Abstract
The protein kinase A (PKA) and the cAMP response element (CRE) binding protein (CREB) signaling pathways mediate plasticity and prosurvival responses in neurons through their ability to regulate gene expression. The PKA–CREB signaling mechanism has been well characterized in terms of nuclear gene expression. We show that the PKA catalytic and regulatory subunits and CREB are localized to the mitochondrial matrix of neurons. Mitochondrial CRE sites were identified by using both serial analyses of chromatin occupancy and chromatin immunoprecipitation. Deferoxamine (DFO), an antioxidant and iron chelator known to inhibit oxidative stress-induced death, activated mitochondrial PKA and increased mitochondrial CREB phosphorylation (Ser-133). DFO increased CREB binding to CRE in the mitochondrial D-loop DNA and D-loop CRE-driven luciferase activity. In contrast, KT5720, a specific inhibitor of PKA, reduced DFO-mediated neuronal survival against oxidative stress induced by glutathione depletion. Neuronal survival by DFO may be, in part, mediated by the mitochondrial PKA-dependent pathway. These results suggest that the regulation of mitochondrial function via the mitochondrial PKA and CREB pathways may underlie some of the salutary effects of DFO in neurons.
Protein kinase A (PKA) and cAMP response element (CRE) binding protein (CREB) play important roles in neuronal plasticity by altering expression of target genes in response to environmental stimuli (1–3). The PKA and CREB pathway is involved in neuronal survival of the central and peripheral nervous systems (4, 5). PKA is a tetramer consisting of two regulatory (Reg) and two catalytic (Cat) subunits. The binding of cAMP to a Reg subunit dissociates the holoenzyme into two free Cat subunits, which induce the phosphorylation of cellular substrates and a Reg subunit dimer. PKA phosphorylates CREB at Ser-133. Phosphorylated CREB (pCREB) subsequently binds to CRE and recruits the CREB binding protein, a transcriptional coactivator, to activate gene expression (6–8). It has been shown that PKA and CREB molecules localize into subcellular organelles (8–13). Interestingly, PKA-anchoring proteins in dendrites have been shown to tether PKAII-α to the outer membrane of mitochondria (8). Furthermore, the localization of the Cat subunit of PKA, and Reg subunit I (RI) or II (RII), has been found in the mitochondrial matrix (6, 9–12). CREB is also present in the mitochondria of the adult rat brain (7). Mitochondrial CREB has been implicated in plasticity-dependent changes associated with behavioral training (8). Thus, the localization of PKA and CREB in the mitochondria raises the possibility that they may play a role in neuronal plasticity and cell survival by regulating mitochondrial function via the mitochondrial PKA and CREB-dependent pathway (13).
Deferoxamine (DFO), an iron chelator and antioxidant, is neuroprotective, reducing the level of toxic radical species such as redox-reactive irons (14, 15). We have previously shown that neuronal protection from oxidative stress-induced apoptosis by DFO is associated with enhanced CREB binding to DNA, increased binding of HIF-1 to the hypoxia response element sequence, and transcriptional activation of glycolytic enzymes (16). This study suggests that DFO directly activates the signal transduction pathway, inducing gene expression as a mechanism for neuronal survival and plasticity. Because of CREB's involvement in the DFO-induced nuclear transcriptional response, we have proposed that DFO may regulate mitochondrial PKA activity, mitochondrial CREB phosphorylation, and subsequent neuronal survival. As such, we examined whether DFO activates mitochondrial PKA and increases mitochondrial CREB phosphorylation in neurons. We show that DFO regulates neuronal survival, in part, through mitochondria-dependent pathways via mitochondrial PKA and CREB activation.
Materials and Methods
Subcellular Fractionation and Isolation of Mitochondria. Mitochondria were isolated from primary cortical neurons and mouse and rat brain tissue by sucrose density gradient centrifugation (7).
Immunofluorescence Staining and Confocal Microscopy. Indirect labeling methods were used to determine PKA-Cat, PKA-Reg, phosphorylated PKA, CREB, and pCREB in cortical neuronal cultures, human, and rat brain tissues (16, 17). MitoTracker red (CMXRos) and DAPI were purchased from Molecular Probes. Images were analyzed by using a confocal microscope (MRC-1024, Bio-Rad). Control experiments were performed in the absence of primary antibody.
Serial Analysis of Chromatin Occupancy (SACO) Library. To identify CREB targets in rat mitochondrial genome, a method combining chromatin immunoprecipitation (ChIP) with a modification of serial analysis of gene expression was performed by using PC12 cells as described (18). Details of the analysis and the raw mitochondrial tag data can be found at http://saco.ohsu.edu.
mtDNA and Protein Cross-Linking and Immunoprecipitation. mtDNA and protein cross-linking and immunoprecipitation analysis for CREB binding to mtDNA were performed by using a ChIP assay kit (Upstate Biotechnology) (19). PCR amplification was carried out for 35 cycles, and PCR products were separated on 2% agarose gels. Three primers were used to amplify the segment flanking the three or two CRE-like sites in the D-loop of mitochondria. Forward primers were 5′-GTGGTGTCATGCATTTGGTATCT-3′ and 5′-ATCAACATAGCCGTCAAGGCATG-3′, and reverse primer was 5′-TCACCGTAGGTGCGTCTAGACTGT-3′.
ChIP of PC12 cells and quantitative real-time PCR were conducted as described (18).
Nuclear DNA Purification. Nuclei were isolated from 1 × 107 PC12 cells as described (20) except that cells were extracted twice with buffer A in the presence of 0.05% Nonidet P-40. High molecular weight genomic DNA was isolated with the Qiagen DNAeasy kit according to the manufacturer's instructions.
EMSAs and Supershift Analysis. We performed EMSAs on mitochondrial and nuclear extracts from cortical neurons by using a 32P-labeled oligonucleotide containing a mouse mitochondrial D-loop CRE-like site (at nucleotide positions 15940–115961; 5′-TCAACATAGCCGTCAAGGCATG-3′) as described (16, 17, 21). Supershifts were performed with CREB and pCREB-specific antibody for Ser-133 residue (Upstate Biotechnology), ATF-1/CREB (25C10G, Santa Cruz Biotechnology), or CREB-1 (24H4B and 240, Santa Cruz Biotechnology).
Plasmids Construction. Dupex of the D-loop sequences containing a CRE site [5′-ACAGTVTAGACGCACCTACGT-3′ (×2)] in the mouse mtDNA were synthesized (Invitrogen). The annealed oligonucleotides were cloned into pGL3 enhancer luciferase reporter vector (Promega).
Results
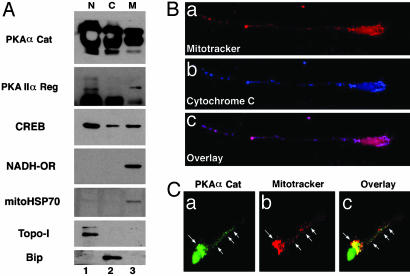
Neuronal Mitochondria Contain PKA and CREB. To determine the presence of PKA-Cat and PKA-Reg subunits in mitochondrial fractions, subcellular fractions were obtained by sucrose density centrifugation from mouse brain tissue (Fig. 1A). Western blot analysis confirmed that PKA-Cat and PKA-Reg subunits are present in pure mitochondrial fractions in mouse brain tissue. PKA-Cat and PKA-Reg subunits were detected at the 36- and 43-kDa positions, respectively. CREB also was detected in the mitochondrial fraction (Fig. 1A). The fact that topoisomerase I and Bip (GRP78) immunoreactivity were found only in the nuclear and the cytoplasmic fractions, respectively, indicates that the mitochondrial fractions were not contaminated with other subcellular organelles (Fig. 1A). We further investigated the subcellular distribution of PKA in primary cultured cortical neurons by confocal microscopy. We stained cortical neurons with MitoTracker to characterize the cellular distribution of mitochondria. Mitochondria were distributed throughout the cell body and also extended into the proximal and distal segments of the dendrites (Fig. 1Ba). To confirm the specificity of mitochondria staining by MitoTracker, we performed immunofluorescence staining with anticytochrome c combined with MitoTracker. Fig. 1Bb shows typical staining of cytochrome c in a cortical neuron. There was a punctate pattern of cytochrome c in the cell cytoplasm. The mitochondria in cortical neurons, displayed by combined MitoTracker and cytochrome c immunostaining, were also punctate in nature. There was absolute colocalization of mitochondrial cytochrome c immunoreactivity to the punctate structures of MitoTracker (Fig. 1Bc). In addition, we examined the subcellular PKA-Cat distribution in primary cultured rat cortical neurons. PKA-Cat immunoreactivity was present in neuronal cell bodies and distributed along the dendrite processes (Fig. 1Ca). PKA punctate-immunoreactivity was prominent in the perinuclear region, as seen when MitoTracker staining was overlaid with PKA. The greatest PKA/MitoTracker double-staining was concentrated in the proximal dendritic arbors (Fig. 1Cc).
Fig. 1.
Mitochondrial localization of PKA-Cat and PKA-Reg subunits. (A) Ultrafractionation of cellular compartments confirmed that PKA-Cat and PKA-Reg subunits are present in pure mitochondrial fractions in mouse brain tissues. PKA-Cat and PKA-Reg subunits were detected at 36- and 43-kDa positions, respectively. The same blot was stripped and reblotted with anti-mitochondrial HSP70 and CREB antibody. N, nuclear fraction; C, cytoplasmic fraction; M, pure mitochondrial fraction. (B) Mitochondria are shown by colocalization of MitoTracker (a, red color) and cytochrome c (b, blue color) in a single neuron. (c) The image (pink color) is derived from superimposing the fluorescence of both a and b. (C) Punctate structures of PKA-Cat (a) are colocalized with MitoTracker (b). White arrows indicate colocalization of PKA and MitoTracker in the perinuclear region and dendritic segment in a single neuron. (c) The overlay image of a and b..
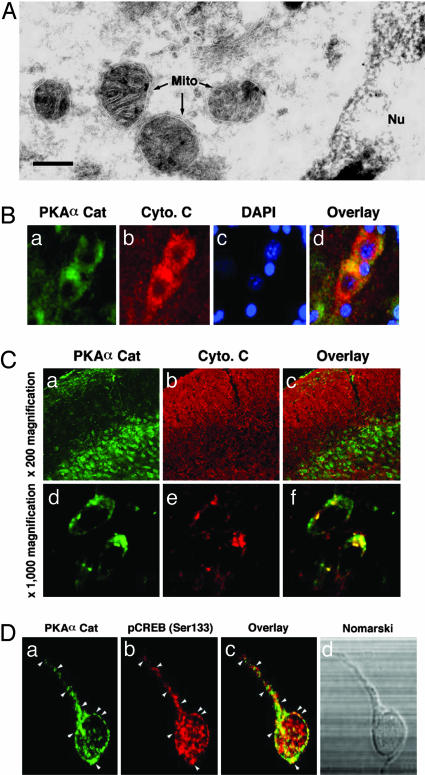
PKA-Cat Subunit Is Localized in the Mitochondrial Matrix of Neurons. To confirm our confocal microscopic observations, we further investigated the ultrastructural distribution of PKA-Cat in subcellular organelles of cortical neurons by using immunogold labeling and transmission EM. Under normal conditions the gold-labeled particles in the embryonic day 17 cortical neurons were found in the mitochondria, nucleus, and membrane vesicles such as the endoplasmic reticulum (Fig. 2A). The gold particles representing PKA-Cat were accumulated in the inner matrix of the mitochondria (Fig. 2A). However, the control staining in the absence of primary antibody did not show any particle in the mitochondria (data not shown). These EM data further support the mitochondrial localization of PKA-Cat and are consistent with the data from ultrafractionation (Fig. 1A) and confocal microscopy (Fig. 1B). Punctate PKA-Cat immunoreactivity was present primarily in extranuclear regions and was colocalized with cytochrome c in both the adult human cortex and rat cortex (Fig. 2 B and C). Furthermore, we found that PKA-Cat colocalized with pCREB(Ser-133) in rat cortical neurons. This colocalization was primarily extranuclear (Fig. 2D).
Fig. 2.
PKA-Cat is localized in the mitochondrial matrix in neurons. (A) Immunogold particles of PKA-Cat are present in the mitochondrial matrix in neurons. Gold particles are seen along portions of mitochondria (M). PKA-Cat visualization in embryonic day 17 rat cortical neurons by immunogold labeling was processed by using anti-PKA polyclonal antibody at 1:10 dilution. (Scale bar: 20 nm.) (B) PKA-Cat (a, green color) colocalizes with cytochrome c (b, red color) in human cerebral cortex neurons. (c) Nuclei are stained with DAPI. (d) The image is derived from superimposing the fluorescence images of a–c. (Scale bar: 10 μm.) (C) PKA-Cat (a and d, green) colocalizes with cytchrome c (b and e, red) in adult rat brain. c and f are overlay images. (D) PKA colocalizes with pCREB in cortical neurons. Cultures were double-stained for PKA (a) and pCREB (b). Arrows indicate punctate structures of colocalization of PKA and pCREB. (c) An overlay of a and b. (d) Nomarski microscopy of neuron observed in a–c.
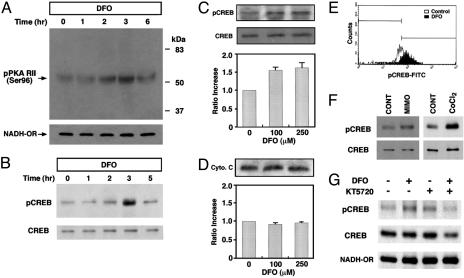
DFO Activates Mitochondrial PKA and the Phosphorylation of Mitochondrial CREB. PKA is one of most well characterized kinases that are responsible for site-specific phosphorylation and activation of CREB in vitro and in vivo. Harada and colleagues (9) found that PKA is anchored to mitochondria through the association of RII subunits with a family of A-kinase-anchoring proteins. In the present study, we found that DFO increases phosphorylation of Reg subunit II of PKA at a site targeted by the Cat subunit (22) (Fig. 3A). Cortical neuron cultures were exposed to 250 μM of DFO and lysates were prepared for Western blot analysis. The size of PKA-Reg subunit II was 51 kDa. The phosphorylation of PKA-Reg subunit II was observed 2–3 h after DFO treatment and was sustained for 6 h. The status of phosphorylation recovered to basal levels 8 h after DFO treatment.
Fig. 3.
DFO induces mitochondrial PKA activation and mitochondrial CREB phosphorylation. (A) DFO transiently increases the level of the phosphorylated Reg subunit II of PKA (PKA RII) in primary cortical neurons. NADH-oxidoreductase (NADH-OR) was detected as a control of protein level in the mitochondrial fraction. (B) DFO induces a transient phosphorylation of mitochondrial CREB. (C) Dose–response relationships for the phosphorylation of CREB by DFO in the mitochondrial fraction (n = 5). pCREB blot was stripped and reprobed with anti-CREB polyclonal (240) antibody. (D) The level of cytochrome c was not changed by DFO (n = 3). (E) In vitro mitochondrial pCREB staining was measured by flow cytometry and confirmed that mitochondrial pCREB was increased by DFO (100 μM). (F) Other iron chelators, mimosine (MIMO) and cobalt chloride (CoCl2), induced the phosphorylation of mitochondrial CREB. CONT, control. (G) Inhibition of PKA blocked the phosphorylation of mitochondrial CREB by DFO.
Our previous and current studies show that iron chelating agents, such as DFO and mimosine, which are known to abrogate apoptosis induced by oxidative stress in cortical neurons, increase CREB–DNA binding activity (16, 23). To address the possibility that DFO induces CREB Ser-133 phosphorylation in the mitochondria, we tested whether DFO treatment altered the level of pCREB in the mitochondrial fraction of cortical neurons (Fig. 3B). CREB phosphorylation was monitored by immunoblotting with a specific antibody for pCREB (Ser-133). The dose-dependent induction of pCREB was studied after treatment of cortical neuron cultures by DFO. Mitochondrial levels of pCREB on Ser-133 rose >2-fold after the treatment of 100 μM and 250 μM DFO (Fig. 3C). There was no change in the total level of CREB in the mitochondria by DFO (Fig. 3 B and C), suggesting that DFO stimulation transiently affects the phosphorylation status of mitochondrial CREB. Thus, phosphorylation of CREB is a specific response to DFO treatment. Although the mitochondrial fraction showed an increase in pCREB, there was no change in the level of cytochrome c (Fig. 3 C and D). In vitro mitochondrial pCREB immunostaining was measured by flow cytometry and confirmed that mitochondrial pCREB was increased by DFO treatment (100 μM) (Fig. 3E). Other iron chelators, such as mimosine and cobalt chloride, also induced the phosphorylation of mitochondrial CREB without any change in the total CREB level (Fig. 3F). To examine whether mitochondrial PKA is directly involved in the DFO-induced phosphorylation of mitochondrial CREB, we pretreated neurons with KT5720 (10 μM), a PKA-specific inhibitor, 30 min before DFO (100 μM) treatment. We found that inhibition of PKA by DFO blocked the phosphorylation of mitochondrial CREB (Fig. 3G).
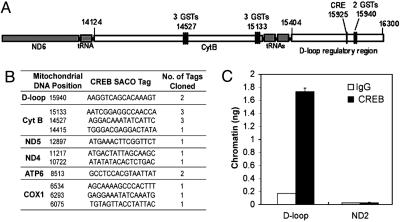
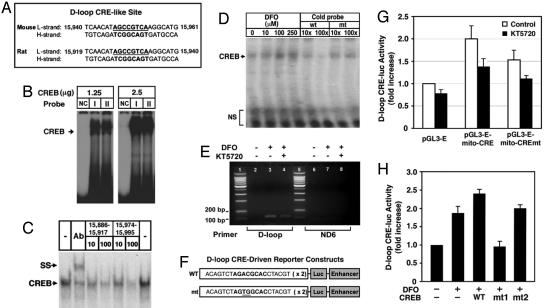
DFO Increases CREB–DNA Binding to the CRE Site in the D-Loop of the Mitochondrial Genome. SACO was used to identify mtDNA sequences that interact with CREB in rat PC12 cells (18) (Fig. 4). SACO generates a library of 21-bp tags from DNA regions cross-linked to CREB in vivo. Clustering of tags predicts novel Reg regions, and 100% of a selection of these regions found in the cellular genome were confirmed in repeat ChIPs. Because some 21-bp tags were also present in the cellular genome, we only used tags that were found uniquely in the rat mitochondrial genome (GenBank accession no. NC 001665). This analysis led to the identification of a cluster of tags surrounding the mitochondrial D-loop transcriptional Reg region and several smaller clusters of tags (Fig. 4A). Fig. 4B presents mitochondrial CRE-like sites found by SACO and ChIP. The D-loop Reg DNA was quantitatively immunoprecipitated by anti-CREB antibody (Fig. 4C). A region in the distal mitochondrial ND2 gene was not significantly enriched in CREB ChIPs. We also confirmed that this DNA is not present in rat cellular chromatin by quantitative real-time PCR of nuclear DNA (data not shown). Interestingly, there is a CRE sequence present in the D-loop that is conserved in rodents (Fig. 5A). We performed EMSA to examine whether CREB binds to mitochondrial CRE sites in the H-strand of the D-loop in the mouse mitochondrial genome. Purified CREB protein was found to bind to mitochondrial CRE-like sites in a dose-dependent manner (Fig. 5B). Mitochondrial extracts from rat embryonic cortical neurons showed CREB–DNA binding activities at mitochondrial CRE-like sites in the D-loop region (Fig. 5C). The addition of pCREB antibody during in vitro DNA binding resulted in slowing the mobility of the DNA–protein–antibody complex in the gel, representing the specificity of CREB binding to mitochondrial CRE sites (Fig. 5C). As expected, DFO increased CREB binding to mitochondrial CRE in a dose-dependent manner (Fig. 5D). We performed ChIP analysis to verify the presence of mitochondrial CREs by endogenous CREB protein, using mouse primary cortical neurons in the presence of DFO and/or the PKA inhibitor KT5720 (Fig. 5E). Basal CREB binding to the D-loop DNA was low in mouse primary neurons. Interestingly, DFO enhanced CREB binding, and KT5720 partially diminished the binding (Fig. 5E). Although these data show that mitochondrial CREB can bind to CRE-like sites in the mouse mitochondrial D-loop, it does not necessarily demonstrate that CREB regulates mitochondrial gene expression. To address this issue, we constructed a CRE luciferase reporter vector containing CRE-like sites found in the D-loop of mouse mtDNA (Fig. 5F). DFO treatment resulted in a 2-fold increase in D-loop CRE luciferase reporter activity, and KT5720 inhibited 60% of the DFO-induced luciferase activity (Fig. 5G). A mutation of CREB canonical sequence abrogated 50% of DFO-induced D-loop CRE luciferase activity in the absence of the PKA inhibitor, suggesting that other transcription factors may be responsible for the D-loop CRE luciferase activity. Furthermore, WT CREB transfection increased D-loop–CRE luciferase activity 2.5-fold, and mitochondrial CREB (Ser-133A) abrogated the mitochondrial CRE luciferase activity induced by DFO and CREB (Fig. 5H). However, mitochondrial CREB (Ser-142/144A) did not abrogate D-loop–CRE luciferase activity in the presence of DFO. These data suggest that PKA-dependent phosphorylation of the Ser-133 residue in the CREB molecule is critical for DFO-induced mitochondrial D-loop CRE luciferase activity.
Fig. 4.
Confirmation of CREB binding sites in mitochondrial genome of PC12 cells by SACO and ChIP. (A) A diagram depicting the clustering of CREB SACO tags in the rat mitochondrial D-loop region and the adjacent cytochrome B gene. The positions of tags and genes in the mitochondrial genome is based on National Center for Biotechnology Information accession no. NC001665. (B) A summary of rat mitochondrial CREB loci based on their position, sequence, and number of tags identified. The labels on the left indicate the mitochondrial genes in which the tags cluster. Sequences represent L-strand of rat mtDNA. Cyt B, cytochrome B; ND 4 and 5, NADH dehydrogenase 4 and 5, ATP6, ATPase 6; COX1, complex 1. (C) PC12 cells were subjected to a ChIP assay using an anti-CREB antibody or nonspecific IgG as described in Materials and Methods. Levels of immunoprecipitated DNA encompassing the mitochondrial D-loop or the ND2 gene were assessed by real-time quantitative PCR. Data from triplicate determinations were normalized for amplicon size. Error bars denote SEM.
Fig. 5.
DFO modulates CREB binding to D-loop CRE sequence, D-loop CRE-reporter activity, and mitochondrial function. (A) Mouse and rat mtDNA contain CRE-like site. (B) Purified CREB binds to a mitochondrial CRE in the D-loop region of mitochondrial genome. I; mouse mitochondrial CRE, II; rat mitochondrial CRE; NC, negative control probe (L-strand, 5′-TGGCCACAGCACTTAAAG-3′). (C) Mitochondrial fractions from rat brain show mitochondrial CRE (rat) binding activity. Supershift (SS) analysis using a pCREB-specific antibody confirmed the specificity of CREB binding. NS, nonspecific bindings. (D) DFO increases CREB DNA binding activity to rat mitochondrial CRE. The mitochondrial fraction of rat primary cortical neurons was used for EMSA in vitro. (E) DFO modulates in vivo CREB binding to the mitochondrial D-loop DNA in mouse primary cultures. A modified ChIP method was used for detection of in vivo binding of CREB to mtDNA as described in Materials and Methods. (F) Construction of mitochondrial CRE-driven reporter vectors. (G) PKA inhibition abrogates DFO-induced mitochondrial CRE reporter activity. (H) DFO regulates CREB phosphorylation-dependent mitochondrial CRE-reporter activities. HT-22 cells were transiently transfected with pGL3E and pGL3E–D-loop CRE with WT or mutant (mt) CREB. mt1, Ser-133A CREB; mt2, Ser-142/144A CREB. Luciferase activity was normalized to the protein concentration of each sample. Data are the mean ± SE of three separate experiments.
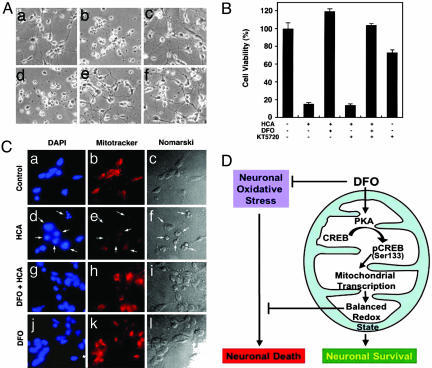
PKA Pathway Is Involved in DFO-Induced Neuronal Protection. To examine whether PKA participates in DFO-induced neuronal protection, we examined the role of the PKA inhibitor KT5720. At 100 μM concentration, DFO completely inhibited homocysteate (HCA)-induced neuronal cell death (Fig. 6A). KT5720 (5 μM) was added to cultures for 16 h, with or without DFO, after the addition of HCA. We found that inhibition of PKA activity significantly reduced the DFO-mediated neuronal viability. These results suggest that the PKA pathway mediates, in part, DFO-induced neuronal protection in response to oxidative stress (Fig. 6B).
Fig. 6.
PKA pathway is involved in neuronal protection by DFO. (A) Cortical neurons treated with HCA, DFO, and KT5720. Cells were pretreated with KT5720 (10 μM) 30 min before DFO (100 μM) treatment. (a) Control. (b) HCA. (c) DFO plus HCA. (d) KT5720 plus DFO. (e) KT5720 plus DFO plus HCA. (f) KT5720. (B) The PKA inhibitor KT5720 partially abrogates the protective function of DFO in HCA-treated cortical neurons. (C) DFO prevents loss of mitochondrial membrane potential induced by oxidative stress in cortical neurons. Cortical neuron cultures were exposed to 1 mM of HCA for 18 h with or without DFO (100 μM). Cultures were stained for nucleus with DAPI (a, d, g, and j) and for mitochondria with MitoTracker (b, e, h, and k). (a–c) Control. (d–f) HCA treatment. (g–i) DFO plus HCA treatment. (j–l) DFO treatment. White arrows in d, e, and f indicate the apoptotic cells induced by HCA. (c, f, i, and l) The Nomarski view of cortical neurons. (D) A proposed scheme of DFO-mediated neuronal survival through the mitochondrial PKA and CREB-dependent pathway.
Protection of Cortical Neurons by DFO Reflects Mitochondrial Function. The mitochondrial membrane potential is important in response to certain cell death signals (24). Using DAPI and Mito-Tracker probe (CMXRos), the nucleus and the mitochondria were labeled in the presence or absence of HCA and DFO. Control cells showed intact nuclear DAPI staining (Fig. 6Ca). The fluorescence staining pattern of active mitochondria in control cells was concentrated in the peri-nuclear region (Fig. 6Cb). In the presence of HCA for 18 h, cortical neurons showed apoptotic nuclear fragments (Fig. 6Cd) and the loss of mitochondrial membrane potential (Fig. 6Ce). DFO treatment not only restored the mitochondrial membrane potential (Fig. 6Ch), but also reduced nuclear fragmentation (Fig. 6Cg) in HCA-treated cortical neurons. DFO itself did not change the nuclear morphology (Fig. 6Cj) or the degree of mitochondrial fluorescence (Fig. 6Ck). These results strongly suggest an involvement of the mitochondria in HCA-induced cell death.
Discussion
Mitochondria play a central role in apoptosis (25–27). The change in mitochondrial membrane integrity, the activation of caspases and endonucleases by mitochondrial products, and the presence of the antiapoptotic molecule Bcl-2 and its homolog have shown that normal and dysfunctional mitochondrial processes regulate cell survival and death (28, 29). There is increasing evidence that the mitochondrial membrane potential (Δψm) provides a method for determining early events during apoptosis. In addition, mitochondria are the major intracellular source of free radicals. Increased mitochondrial calcium concentrations enhance free-radical generation (30). Furthermore, mtDNA is particularly susceptible to oxidative stress. There is a growing body of evidence that mitochondrial dysfunction, caused by defects in energy metabolism and oxidative damage, is involved in the pathogenesis of age-related neurodegenerative diseases, such as Alzheimer's disease, amyotrophic lateral sclerosis, Huntington's disease, and Parkinson's disease (30–33).
The present study shows that CREB localizes into mitochondria and binds to mitochondrial CRE-like sites. DFO, an antioxidant and iron chelator, phosphorylates mitochondrial CREB in primary cortical neurons through the activation of mitochondrial PKA. In addition, the antiapoptotic effect of DFO against oxidative stress-induced cell death is, in part, mediated by mitochondrial PKA and CREB. Interestingly, recent studies have shown that pCREB (Ser-133) is localized in the mitochondria of the adult rat brain (7). CREB localization in mitochondria has been implicated in plasticity-dependent changes associated with behavioral training (8). Thus, the morphological findings of PKA and CREB in the mitochondria suggest that these proteins may mediate neuronal cell survival by regulating mitochondrial function, thereby altering the response to various stimuli (6–8, 10). To further examine this relationship, we tested whether the change in PKA activation and pCREB levels induced by iron chelators accompanies a change in mitochondrial gene expression by using D-loop CRE-driven reporter analysis in neuronal cells. Because the iron metabolic pathways between the cytoplasm and the mitochondrial matrix present a number of important roles associated with the redox status in neuronal cells, deciphering the pathway of PKA activation and CREB phosphorylation by ion chelators may provide mechanistic information that is relevant to neuronal survival. Activation of mitochondrial PKA by DFO results in an increase of mitochondrial CREB-mediated transcriptional activity through the phosphorylation at Ser-133. The importance of CREB Ser-133 phosphorylation after growth factor stimulation, such as by insulin and nerve growth factor, has been well characterized by experiments in which the mutation of Ser-133 to Ala blocked growth factor responsiveness (5, 13). Similarly, DFO-induced CREB-mediated mitochondrial D-loop luciferase activity was abrogated by a mutation in Ser-133 to Ala, but not by Ser-142/144 to Ala.
Previous studies using transgenic and knockout mice show that CREB and its paralog, CREM, are important for cell survival (34, 35). CREB has been also shown to mediate cell survival in response to growth factors against several apoptotic stimuli (5). In addition, the overexpression of the dominant negative CREB transgene induces apoptosis in immune cells after growth factor stimulation (36). One mechanism by which CREB prevents apoptosis proceeds through up-regulation of Bcl-2 expression. The promoter region of Bcl-2 contains a CRE site, and CREB has been identified as an important regulator of Bcl-2 expression (5, 27, 28, 37). Current evidence suggests that CREB may directly mediate mitochondrial transcription (38, 39). In addition, it has been proposed that CREB influences mitochondrial gene expression in neurons (7, 8). Our data further support the hypothesis that mitochondrial CREB may be involved in mitochondrial gene expression and regulation of neuronal cell function. Otherwise, considering the architectural role for mitochondrial transcription factor A in the maintenance of mtDNA and its role in transcription activation, CREB may also play a similar role to stabilize mtDNA (40–42).
Oxidative stress induces mitochondrial dysfunction (24–26, 31). The membrane potential of mitochondria (Δψm) is likely to be a determining factor for apoptotic or survival signals in many cell types via external stimulation (43). DFO increases mitochondrial membrane potential, inhibits cytochrome c release and caspase-3 activation, and prevents mitochondrial damage induced by cocaine (24). We previously proposed that DFO could reduce the ambient free-radical burden in neuronal mitochondria in response to oxidative stress by inducing glycolytic enzyme and prosurvival gene expression (16). Therefore, our current findings point to a direct mechanism by which DFO modulates mitochondria function via mitochondrial PKA and mitochondrial CREB activation. DFO also protects neurons against oxidative stress and mitochondrial toxin (Fig. 6D and Fig. 7, which is published as supporting information on the PNAS web site). The enhancement of CREB binding to the mitochondrial genome by DFO may increase mitochondrial genome stability and maintain normal mitochondrial respiratory protein expression and membrane potential to abrogate cellular oxidative stress in neurons (44). Our finding that PKA and CREB localize in the mitochondria and are regulated by the antioxidant DFO raises the possibility that neuronal survival may additionally be affected by the regulation of mitochondrial function through the PKA and CREB signaling pathways.
In summary, antioxidants activate mitochondrial PKA and increase mitochondrial CREB binding to CRE sites in the mitochondrial genome. As such, mitochondrial PKA and CREB have the ability to modulate mitochondrial function and neuronal survival.
Supplementary Material
Acknowledgments
We thank T. Flood and A. Suchy-Dicey for help in the preparation of the manuscript. H.R. and R.J.F are grant awardees of the Jerry McDonald Huntington's Disease Foundation of the Boston University School of Medicine. This work was supported by National Institutes of Health Grant NS52724-01 (to H.R.), National Institutes of Health Grant P30 AG13846 (to J.L.), National Institutes of Health Grant NS045806 (to R.J.F.), National Institutes of Health Grant NS40591 (to R.R.R.), the Huntington's Disease Society of America (H.R. and R.J.F.), the High Q Foundation (H.R. and R.J.F.), and the Department of Veterans Affairs (R.J.F.).
Author contributions: H.R. designed research; H.R., J.L., and S.I. performed research; H.R., J.L., and S.I. contributed new reagents/analytic tools; H.R., R.R.R., and R.J.F. analyzed data; and H.R. and R.J.F. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PKA, protein kinase A; CRE, cAMP response element; CREB, CRE binding protein; pCREB, phosphorylated CREB; DFO, deferoxamine; Cat, catalytic; Reg, regulatory; SACO, serial analysis of chromatin occupancy; ChIP, chromatin immunoprecipitation; HCA, homocysteate.
References
- 1.Shaywitz, A. J. & Greenberg, M. E. (1999) Annu. Rev. Biochem. 68, 821–861. [DOI] [PubMed] [Google Scholar]
- 2.Mayr, B. & Montminy, M. (2001) Nat. Rev. Mol. Cell. Biol. 8, 599–609. [DOI] [PubMed] [Google Scholar]
- 3.Lonze, B. E. & Ginty, D. D. (2002) Neuron 35, 605–623. [DOI] [PubMed] [Google Scholar]
- 4.Paolillo, M., Feliciello, A., Porcellini, A., Garbi, C., Bifulco, M., Schinelli, S., Ventra, C., Stabile, E., Ricciardelli, G., Schettini, G. & Avvedimento, E. V. (1999) J. Biol. Chem. 274, 6546–6552. [DOI] [PubMed] [Google Scholar]
- 5.Riccio, A., Ahn, S., Davenport, C. M., Blendy, J. A. & Ginty, D. D. (1999) Science 286, 2358–2361. [DOI] [PubMed] [Google Scholar]
- 6.Sardanelli, A. M., Technikova-Dobrova, Z., Scacco, S. C., Speranza, F. & Papa, S. (1995) FEBS Lett. 377, 470–474. [DOI] [PubMed] [Google Scholar]
- 7.Cammarota, M., Paratcha, G., Bevilaqua, L. R., Levi de Stein, M., Lopez, M., Pellegrino de Iraldi, A., Izquierdo, I. & Medina, J. H. (1999) J. Neurochem. 72, 2272–2277. [DOI] [PubMed] [Google Scholar]
- 8.Bevilaqua, L. R., Cammarota, M., Paratcha, G., de Stein, M. L., Izquierdo, I. & Medina, J. H. (1999) Eur. J. Neurosci. 11, 3753–3756. [DOI] [PubMed] [Google Scholar]
- 9.Harada, H., Becknell, B., Wilm, M., Mann, M., Huang, L. J., Taylor, S. S., Scott, J. D. & Korsmeyer, S. J. (1999) Mol. Cell 3, 413–422. [DOI] [PubMed] [Google Scholar]
- 10.Sardanelli, A. M., Technikova-Dobrova, Z., Speranza, F., Mazzocca, A., Scacco, S. & Papa, S. (1996) FEBS Lett. 396, 276–278. [DOI] [PubMed] [Google Scholar]
- 11.Papa, S., Sardanelli, A. M., Cocco, T., Speranza, F., Scacco, S. C. & Technikova-Dobrova, Z. (1996) FEBS Lett. 379, 299–301. [DOI] [PubMed] [Google Scholar]
- 12.Papa, S., Sardanelli, A. M., Scacco, S. & Technikova-Dobrova, Z. (1999) FEBS Lett. 444, 245–249. [DOI] [PubMed] [Google Scholar]
- 13.Mootha, V. K., Bunkenborg, J., Olsen, J. V., Hjerrild, M., Wisniewski, J. R., Stahl, E., Bolouri, M. S., Ray, H. N., Sihag, S., Kamal, M., et al. (2003) Cell 115, 629–640. [DOI] [PubMed] [Google Scholar]
- 14.Farinelli, S. E. & Greene, L. A. (1996) J. Neurosci. 16, 1150–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiueh, C. C. (2001) Pediatr. Neurol. 25, 138–147. [DOI] [PubMed] [Google Scholar]
- 16.Zaman, K., Ryu, H., Hall, D., O'Donovan, K., Lin, K. I., Miller, M. P., Marquis, J. C., Baraban, J. M., Semenza, G. L. & Ratan, R. R. (1999) J. Neurosci. 19, 9821–9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ryu, H., Lee, J., Olofsson, B. A., Mwidau, A., Deodeoglu, A., Escudero, M., Flemington, E., Azizkhan-Clifford, J., Ferrante, R. J. & Ratan, R. R. (2003) Proc. Natl. Acad. Sci. USA 100, 4281–4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Impey, S., McCorkle, S. R., Cha-Molstad, H., Dwyer, J. M., Yochum, G. S., Bose, J. M., McWeeney, S., Dunn, J. J., Mandel, G. & Goodman, R. H. (2004) Cell 119, 1041–1054. [DOI] [PubMed] [Google Scholar]
- 19.Cha-Molstad, H., Keller, D. M., Yochum, G. S., Impey, S. & Goodman, R. H. (2004) Proc. Natl. Acad. Sci. USA 101, 13572–13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schindler, C., Fu, X. Y., Importa, T., Aebersold, R. & Darnell, J. E., Jr. (1992) Proc. Natl. Acad. Sci. USA 89, 7836–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ryu, H., Lee, J., Zaman, K., Kubilis, J., Ferrante, R. J., Ross, B. D., Neve, R. & Ratan, R. R. (2003) J. Neurosci. 23, 3597–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takio, K., Smith, S. B., Krebs, E. G., Walsh, K.A. & Titani, K. (1982) Proc. Natl. Acad. Sci. USA 79, 2544–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ratan, R. R., Murphy, T. H. & Baraban, J. M. (1994) J. Neurosci. 14, 4385–4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaragoza, A., Diez-Fernandez, C., Alvarez, A. M., Andres, D. & Cascales, M. (2001) Br. J. Pharmacol. 132, 1063–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green, D. R. & Reed, J. C. (1998) Science 281, 1309–1312. [DOI] [PubMed] [Google Scholar]
- 26.Kroemer, G., Dallaporta, B. & Resche-Rigon, M. (1998) Annu. Rev. Physiol. 60, 619–642. [DOI] [PubMed] [Google Scholar]
- 27.Susin, S. A., Zamzami, N. & Kroemer, G. (1998) Biochim. Biophys. Acta 1366, 151–165. [DOI] [PubMed] [Google Scholar]
- 28.Pugazhenthi, S., Miller, E., Sable, C., Young, P., Heidenreich, K. A., Boxer, L. M. & Reusch, J. E. (1999) J. Biol. Chem. 274, 27529–27535. [DOI] [PubMed] [Google Scholar]
- 29.Pugazhenthi, S., Nesterova, A., Sable, C., Heidenreich, K. A., Boxer, L. M., Heasley, L. E. & Reusch, J. E. (2000) J. Biol. Chem. 275, 10761–10766. [DOI] [PubMed] [Google Scholar]
- 30.Tritschler, H. J., Packer, L. & Medori, R. (1994) Biochem. Mol. Biol. Int. 34, 169–181. [PubMed] [Google Scholar]
- 31.Beal, M. F. (1995) Ann. Neurol. 38, 357–366. [DOI] [PubMed] [Google Scholar]
- 32.Borthwick, G. M., Johnson, M. A., Ince, P. G., Shaw, P. J. & Turnbull, D. M. (1999) Ann. Neurol. 46, 787–790. [DOI] [PubMed] [Google Scholar]
- 33.Chandrasekaran, K., Giordano, T., Brady, D. R., Stoll, J., Martin, L. J. & Rapoport, S. I. (1994) Brain Res. Mol. Brain Res. 24, 336–340. [DOI] [PubMed] [Google Scholar]
- 34.Blendy, J. A., Kaestner, K. H., Weinbauer, G. F., Nieschlag, E. & Schutz, G. (1996) Nature 380, 162–165. [DOI] [PubMed] [Google Scholar]
- 35.Nantel, F., Monaco, L., Foulkes, N. S., Masquilier, D., LeMeur, M., Henriksen, K., Dierich, A., Parvinen, M. & Sassone-Corsi, P. (1996) Nature 380, 159–162. [DOI] [PubMed] [Google Scholar]
- 36.Barton, K., Muthusamy, N., Chanyangam, M., Fischer, C., Clendenin, C. & Leiden, J. M. (1996) Nature 379, 81–85. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, B. E., Mochon, E. & Boxer, L. M. (1996) Mol. Cell. Biol. 16, 5546–5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crino, P., Khodakhah, K., Becker, K., Ginsberg, S., Hemby, S. & Eberwine, J. (1998) Proc. Natl. Acad. Sci. USA 95, 2313–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki, T., Usuda, N., Ishiguro, H., Mitake, S., Nagatsu, T. & Okumura-Noji, K. (1998) Brain Res. Mol. Brain Res. 61, 69–77. [DOI] [PubMed] [Google Scholar]
- 40.Takamatsu, C., Umeda, S., Ohsato, T., Ohno, T., Abe, Y., Fukuoh, A., Shinagawa, H., Hamasaki, N. & Kang, D. (2002) EMBO Rep. 3, 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alam, T. I., Kanki, T., Muta, T., Ukaji, K., Abe, Y., Nakayama, H., Takio, K., Hamasaki, N. & Kang, D. (2003) Nucleic Acids Res. 31, 1640–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanki, T., Ohgaki, K., Gaspari, M., Gustafsson, C. M., Fukuoh, A., Sasaki, N., Hamasaki, N. & Kang, D. (2004) Mol. Cell. Biol. 24, 9823–9834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds, I. J. (1999) Ann. N.Y. Acad. Sci. 893, 33–41. [DOI] [PubMed] [Google Scholar]
- 44.Shackelford, R. E., Manuszak, R. P., Johnson, C. D., Hellrung, D. J., Steele, T. A., Link, C. J. & Wang, S. (2003) DNA Repair 2, 971–981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.