′ (Fig. 2C). The motif is formed between neighboring chains throughout the ccCor1 structure (Fig. 1).
Fig. 1.
X-ray crystal structure of ccCor1. Side view with the N terminus on top. Side chains and backbones are shown as sticks and cartoon representation, respectively. Oxygen and nitrogen atoms are colored in red and blue, and carbon atoms are shown in cyan, green, and yellow for monomers A, B, and C, respectively. Residues Arg-450–Glu-455 that form the structural motif are shown with van der Waals spheres. For the sequence of ccCor1 see Fig. 4.
Fig. 2.
Prominent structural motif seen in the ccCor1 trimer. (A) Side view of the salt-bridge network (indicated by yellow dots) formed between Arg-450, Asp-452′, and Glu-455′ and the water-mediated hydrogen bond between Glu-455′:Oε2 and Arg-450:O. (B) End-on view of the a3 layer showing the shielding of the Leu-451 residues from solvent by the aliphatic side-chain moieties of Arg-450. (C) End-on view of the d3 layer showing the hydrophobic packing between the Leu-454 and the aliphatic side-chain moieties of the Glu-455 residues. Side chains of residues are shown as stick representation and van der Waals spheres (B and C), the water molecule as a small red sphere (A), and the three monomers are shown as Cα traces. Colors of atoms and monomers are the same as in Fig. 1.
The Network of Interhelical Salt Bridges Determines the Trimeric Structure of ccCor1. Modeling studies (data not shown) indicated that both the characteristic bifurcated interaction seen in the ccCor1 trimer between Arg-450 and Glu-455 and the hydrophobic packing of their side chains against the core cannot be formed by interchanging the positions of these residues. Modeling of ccCor1 in the two-stranded state based on a superposition of monomers A and B on the WT GCN4-p1 leucine zipper (32) suggests that the triad formed between the Arg-450, Asp-452′, and Glu-455′ side chains cannot be established as a result of the increased distance between these residues in the two-stranded structure. Likewise, residues Leu-451 and Leu-454 would be less well shielded from solvent exposure by the aliphatic side-chain moieties of the Arg-450 and Glu-455 residues, respectively. Both effects are expected to disfavor dimer formation. Modeling of the ccCor1 monomers A and B on the four-stranded GCN4-pLV variant (15) suggests that a salt-bridge network as seen in ccCor1 may sterically be possible. However, as a result of the decreased distance between the core flanking residues in the four-stranded state, optimal packing of the Arg-450 and Glu-455 hydrophobic side-chain moieties against the hydrophobic core is likely to be compromised.
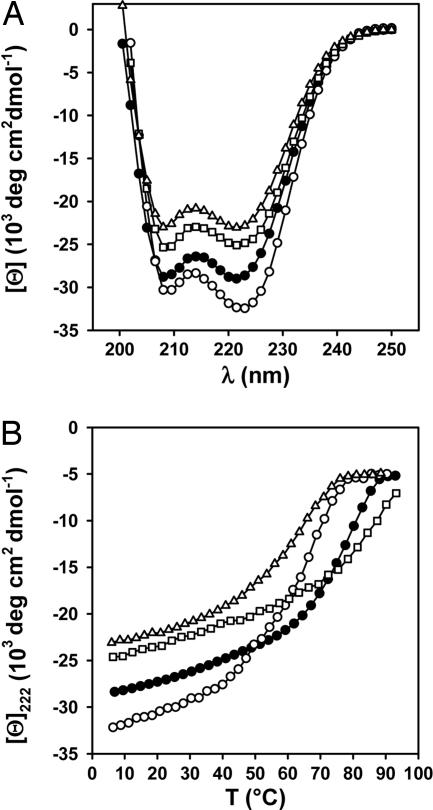
These considerations suggest that the sequence segment Arg-450–Glu-455 is important for determining the trimeric oligomerization state of ccCor1. To test this hypothesis, Arg-450 was replaced by alanine (referred to as ccCor1-R450A) and the structure of the mutant was compared with the WT peptide by CD spectroscopy (Fig. 3 and Table 1), AUC (Table 1 and Fig. 6A, which is published as supporting information on the PNAS web site), and SLS (Table 2). The far-UV CD spectra recorded from ccCor1 and ccCor1-R450A at 5°C are characteristic of proteins with a high helical content. The stabilities of the peptides were assessed by thermal unfolding profiles recorded by CD at 222 nm. Consistent with native-like coiled-coil structures, both peptides revealed reversible sigmoidal unfolding profiles. Sedimentation equilibrium and SLS experiments of ccCor1 revealed the formation of trimers, which agrees with the x-ray crystal structure analysis. In marked contrast, ccCor1-R450A yielded average molecular masses consistent with the formation of tetrameric structures.
Fig. 3.
CD analysis of WT ccCor1 and variants. Far-UV CD spectra recorded at 5°C (A) and thermal unfolding profiles recorded by CD at 222 nm (B) of ccCor1 (•), ccCor1-R450A (○), ccCor1-R450Nle (□), and ccCor1-R450K (▵). The experiments were carried out at 35 μM peptide concentration (monomer) in PBS.
Table 1. CD, sedimentation equilibrium AUC, and SLS data for ccCor1 WT and variants.
| Peptides | [Θ]222* (103 deg·cm2·dmol-1) | Tm,† °C | MWobs/MWmonomer,‡ 50 μM/100 μM/200 μM | SLS,§ kDa |
|---|---|---|---|---|
| ccCor1 | -28.4 | 79 | 3.1/3.1/3.0 | 10.9 (3.8) |
| ccCor1-R450A | -32.2 | 67 | 3.8/3.8/4.0 | 13.8 (3.7) |
| ccCor1-R450Nle | -24.7 | 86 | 4.0/3.9/4.1 | 14.8 (3.8) |
| ccCor1-R450K | -23.0 | 66 | 3.1/3.3/3.8 | n.d. |
[Θ]222 measured at 5 °C at peptide concentrations (monomer) of 35 μM.
Tm determined at peptide concentrations (monomer) of 35 μM.
Average molecular masses determined by AUC.
Average molecular masses determined by SLS. The molecular masses of the monomers are given in parentheses. n.d., not determined.
Table 2. Effect of g Arg to e′ Glu interhelical salt-bridge mutations on the oligomerization state of short three-stranded coiled coils.
| Coiled coil | Mutation | Oligomerization state | Source |
|---|---|---|---|
| TN-C-p1C64S | WT | Trimer | 31 |
| R125A | Trimer/tetramer | This work* | |
| E130L | Trimer/tetramer | 31 | |
| E130A | Trimer/tetramer | 31 | |
| ccMat1 | WT | Trimer | 30, 34 |
| R487K | Trimer/tetramer | This work* | |
| R487Q | Tetramer | 34 | |
| ccβ-p | WT | Trimer | This work† |
| R8K | Dimer | This work† | |
| p1 | WT | Trimer‡ | 45 |
| R6A/E11A | Octamer‡ | 46 |
See Fig 6.
Sedimentation equilibrium AUC at 5 °C of 0.2-0.5 mg/ml peptide solution yielded averaged molecular mass of 6.0 kDa for ccβ-p (monomer mass is 2.1 kDa) and 4.2 kDa for ccβ-pR8K (monomer mass is 2.05 kDa).
Oligomerization states as seen in crystal structures. The oligomerization state in solution was found to depend on ionic strength.
In the trimer, neither a lysine (because of its restricted salt bridge-forming potential) nor an aspartate (because of the shortened length of its side chain) are expected to substitute for Arg-450 and Glu-455, respectively. To test whether the guanidinium group is important for determining trimer assembly, two further ccCor1 mutants were prepared in which Arg-450 was replaced by either the arginine isostere norleucine (referred to as ccCor1-R450Nle) or by lysine (referred to as ccCor1-R450K). CD experiments carried out for both mutants were consistent with the formation of stable coiled-coil structures (Fig. 3 and Table 1). Biophysical studies revealed the formation of tetramers of ccCor1-R450Nle, and ccCor1-R450K was found to be in a concentration-dependent equilibrium between trimers and tetramers (Table 1).
These findings demonstrate that the distinct electrostatic interaction network formed between the guanidinium group of Arg-450 and the Asp-452′ and Glu-455′ side chains controls the three-stranded coiled-coil formation of ccCor1.
The Sequence Motif R-h-x-x-h-E Is Conserved Among Parallel Trimeric Coiled Coils. As shown in the sequence alignments in Fig. 4 and Fig. 7, which is published as supporting information on the PNAS web site, potential interhelical g Arg to e′ Glu salt bridges are found in many other short (15–50 aa in length) parallel trimeric coiled-coil domains of intracellular, extracellular, transmembrane, viral, and synthetic proteins. Several members of these autonomous coiled coils are well characterized (9, 30, 31, 33–43). Based on the alignment, the sequence motif R1-h2-x3-x4-h5-E6 can be deduced (R = Arg; E = Glu, L = Leu; h1 = Ile, Leu, Val, Met; h2 = Leu, Ile, Val; x = any amino acid residue). Notably, although present in ccCor1 and many other coiled coils, several sequences contain a nonacidic residue at position 3, suggesting that the interhelical electrostatic interaction with the arginine at position 1 (Fig. 2 A) is not strictly necessary for determining a trimeric structure.
Fig. 4.
The ccCor1 sequence Arg-450–Glu-455 is conserved in intracellular (I), extracellular (II), transmembrane (III), viral (IV), and synthetic (V) proteins containing short three-stranded parallel coiled-coil domains. Sequence alignments of mouse coronin 1A (mCor-1A) with the coiled-coil domains of human heat shock factor-binding protein 1 (hHSBP1), chicken tenascin C (chTN-C), human matrilin-1 (hMat-1), human type XVII collagen a1 (hXVII), influenza hemagglutinin (HA2), HIV-1 envelope glycoprotein (HIVgp41), and the synthetic ccβ-p coiled coil are shown. Heptad repeats are shown in blocks of seven amino acids, and residues at positions a and d are underlined. The conserved residues forming the trimerization motif in ccCor1 (Fig. 2) are shown in bold and colored according to their physicochemical properties: blue, positively charged; red, negatively charged; green, hydrophobic. The deduced consensus sequence is shown at the bottom. For a full alignment, see Fig. 7.
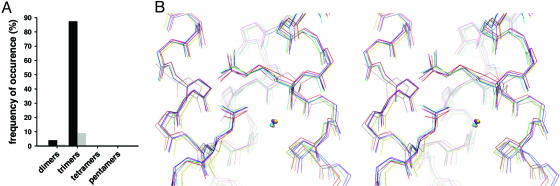
To further assess the statistical significance of the sequence motif, high-resolution structures of short coiled coils were analyzed by cross-referencing the SOCKET database (14) against a PROSITE PDB hit list obtained with the sequence pattern R-[ILVM]-X-X-[ILV]-E (for details, see Materials and Methods). The search revealed that the trimerization motif is found in 86% of all short, autonomous, and parallel three-stranded coiled coils (Fig. 5A). In contrast, the sequence pattern is rarely observed in short antiparallel trimers (9%), parallel and antiparallel dimers (4% and 0.7%, respectively), and not at all in tetramers and pentamers. As the only identified antiparallel trimer (Stat3β, PDB ID code 1bg1) folds into an intramolecular coiled coil and thus cannot be considered as an oligomerization domain the frequency of occurrence in antiparallel trimers for the present database analysis is actually nil. In the few dimers the frequency is also <4% when taking into account that the motif is located at the very N terminus of the structures where it does not display optimal knobs-into-holes packing. In one of these examples, GCN4-p1, the occurrence of the motif might be responsible for the dimer–trimer equilibrium seen for Asn-16 mutants (15). Not surprisingly, the trimerization motif is seen in crystal structures of three-stranded GCN4-p1 variants (see, for example, PDB ID code 1ij3).
Fig. 5.
The trimerization motif is structurally conserved. (A) Frequency of occurrence of the sequence pattern R-[ILVM]-X-X-[ILV]-E in a structurally verified set of short (≤50 amino acid residues) parallel (black bars) and antiparallel (gray bars) coiled coils. (B) Superimposition of trimerization motifs found in ccCor1 (red), simian immunodeficiency virus gp41 (blue; PDB ID code 1qbz), visna TM (yellow, PDB ID code 1jek), ccβ-p (magenta; PDB ID code 1s9z), p4 (cyan; PDB ID code 1kyc), and influenza hemagglutinin (salmon; PDB ID code 1eo8) is shown in stereo and in line representation.
As shown in Fig. 5B, the conformations of side chains at pattern positions 1, 2, 5, and 6 as well as the position of the water molecule between the Arg-450:O and Glu-455′:Oε2 atoms seen in crystal structures of various parallel three-stranded coiled-coil structures can be superimposed, demonstrating that the motif is also structurally conserved.
The Conserved Sequence Motif R-h-x-x-h-E Defines a Determinant for Trimer Formation of Short Coiled Coils. To validate the importance of the sequence pattern for determining trimeric coiled-coil formation, mutant polypeptide chains of the well characterized three-stranded coiled-coil domains of tenascin-C, matrilin-1, and ccβ-p were prepared in which the arginine residue at position 1 was replaced by alanine (for sequences, see Fig. 4). These mutants are referred to as TN-C-p1C64S/R125K, ccMat1-R487A, and ccβ-pR8K. Their oligomerization states (Table 2) were assessed either by cross-linking of the flanking cysteine residues after redox shuffling (TN-C-p1C64S/R125K and ccMat1-R487A) or AUC equilibrium experiments (ccβ-pR8K).
Replacement of Arg-125 by lysine had a dramatic effect on the oligomerization state of TN-C-p1C64S. When analyzed by SDS/PAGE under nonreducing conditions and compared with the TN-C-p1C64S protein (31), TN-C-p1C64S/R130K revealed the appearance of a new disulfide-linked species corresponding in size to a tetramer (Fig. 6B). The same gel pattern has been observed previously for mutations targeting the partner Glu-130 residue of the potential interhelical salt bridge (31), demonstrating the importance of this interaction for determining a trimeric oligomerization state of tenascin-C. The ccMat1-R487A peptide also populates disulfide-linked four-stranded structures although to a lesser extent than the TN-C-p1C64S mutants (Fig. 6C). A complete shift from trimer to tetramer, however, has been observed previously by mutating Arg-487 to glutamine (34). Interestingly, matrilin-3 is the only member of this family of proteins that contains a lysine instead of an arginine residue at the corresponding position. Consistent with our findings, matrilin-3 folds into four-stranded coiled-coil structures (30). A switch in oligomerization state from trimer to dimer is found for the ccβ-pR8K variant (Table 2). Whereas the preferred formation of tetramers over dimers in coronin, tenascin, and matrilin interhelical salt bridge mutants is unclear at present, the dimeric nature of ccβ-pR8K can be explained by the distribution of isoleucine and leucine residues at the core positions (15, 16).
Together, these findings demonstrate that the conserved motif represents a universal determinant for parallel trimer formation of short coiled coils.
Discussion
It is generally acknowledged that the identities of hydrophobic and polar core residues play an important role in defining the specificity of coiled coils. Here, we show how a distinct structural motif determines the topology of short (≤50 aa) autonomous coiled-coil domains by dominating the influence of core residues. Our search in a structurally verified set of coiled coils revealed that the trimerization motif is highly represented in parallel three-stranded coiled coils, whereas it is very rarely observed in dimers and antiparallel trimers and not at all in tetramers and pentamers. The frequencies of occurrence of the motif (Fig. 5A) are comparable or even higher than those of general and well established determinants of coiled-coil specificity such as the distribution of β-branched residues (in particular, isoleucine) at the hydrophobic a and d core positions or the presence of asparagine and lysine at heptad position a (15–17, 24).
The sequence pattern R1-h2-x3-x4-h5-E6 forming the motif is functionally conserved in many diverse protein families harboring short three-stranded coiled-coil oligomerization domains (Table 2). As illustrated in Figs. 4 and 7, positions 1 and 6 potentially forming the bifurcated interhelical salt bridge network between arginine and glutamate are strictly conserved. Positions 2 and 5 are occupied by hydrophobic residues that are compatible with the acute core packing geometry of three-stranded coiled coils (15, 16). The structural characteristics of their side chain favor tight packing with the aliphatic moieties of the neighboring arginine and glutamate residues that participate in the formation of the hydrophobic core. Thus the trimerization driving potential of the motif can be explained by optimal side-chain–side-chain interactions. Similar to contacts stabilizing thermostable proteins (44), the motif encompasses both networks of surface salt bridges and optimal internal hydrophobic packing interactions.
The trimerization driving potential of the motif is further underscored by two recently reported de novo designs, ccβ-p (9) and p1 (45, 46). Based on the rational incorporation of isoleucine and leucine at the a and d core positions, respectively, these coiled coils were both expected to form two-stranded (15, 16) instead of the experimentally verified parallel trimeric structures. Mutations targeting the glutamate and/or arginine of the interhelical salt bridge resulted in a switch of the oligomerization state leading either to a two-stranded coiled-coil structure (ccβ-p) or an octameric helical bundle devoid of any coiled-coil interactions (p1; see Table 2). These examples demonstrate the potential of the trimerization motif to dominate the effect of the hydrophobic core residues on the structure of short coiled-coil domains.
The findings of our study have several implications. Although a variety of methods are available to predict coiled-coil sequences with a high degree of confidence, the reliable prediction of their oligomeric structure remains difficult. This difficulty is exemplified by the finding that like ccCor1, many three-stranded coiled-coil domains shown in Fig. 7 are predicted to favor a dimeric structure demonstrating that methods based on statistical occurrence of residues in databases of two- and three-stranded coiled coils alone are not necessarily sufficient to recognize the actual oligomerization state. The implementation of sequence-to-structure rules, like the one described in this study, should significantly improve existing algorithms and is thus expected to contribute toward predicting the structure and function of coiled-coil proteins.
For potential protein engineering, biotechnological, basic research, biomaterial, and medical applications using short coiled coils, knowledge of the factors that determine structural uniqueness of the engineered protein systems is essential. Short de novo-designed coiled coils, however, frequently display a mixture of oligomerization states (19). Likewise, many mutations of natural coiled-coil proteins also result in a loss in oligomerization state specificity (18). These observations indicate that the thermodynamic stabilities of different oligomeric coiled-coil structures are similar. The incorporation or omission of the trimerization motif into designed trimeric and dimeric/tetrameric coiled-coil sequences, respectively, is expected to improve the specificity of the resulting de novo structures and as a result the activities of engineered proteins.
Our observations should also be of interest for developing strategies against viral infection. The interhelical interaction between position 1 arginine and position 6 glutamate residues of the trimerization motif seen in the crystal structures of several three-stranded coiled-coil-containing viral envelope proteins (Fig. 5B) plays a role in HIV infectivity. In single and double mutants where the bifurcated interhelical salt bridge between Arg-579 and Glu-584 of HIV-1 gp41 (Figs. 4 and 5B) has been removed, fusion activity was completely abolished despite incorporation of some of the mutant envelope glycoproteins into virions (47, 48). Synthetic peptides, corresponding to the N and C helices of the HIV-1 gp41 envelope glycoprotein, are potent inhibitors of HIV-1 membrane fusion. Not surprisingly, engineered molecules that present N peptides in three-stranded coiled-coil conformations were found to be more potent inhibitors than the unstructured N peptide monomers themselves (2). To our knowledge, however, none of these peptides contain the conserved sequence pattern identified in this study. Together, these observations indicate that peptides containing the trimerization motif might provide favorable templates from which to develop even more potent HIV-1 peptide inhibitors.
Supplementary Material
Acknowledgments
We thank Drs. T. Tomizaki and C. Schulze-Briese from the Swiss Light Source for excellent technical assistance; Dr. J. Missimer for careful reading of the manuscript; Mr. S. Patel for technical assistance; and the Institute for Molecular Biology and Biophysics of Eidgenössiche Technische Hochschule Zürich for access to the CD spectropolarimeter. This work was supported by grants from the Swiss National Science Foundation (to M.O.S. and J.P.). R.A.K. is a Wellcome Trust Senior Research Fellow in Basic Biomedical Science.
Author contributions: R.A.K., J.P., and M.O.S. designed research; D.K., P.P., S.H., and M.O.S. performed research; D.A. and A.L. contributed new reagents/analytic tools; R.A.K., D.K., F.K.W., and M.O.S. analyzed data; and R.A.K. and M.O.S. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: AUC, analytical ultracentrifugation; ccCor1, coiled-coil domain of coronin 1; PDB, Protein Data Bank; SLS, static light scattering.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 2akf).
References
- 1.Kohn, W. D. & Hodges, R. S. (1998) Trends Biotechnol. 16, 379–389. [Google Scholar]
- 2.Eckert, D. M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 11187–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho, C. H., Kammerer, R. A., Lee, H. J., Steinmetz, M. O., Ryu, Y. S., Lee, S. H., Yasunaga, K., Kim, K. T., Kim, I., Choi, H. H., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 5547–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petka, W. A., Harden, J. L., McGrath, K. P., Wirtz, D. & Tirrell, D. A. (1998) Science 281, 389–392. [DOI] [PubMed] [Google Scholar]
- 5.Ryadnov, M. G., Ceyhan, B., Niemeyer, C. M. & Woolfson, D. N. (2003) J. Am. Chem. Soc. 125, 9388–9394. [DOI] [PubMed] [Google Scholar]
- 6.Muller, K. M., Arndt, K. M. & Alber, T. (2000) Methods Enzymol. 328, 261–282. [DOI] [PubMed] [Google Scholar]
- 7.Rieker, J. D. & Hu, J. C. (2000) Methods Enzymol. 328, 282–296. [DOI] [PubMed] [Google Scholar]
- 8.Arndt, K. M., Muller, K. M. & Pluckthun, A. (2001) J. Mol. Biol. 312, 221–228. [DOI] [PubMed] [Google Scholar]
- 9.Kammerer, R. A., Kostrewa, D., Zurdo, J., Detken, A., Garcia-Echeverria, C., Green, J. D., Muller, S. A., Meier, B. H., Winkler, F. K., Dobson, C. M., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 4435–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Severin, K., Lee, D. H., Kennan, A. J. & Ghadiri, M. R. (1997) Nature 389, 706–709. [DOI] [PubMed] [Google Scholar]
- 11.Contegno, F., Cioce, M., Pelicci, P. G. & Minucci, S. (2002) Proc. Natl. Acad. Sci. USA 99, 1865–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, C., Stewart, R. J. & Kopecek, J. (1999) Nature 397, 417–420. [DOI] [PubMed] [Google Scholar]
- 13.Lupas, A. N. & Gruber, M. (2005) Adv. Protein Chem. 70, 37–38. [DOI] [PubMed] [Google Scholar]
- 14.Walshaw, J. & Woolfson, D. N. (2001) J. Mol. Biol. 307, 1427–1450. [DOI] [PubMed] [Google Scholar]
- 15.Harbury, P. B., Zhang, T., Kim, P. S. & Alber, T. (1993) Science 262, 1401–1407. [DOI] [PubMed] [Google Scholar]
- 16.Harbury, P. B., Kim, P. S. & Alber, T. (1994) Nature 371, 80–83. [DOI] [PubMed] [Google Scholar]
- 17.Vinson, C., Myakishev, M., Acharya, A., Mir, A. A., Moll, J. R. & Bonovich, M. (2002) Mol. Cell. Biol. 22, 6321–6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akey, D. L., Malashkevich, V. N. & Kim, P. S. (2001) Biochemistry 40, 6352–6360. [DOI] [PubMed] [Google Scholar]
- 19.Tripet, B., Wagschal, K., Lavigne, P., Mant, C. T. & Hodges, R. S. (2000) J. Mol. Biol. 300, 377–402. [DOI] [PubMed] [Google Scholar]
- 20.Berger, B., Wilson, D. B., Wolf, E., Tonchev, T., Milla, M. & Kim, P. S. (1995) Proc. Natl. Acad. Sci. USA 92, 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lupas, A., Van Dyke, M. & Stock, J. (1991) Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- 22.Parry, D. A. (1982) Biosci. Rep. 2, 1017–1024. [DOI] [PubMed] [Google Scholar]
- 23.Wolf, E., Kim, P. S. & Berger, B. (1997) Protein Sci. 6, 1179–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolfson, D. N. & Alber, T. (1995) Protein Sci. 4, 1596–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conway, J. F. & Parry, D. A. (1991) Int. J. Biol. Macromol. 13, 14–16. [DOI] [PubMed] [Google Scholar]
- 26.Conway, J. F. & Parry, D. A. (1990) Int. J. Biol. Macromol. 12, 328–334. [DOI] [PubMed] [Google Scholar]
- 27.Singh, M., Berger, B. & Kim, P. S. (1999) J. Mol. Biol. 290, 1031–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, M., Berger, B., Kim, P. S., Berger, J. M. & Cochran, A. G. (1998) Proc. Natl. Acad. Sci. USA 95, 2738–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gatfield, J., Albrecht, I., Zanolari, B., Steinmetz, M. O. & Pieters, J. (2005) Mol. Biol. Cell 16, 2786–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank, S., Schulthess, T., Landwehr, R., Lustig, A., Mini, T., Jeno, P., Engel, J. & Kammerer, R. A. (2002) J. Biol. Chem. 277, 19071–19079. [DOI] [PubMed] [Google Scholar]
- 31.Kammerer, R. A., Schulthess, T., Landwehr, R., Lustig, A., Fischer, D. & Engel, J. (1998) J. Biol. Chem. 273, 10602–10608. [DOI] [PubMed] [Google Scholar]
- 32.O'Shea, E. K., Klemm, J. D., Kim, P. S. & Alber, T. (1991) Science 254, 539–544. [DOI] [PubMed] [Google Scholar]
- 33.Tai, L. J., McFall, S. M., Huang, K., Demeler, B., Fox, S. G., Brubaker, K., Radhakrishnan, I. & Morimoto, R. I. (2002) J. Biol. Chem. 277, 735–745. [DOI] [PubMed] [Google Scholar]
- 34.Beck, K., Gambee, J. E., Kamawal, A. & Bachinger, H. P. (1997) EMBO J. 16, 3767–3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlinden, A., Smith, T. A., Sandell, L. J., Ficheux, D., Parry, D. A. & Hulmes, D. J. (2003) J. Biol. Chem. 278, 42200–42207. [DOI] [PubMed] [Google Scholar]
- 36.Latvanlehto, A., Snellman, A., Tu, H. & Pihlajaniemi, T. (2003) J. Biol. Chem. 278, 37590–37599. [DOI] [PubMed] [Google Scholar]
- 37.Lu, M., Blacklow, S. C. & Kim, P. S. (1995) Nat. Struct. Biol. 2, 1075–1082. [DOI] [PubMed] [Google Scholar]
- 38.Blacklow, S. C., Lu, M. & Kim, P. S. (1995) Biochemistry 34, 14955–14962. [DOI] [PubMed] [Google Scholar]
- 39.Carr, C. M. & Kim, P. S. (1993) Cell 73, 823–832. [DOI] [PubMed] [Google Scholar]
- 40.Yang, Z. N., Mueser, T. C., Kaufman, J., Stahl, S. J., Wingfield, P. T. & Hyde, C. C. (1999) J. Struct. Biol. 126, 131–144. [DOI] [PubMed] [Google Scholar]
- 41.Malashkevich, V. N., Singh, M. & Kim, P. S. (2001) Proc. Natl. Acad. Sci. USA 98, 8502–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burkhard, P., Ivaninskii, S. & Lustig, A. (2002) J. Mol. Biol. 318, 901–910. [DOI] [PubMed] [Google Scholar]
- 43.Supekar, V. M., Bruckmann, C., Ingallinella, P., Bianchi, E., Pessi, A. & Carfi, A. (2004) Proc. Natl. Acad. Sci. USA 101, 17958–17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaenicke, R. & Bohm, G. (1998) Curr. Opin. Struct. Biol. 8, 738–748. [DOI] [PubMed] [Google Scholar]
- 45.Burkhard, P., Meier, M. & Lustig, A. (2000) Protein Sci. 9, 2294–2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meier, M., Lustig, A., Aebi, U. & Burkhard, P. (2002) J. Struct. Biol. 137, 65–72. [DOI] [PubMed] [Google Scholar]
- 47.Weng, Y., Yang, Z. & Weiss, C. D. (2000) J. Virol. 74, 5368–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weng, Y. & Weiss, C. D. (1998) J. Virol. 72, 9676–9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.